Abstract
Multi-atlas segmentation provides a general purpose, fully automated class of techniques for transferring spatial information from an existing dataset (“atlases”) to a previously unseen context (“target”) through image registration. The method used to combine information after registration (“label fusion”) has a substantial impact on the overall accuracy and robustness. In practice, weighted voting techniques have dramatically outperformed algorithms based on statistical fusion (i.e., algorithms that incorporate rater performance into the estimation process — STAPLE). We posit that a critical limitation of statistical techniques (as generally proposed) is that they fail to incorporate intensity seamlessly into the estimation process and models of observation error. Herein, we propose a novel statistical fusion algorithm, Non-Local STAPLE, which merges the STAPLE framework with a non-local means perspective. Non-Local STAPLE (1) seamlessly integrates intensity into the estimation process, (2) provides a theoretically consistent model of multi-atlas observation error, and (3) largely bypasses the need for group-wise unbiased registrations. We demonstrate significant improvements in two empirical multi-atlas experiments.
Keywords: Simultaneous Truth And Performance Level Estimation (STAPLE), Statistical Label Fusion, Rater Models, Multi-Atlas Segmentation
1 Introduction
The de facto standard baseline for large-scale, consistent, and robust segmentation is to perform a multi-atlas segmentation in which a collection of canonical atlases (with labels) are registered to a target-of-interest [1, 2]. Here, we focus on the problem of resolving voxelwise conflicts between the registered atlases (i.e., “label fusion”).
Voting fusion strategies (e.g., a majority vote) have long provided robust segmentations. Recently, weighted voting using global [3], local [4], semi-local [5] and non-local [6] intensity similarities between the atlases and the target have demonstrated significant improvements in segmentation accuracy. Particularly for neurological applications, highly local weights have provided the most consistent and accurate segmentation estimates [4, 5].
In contrast to voting, statistical fusion strategies (e.g., Simultaneous Truth And Performance Level Estimation, STAPLE [7]) directly integrate a model of rater behavior (i.e., labeling error probabilities). Despite elegant theory and success with human raters, applications to the multi-atlas context have proven problematic [5, 8, 9]. In response, a myriad of variations have been proposed to account for spatially varying difficulty [10] and performance [8, 9]. Nevertheless, seamless integration of exogenous intensity information into the STAPLE context has proven difficult — efforts have relied upon ignoring voxels based on intensity similarities [9]. As a result, statistical fusion strategies are often less accurate in clinical multi-atlas applications.
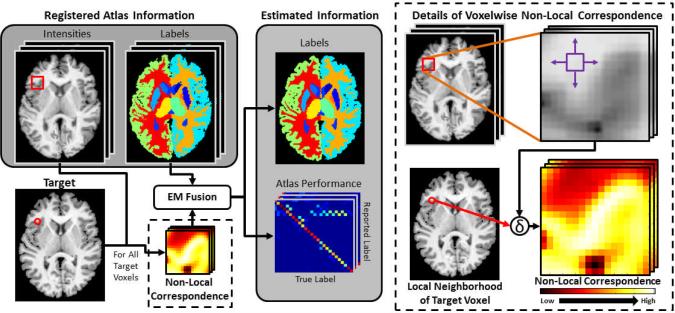
Herein, we propose a novel statistical fusion algorithm (Non-Local STAPLE — NLS) that merges the STAPLE framework with a non-local means perspective [11]. NLS models the registered atlases as collections of volumetric patches containing both intensity and label information and uses the non-local criteria [6, 11] to resolve imperfect correspondence (Figure 1). Through this reformulation, we seamlessly integrate exogenous intensity information into the estimation process to provide a theoretically consistent model of multi-atlas observation error. We derive the theoretical basis governing NLS, demonstrate significant improvement over premier fusion algorithms on two distinct datasets (CT thyroid and MR whole-brain segmentation), and assess the sensitivity of NLS to the various model parameters.
Fig. 1.
Flowchart of the Non-Local STAPLE (NLS) algorithm. NLS uses the atlas and target intensities to construct a non-local correspondence and integrates this into the estimation process. Point-wise correspondence is constructed in a traditional non-local means approach.
