Abstract
Background
The IG20/MADD gene is overexpressed in thyroid cancer tissues and cell lines, and can contribute to tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) resistance. The ability of the MADD protein to resist TRAIL-induced apoptosis is dependent upon its phosphorylation by Akt. Interestingly, while TRAIL induces a significant reduction in the levels of phospho-Akt (pAkt) and phospho-MADD (pMADD) in TRAIL-sensitive cells, it fails to do so in TRAIL-resistant cells. In this study, we investigated if MADD phosphorylation by Akt was contributing to TRAIL resistance in thyroid cancer cells.
Methods
We determined the susceptibility of different thyroid cancer cell lines to TRAIL-induced apoptosis by fluorescence-activated cell sorting (FACS) analysis. We tested for various TRAIL resistance factors by FACS analyses or for IG20/MADD expression by quantitative reverse transcription–polymerase chain reaction. We determined the levels of pAkt and pMADD upon TRAIL treatment in thyroid cancer cells by Western blotting. We tested if down-modulation of IG20/MADD gene expression using shRNA or phosphorylation using a dominant negative Akt (DN-Akt) or pretreatment with LY294002, a PI3 kinase inhibitor, could help overcome TRAIL resistance.
Result
BCPAP and TPC1 cells were susceptible, while KTC1 and FTC133 cells were resistant, to TRAIL-induced apoptosis. The differential susceptibility to TRAIL was not related to the levels of expression of death receptors, decoy receptors, or TRAIL. KTC1 and FTC133 cells showed higher levels of IG20/MADD expression relative to BCPAP and TPC1, and were rendered susceptible to TRAIL treatment upon IG20/MADD knockdown. Interestingly, upon TRAIL treatment, the pAkt and pMADD levels were reduced in TRAIL-sensitive BCPAP and TPC1 cells, while they remained unchanged in the resistant KTC1 and FTC133 cells. While expression of a constitutively active Akt in BCPAP and TPC1 cells rendered them resistant to TRAIL, pretreating KTC1 and FTC133 cells with LY294002 rendered them TRAIL-sensitive. Moreover, expression of a DN-Akt in KTC1 and FTC133 cells reduced the levels of pAkt and pMADD and sensitized them to TRAIL-induced apoptosis.
Conclusion
Our results show that pMADD is an important TRAIL resistance factor in certain thyroid cancer cells and suggest that down-modulation of either IG20/MADD expression or phosphorylation can render TRAIL-resistant thyroid cancer cells sensitive to TRAIL.
Introduction
A majority of patients with thyroid cancer who undergo appropriate treatment (thyroidectomy and radioactive iodine [RAI] treatment) have an excellent outcome. However, ∼10% of patients with well-differentiated thyroid cancer (papillary thyroid cancer [PTC] and follicular thyroid cancer [FTC]) are refractory to the conventional therapy because the tumors lose their ability to take up RAI or become poorly differentiated or dedifferentiated, resulting in recurrent disease and death (1). Alternatively, drugs directed to specific targets, such as kinase inhibitors, can achieve, in most cases, only a partial response and the toxicities may be significant (2). Therefore, a treatment that can selectively kill cancer cells with little or no effect on normal cells is highly desired. The tumor necrosis factor–related apoptosis-inducing ligand (TRAIL) is one such potential therapeutic agent (3).
TRAIL can selectively kill tumor cells, while sparing most of the normal cells. Although the specific reason for this selective effect on cancer versus normal cells is not known, it is thought to be due to differential expression of death receptors (DRs) and decoy receptors (DcRs). TRAIL binds to DRs and causes DR oligomerization, which results in the formation of the death-inducing signaling complex (DISC) via the recruitment of Fas-associated death domain (FADD) protein and procaspase-8 to the DR. Procaspase-8 undergoes proximity-induced activation to form active caspase-8, which activates caspase-3 and results in apoptosis (4). In spite of its selective apoptotic effect on cancer cells, the clinical utility of TRAIL in cancer treatment has been limited due to the expression of TRAIL resistance factors (5–7).
We have identified the human IG20 (insulinoma–glucagonoma clone 20 and also known as MADD) gene as a TRAIL resistance factor in different cancers (8–14). The IG20/MADD gene can encode four different isoforms in non-neuronal cells (15), and two additional isoforms in select neuronal cells (13). The non-neuronal isoforms are characterized by the differential expression of exon 13L and 16 in that IG20pa splice variant expresses both exons, while MADD, IG20-SV2, and DENN-SV splice variants* fail to express exon 16, 13, or both, respectively (15). Using exon-specific shRNAs and expression of individual exogenous splice isoforms, in the absence of all endogenous isoforms of IG20, we have conclusively demonstrated that only the MADD isoform, and not the other isoforms of the IG20 gene, can confer resistance to spontaneous as well as TRAIL-induced apoptosis in a variety of cancer cells, including thyroid cancer cells (10,14).