2 Theory
Consider a target gray-level image represented as a vector, . Let T ∈ LN×1 be the latent representation of the true target segmentation, where L = {0, ..., L – 1} is the set of possible labels. Consider a collection of R registered atlases with associated intensity values, , and label decisions, D ∈ LN×R. Let parameterize the raters (registered atlases) performance level. Each element of θ, θjs′s, represents the probability that rater j observes label s′ given that the true label is s at a given target voxel and the corresponding voxel on the associated atlas — i.e., , where i* is the voxel on atlas j that corresponds to target voxel i. Throughout, the index variables i, i* and i′ will be used to iterate over the voxels, s and s′ over the labels, and j over the registered atlases.
2.1 The Non-Local STAPLE Algorithm
NLS uses an Expectation-Maximization (EM) approach to estimate the true latent segmentation based on the target intensities, atlas information, and the rater performance level parameters (Figure 1). Estimation of the true segmentation (E-Step) follows [7]. Let , where Wsi represents the probability that the true label associated with voxel i is label s. Using a Bayesian expansion and conditional independence between the raters, the solution for W on iteration k is
| (1) |
where f(Ti = s) is a voxelwise a priori distribution of the underlying segmentation, and Di*j is the label decision by atlas j and corresponding voxel i*.
In NLS, we assume that we do not know which voxel i* on atlas j corresponds with voxel i on the target. We approximate the expansion with the expected value of Eq. 1 based on the probability of correspondence across the images and an assumption of conditional independence between the labels and intensity:
| (2) |
where is the search neighborhood of voxel i, and f(Ai′j|Ii) is the probability of the non-local correspondence between the target at voxel i and voxel i′ on atlas j. We use a standard non-local means approach to define:
| (3) |
where is the set of intensities in the patch neighborhood of a given intensity location, dii′ is the Euclidean distance between voxels i and i′ in image space, and σi and σd are the standard deviations of the assumed Gaussian distributions governing the intensity similarity and the Euclidean distance-based decay, respectively. Lastly, Zα is a partition function that enforces the constraint that .
Finally, we revisit Eq. 2 and, using the fact that s′ = Di′j, find that the final representation for the voxelwise label probabilities is
| (4) |
The estimate of the performance level parameters (M-Step) is obtained by finding the parameters that maximize the expected value of the conditional log likelihood function found in Eq. 4.
| (5) |
Noting the constraint that each row of the rater performance matrix must sum to one to be a valid probability mass function (i.e., ), we can maximize the performance level parameters by formulating the constrained optimization problem using a LaGrange multiplier. After taking the element-wise partial derivative and using the constraint that , the performance update becomes
| (6) |
2.2 Initialization and Convergence
As is typical [7], NLS was initialized with performance parameters equal to 0.95 along the diagonal and randomly setting the off-diagonal elements to fulfill the required constraints. For all presented experiments, the voxelwise label prior, f(Ti = s), was initialized using the probabilities from a “weak” log-odds majority vote (i.e., decay coefficient set to 0.5) [5], and the search neighborhood, , was initialized to an 11 × 11 × 11 window centered at the target voxel of interest. Several values for the patch neighborhood, , are considered in this manuscript all of which are centered at the voxels of interest. Unless otherwise noted, the values of the standard deviation parameters, σi and σd, were set to 0.1 and 2, respectively. Lastly, convergence of the algorithm was detected when the average change in the on-diagonal elements of the performance level parameters fell below 10–4.
3 Methods and Results
As benchmarks, we compare to a log-odds majority vote (MV) [5], a locally weighted vote (LWV) [5], and STAPLE [7]. For the voting algorithms, the implementation was the same as suggested in [5]. Note that LWV has a parameter that is essentially equivalent to the NLS parameter σi. For fairness of comparison, this parameter was set to the same value (herein, 0.1) for both algorithms. STAPLE was initialized using the same value for f(Ti = s) as NLS. For both STAPLE and NLS, “consensus voxels” (herein, voxels where maxs f(Ti = s) > 0.95) were ignored. For all experiments, the atlases were intensity normalized to the 25th and 75th percentiles. All pair-wise registrations were performed using an initial affine registration followed by a non-rigid procedure (Adaptive Bases Algorithm [12]). After registration the images were cropped to obtain a reasonable region of interest. Quantitative accuracy was primarily assessed using the Dice Similarity Coefficient (DSC).
3.1 Thyroid Multi-Atlas Segmentation
First, we analyzed the fusion accuracy on an empirical multi-atlas approach for thyroid segmentation using a collection of 15 head and neck atlases. The computed tomography (CT) images used in this experiment were collected from consenting patients who underwent intensity-modulated radiation therapy. The patients were injected with 80mL of Optiray 320, a 68% iversol-based nonionic contrast agent. Each image has a voxel size of 1 × 1 × 3 mm3. We performed a leave-one-out cross-validation experiment (i.e., 14 atlases per segmentation estimate) to assess fusion accuracy. NLS was run using various patch neighborhood sizes (1 × 1 × 1, 3 × 3 × 3, 5 × 5 × 3, and 7 × 7 × 3).
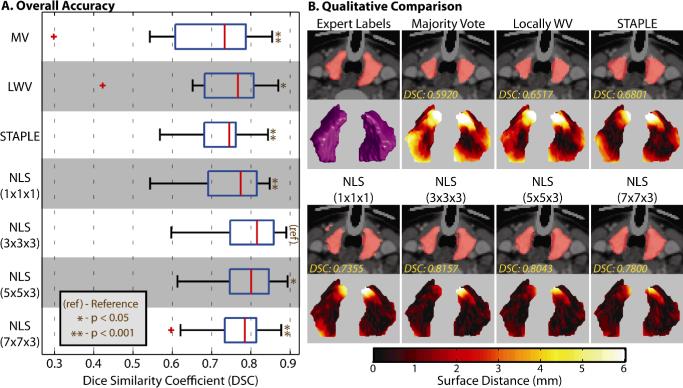
NLS substantially improved thyroid segmentation accuracy with the 3 × 3 × 3 patch neighborhood significantly outperforming all other algorithms (p < .05, Figure 2A). Median DSC performance was improved by 0.05 over LWV and 0.08 over STAPLE. The quantitative results seen in Figure 2A show the accuracy (in terms of the DSC) of the considered algorithms across the 15 atlases. Note the significant outliers in the results for the voting-based algorithms. Qualitative results can be seen in Figure 2B, where, for all considered algorithms, a representative slice and a 3D rendering of the point-wise surface distance error is presented. The various estimations from NLS are all qualitatively superior to the other benchmarks, as they more accurately estimated the underlying shape and size and resulted in substantial reductions in point-wise surface distance error. For small patch neighborhoods (e.g., 1 × 1 × 1), it is evident that high quality boundaries are estimated but “speckle noise” is more likely to be apparent. Alternatively, for larger windows, estimations are smoother but sacrifice the high quality boundary estimation.
Fig. 2.
Results of the empirical multi-atlas segmentation of the thyroid. The quantitative results (A) show that NLS provides significant improvement, with a 3 × 3 × 3 patch neighborhood significantly outperforming all other algorithms. The qualitative results (B) demonstrate that NLS provides improvement in terms of shape, boundary and point-wise surface distance error.
3.2 Whole-Brain Multi-Atlas Segmentation
Second, we examine fusion accuracy on an empirical experiment for whole-brain segmentation. A collection of 15 brains (OASIS, www.oasis-brains.org) were manually labeled (www.braincolor.org) by an expert anatomist. For each atlas a collection 26 labels were considered ranging from large structures (e.g. cortical gray matter) to smaller deep brain structures. All images were 1mm isotropic. To assess overall accuracy, we performed a cross-validation experiment using 5 to 14 atlases per target atlas. The per-label accuracy was assessed using 5 atlases per target. Lastly, the sensitivity of NLS with respect to the parameters σi and σd was assessed. Due to the large number of labels and limited atlases, STAPLE results were poor (not shown).