MADD binds to the cytoplasmic tail of the DRs and prevents DISC formation by blocking FADD from binding to DRs. Since many cancer cells express higher levels of MADD, but are susceptible to TRAIL, we investigated how the antiapoptotic effect of MADD is overcome in TRAIL-susceptible cells. More recently, we showed that the MADD function is physiologically regulated by Akt. While Akt phospho-MADD (pMADD) can bind to the DRs and prevent FADD recruitment, the non-pMADD cannot bind to DRs, and thus, allows FADD recruitment to DRs followed by the formation of the DISC, which eventually results in apoptosis. Interestingly, in TRAIL-susceptible cells, TRAIL treatment resulted in reduced levels of phospho-Akt (pAkt) and pMADD; however, in TRAIL-resistant cells the pAkt and pMADD levels remained unchanged (9).
Earlier, we showed that IG20/MADD is overexpressed in thyroid cancer tissues and in WRO and FRO cell lines compared to normal thyroid tissues (10). The WRO cells, but not the FRO cells, were susceptible to TRAIL-induced apoptosis and became even more susceptible upon IG20/MADD knockdown (10). This very limited study indicated the potential utility of knocking down IG20/MADD in rendering thyroid cancer cells susceptible to TRAIL; it also pointed to the potential limitation arising from the heterogeneity of thyroid cancer cells of different origin in their responses to TRAIL treatment (10).
To understand the underlying molecular basis for TRAIL susceptibility and resistance in thyroid cancer cells, we investigated four additional thyroid cancer cell lines (i.e., BCPAP, TPC1, KTC1, and FTC133) for their susceptibility to TRAIL. More importantly, we examined the role of Akt pMADD in TRAIL resistance and explored the potential utility of down-modulating either MADD expression using a MADD-specific shRNA or MADD phosphorylation using LY294002 pretreatment or a dominant-negative Akt (DN-Akt) in overcoming TRAIL resistance in thyroid cancer cells.
Materials and Methods
Thyroid cancer cell lines and normal thyroid tissues
We used a panel of four thyroid cancer cell lines; namely, BCPAP, TPC1, KTC1, and FTC133, which were kindly provided by Dr. Bryan Haugen, Division of Endocrinology, Diabetes, and Metabolism, the University of Colorado Denver, Aurora, Colorado, CO. The above cell lines have been genetically fingerprinted and verified to be unique (16). The BCPAP cells were cultured in Dulbecco's modified Eagle's medium (DMEM) and the TPC1 cells were cultured in RPMI 1640 containing 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, and 0.1 mM non-essential amino acid (NEAA). The KTC1 cells were cultured in RPMI 1640 with 10% FBS and 0.1 mM NEAA, and the FTC133 cells were cultured in the DMEM/HAM-F12 1:1 medium supplemented with 10% FBS. All culture media were supplemented with 100 units/mL penicillin-G and 100 g/mL streptomycin, and the cells were maintained at 37°C in a humidified atmosphere with 5% CO2. Normal human thyroid tissues were obtained and RNA extracted as described before (10). The study using human tissues was approved by the institutional review board at the University of Illinois at Chicago.
Antibodies
Antibodies against phospho-ERK, total ERK, pAkt, and total Akt were purchased from Cell Signaling Technology, Inc. Production and characterization of the anti-MADD and anti-pMADD antibodies have been described previously (9). The goat anti-mouse IgG1 peroxidase-conjugated secondary antibody was obtained from Caltag Laboratories and the anti-rabbit peroxidase-conjugated polyclonal secondary antibody was purchased from GE Healthcare. The donkey anti-goat or the goat anti-rabbit IgG antibodies were purchased from Jackson Immunoresearch Laboratories. The HRP-conjugated anti-β-actin antibody used as loading control was purchased from Sigma-Aldrich. Phycoerythrin (PE)-conjugated mouse IgG isotype control, anti-DR4, anti-DR5, anti-DcR1, anti-DcR2, and anti-TRAIL antibodies were purchased from eBioscience.