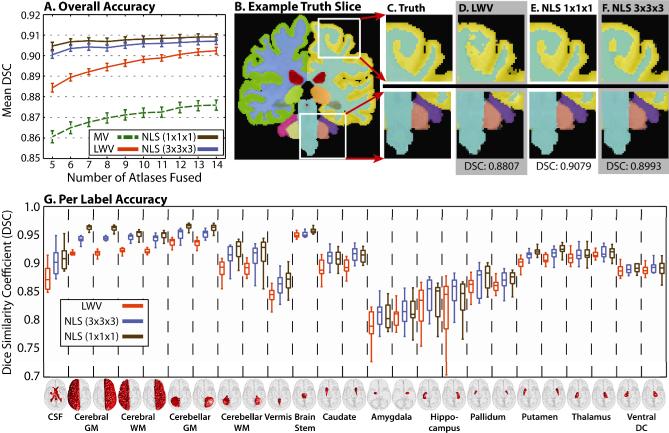
The results of the cross-validation (Figure 3) demonstrate that NLS provides consistent improvement in segmentation accuracy. For overall accuracy (reported as mean DSC, Figure 3A), NLS resulted in significant improvement (p < 0.05) over the other algorithms regardless of the number of atlases and provided estimates that are less dependent upon the number of atlases fused. Unlike the thyroid results, a single voxel neighborhood resulted in consistent improvement over larger neighborhood sizes. NLS using a single voxel neighborhood resulted in qualitatively more accurate segmentations (Figure 3B-3F). The per-label results (Figure 3G) demonstrate that, particularly for the larger labels, the NLS estimates are vastly superior to a locally weighted vote. NLS using a 1 × 1 × 1 patch neighborhood resulted in significantly superior (p < 0.05) results over LWV on 23 out of 26 labels and for 16 out of 26 labels over NLS using a 3 × 3 × 3 patch neighborhood. Neither MV nor LWV was significantly superior to either NLS approach for any label.
Fig. 3.
Results from the empirical whole-brain experiment. Overall (A) and qualitative (B-F) results show NLS (with a single voxel neighborhood) significantly outperforming the other algorithms. Per-label results (G) show consistent improvement regardless of label size or location.
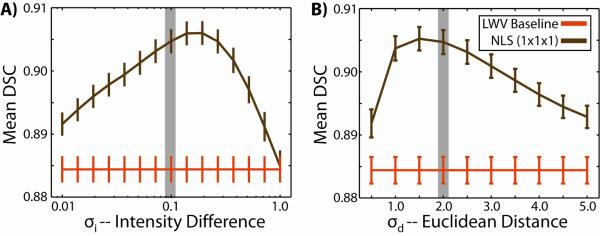
Lastly, the sensitivity of NLS to two of the model parameters, σi and σd (Eq. 3), can be appreciated in Figure 4. NLS accuracy decreases for σi values that result in segmentations that are overly noisy (small σi values) or overly smooth (larger σi values) (Figure 4A). Note that the value of this parameter is largely dependent upon the intensity normalization process (i.e., the relative distribution of atlas and target intensities). NLS sensitivity to σd, which can be interpreted as a proxy for sensitivity to search window size, is presented in Figure 4B. For values that are too small, accuracy sharply diminishes as too few voxels are used in constructing the non-local correspondence. Alternatively, values that are too large result in the inclusion of regions of the image that are not anatomically indicative of the label of interest.
Fig. 4.
Sensitivity to NLS model parameters. The sensitivity of NLS to σi (A) and σd (B) demonstrate degraded performance for values that are either too small or too large. Regardless, consistent improvement over a locally weighted vote is achieved. Gray outlines indicate the values used in the previously presented experiments.
4 Discussion
Non-Local STAPLE represents the first statistical fusion algorithm that (1) creates a cohesive theoretical model specifically targeting registered atlas observation behavior, and (2) incorporates intensity seamlessly into the core of the fusion framework's rater model. Both of these goals are accomplished through the reformulation of the STAPLE algorithm from a non-local means perspective and the integration of the concept of non-local correspondence into the estimation process. NLS models atlas observation behavior by learning which label would have been observed, given perfect correspondence between the target and the atlases. NLS overcomes several of the current obstacles that plague both the accuracy and theoretical underpinning of label fusion algorithms. We demonstrated superior performance over premier label fusion algorithms on two empirical multi-atlas experiments for segmentation of the thyroid (Figure 2) and whole-brain segmentation (Figures 3 and 4).
While the sensitivity of NLS is assessed with respect to the number of atlases fused (Figure 3A) and the parameters σi and σd (Figure 4), several questions still persist in order to understand the optimality of the algorithm. For example, the effect of using an alternative similarity metric (e.g., normalized correlation coefficients or mutual information) to the assumed Gaussian difference model presented here (Eq. 3) need to be investigated. Alternative similarity measures may dramatically lessen the impact of noise in the intensity images and the need for accurate intensity normalization between the target and the atlases. Additionally, automated techniques for determining optimal window sizes (i.e. and ) and initialization strategies would provide valuable advancements for the applicability of NLS to new problem spaces.
Recently, several advancements to the STAPLE framework have been suggested to account for spatially varying labeling difficulty [2, 10] and rater performance [8, 9]. We propose that these advancements could be integrated in a straightforward manner into the current theoretical formulation governing NLS. Further investigation into their applicability to the NLS framework represents fascinating areas of continuing research. Additionally, while not presented here, future investigation into the relationship between NLS and non-local voting based algorithms [6] is critical to understanding the importance of integrating performance level estimates into the multi-atlas estimation process. Lastly, integration of Markov Random Fields [5, 7] and global/local atlas pre-selection [9] into NLS could provide valuable benefits in terms of segmentation accuracy (e.g., limiting the effects in Figure 2B).
Acknowledgments
This work was supported in part by NIH/NINDS 1R01EB006136, 1R01EB006193, 1R03EB012461, and 1R21NS064534. The authors thank Dr. Andrew Worth and Dr. Benoit Dawant for their expertly labeled datasets.
References
- 1.Heckemann RA, et al. Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage. 2006;33:115–126. doi: 10.1016/j.neuroimage.2006.05.061. [DOI] [PubMed] [Google Scholar]
- 2.Rohlfing T, et al. Performance-based classifier combination in atlas-based image segmentation using expectation-maximization parameter estimation. Medical Imaging, IEEE Transactions on. 2004;23:983–994. doi: 10.1109/TMI.2004.830803. [DOI] [PubMed] [Google Scholar]
- 3.Artaechevarria X, et al. Combination strategies in multi-atlas image segmentation: Application to brain MR data. Medical Imaging, IEEE Transactions on. 2009;28:1266–1277. doi: 10.1109/TMI.2009.2014372. [DOI] [PubMed] [Google Scholar]
- 4.Isgum I, et al. Multi-atlas-based segmentation with local decision fusion—Application to cardiac and aortic segmentation in CT scans. Medical Imaging, IEEE Transactions on. 2009;28:1000–1010. doi: 10.1109/TMI.2008.2011480. [DOI] [PubMed] [Google Scholar]
- 5.Sabuncu MR, et al. A generative model for image segmentation based on label fusion. Medical Imaging, IEEE Transactions on. 2010;29:1714–1729. doi: 10.1109/TMI.2010.2050897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coupé P, et al. Patch-based segmentation using expert priors: Application to hippocampus and ventricle segmentation. NeuroImage. 2011;54:940–954. doi: 10.1016/j.neuroimage.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 7.Warfield SK, et al. Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. Medical Imaging, IEEE Transactions on. 2004;23:903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asman A, Landman B. Information Processing in Medical Imaging (IPMI) Springer; 2011. Characterizing spatially varying performance to improve multi-atlas multi-label segmentation. pp. 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weisenfeld N, Warfield S. Learning likelihoods for labeling (L3): a general multi-classifier segmentation algorithm. Medical Image Computing and Computer-Assisted Intervention–MICCAI 2011. 2011:322–329. doi: 10.1007/978-3-642-23626-6_40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asman A, Landman B. Robust Statistical Label Fusion through Consensus Level, Labeler Accuracy and Truth Estimation (COLLATE). Medical Imaging, IEEE Transactions on. 2011;30:1779–1794. doi: 10.1109/TMI.2011.2147795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buades A, et al. Computer Vision and Pattern Recognition (CVPR) Vol. 62. IEEE; 2005. A non-local algorithm for image denoising. pp. 60–65. [Google Scholar]
- 12.Rohde GK, et al. The adaptive bases algorithm for intensity-based nonrigid image registration. Medical Imaging, IEEE Transactions on. 2003;22:1470–1479. doi: 10.1109/TMI.2003.819299. [DOI] [PubMed] [Google Scholar]