Quantitative real-time polymerase chain reaction
Quantitative real-time reverse transcription–polymerase chain reaction (qRT-PCR) was carried out using TaqMan® one-step RT-PCR Master Mix reagents (Applied Biosystems) according to the manufacturer's protocol. Briefly, 100 ng of total RNA extracted either from the normal thyroid tissue or from thyroid cancer cell lines using the Trizol® reagent (Invitrogen) was added into 25 μL of the reaction mixture containing 12.5 μL of Master mix, 0.75 μL of 40× MultiScribe, RNase inhibitor mix, 0.75 nM each primer, and 0.25 nM probe. The primer and probe sequences have been reported earlier (8). The RT-PCR was performed as follows: reverse transcription at 48°C for 30 minutes, denaturation at 95°C for 10 minutes followed by 40 cycles of denaturation at 95°C for 15 seconds, annealing at 60°C for 1 minute. TaqMan 18s Ribosomal RNA control reagents (Applied Biosystems) were used for amplifying the endogenous control. The ΔΔCT method was used to calculate the relative expression levels of tested genes versus to 18s rRNA. Data were analyzed using the q-gene program (17).
Reverse transcription–polymerase chain reaction
Total RNA from cell lines was prepared using the Trizol reagent (Invitrogen) and was used for RT-PCR using the Super-Script III One-Step RT-PCR system (Invitrogen) according to the manufacturer's protocol. Briefly, 1 μg RNA from each cell line was reverse transcribed with gene-specific primers for IG20/MADD and GAPDH at 50°C for 30 minutes, followed by 30 cycles of PCR with denaturation at 94°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 1 minute, followed by a final incubation at 72°C for 7 minutes. The primers used to amplify IG20/MADD and GAPDH have been described previously (15). The amplified products were separated using a 2% agarose gel.
Apoptosis assay
Thyroid cancer cells were cultured in six-well plates. To evaluate sensitivity to TRAIL-induced apoptosis, we treated the cells with different concentrations of TRAIL (Peprotech) for 6 hours. In one set of experiments, KTC1 and FTC133 cells were pretreated with 10 μM of LY294002, aPI3K inhibitor (EMD Millipore), for 1 hour. The cells were collected, washed in cold phosphate-buffered saline (PBS), and stained either with 100 nM of tetramethyl-rhodamine methyl ester (TMRM) for 15 minutes at 37°C or with 20 μL of PE-conjugated antibody against activated caspase-3 (Clone C92-605, BD Pharmingen) on ice for 20 minutes. The cells were washed with ice-cold PBS, and then subjected to fluorescence-activated cell sorting (FACS) analysis. The loss of TMRM staining, because of mitochondrial depolarization, or an increase of PE-positive cells was used as a marker of apoptosis.
Lentivirus production and transduction
Subconfluent 293T cells grown in 100-mm plates were cotransfected with 10.8 μg of the lentivirus vector, 0.6 μg of pcRev, 0.6 μg of pcTat, and 0.3 μg of pHIT/G using calcium phosphate. The culture medium was replaced after 16 hours, and the supernatant containing the lentivirus was harvested at 40 hours post-transfection and filtered using a 0.45-μm filter as described previously (13). The SCR lentivirus expresses a scrambled (SCR) shRNA and served as a negative control. The MID shRNA can target exon 15 of the IG20/MADD gene expressed in all its splice isoforms identified to date, and therefore is capable of down-modulating all splice variants expressed in a cell. Thyroid cancer cells were transduced with lentiviruses and the transduction efficiency was monitored through green fluorescent protein (GFP) expression using flow cytometry. Invariably, greater than 85% of the cells were positive. The knock down of IG20/MADD was confirmed by RT-PCR and Western blot at 72 hours post-transduction.
Flow cytometry to detect various cell surface proteins
Cell surface expression of DRs/DcRs was evaluated by flow cytometry as previously described (8,14). For each cell line, 1×106 cells were incubated with 5.0 μg of the appropriate anti-DR/DcR antibody for 45 minutes. Cells were then washed, fixed with 1% formaldehyde-PBS, and analyzed using a Cyan ADP flow cytometer (Beckman Coulter).
Transfection with DN-Akt construct
KTC1 or FTC133 cells were plated into six-well plates at 1.2×105 cells/well and cultured overnight. These cells were transfected with either pcDNA, which served as a control, or a DN-Akt vector (0.4 μg each) using the Qiagen effectene transfection reagent. One set of cells was cotransfected with a vector carrying the GFP protein (EGFP) at a ratio of 1:1 to ensure that the transfection efficiency was greater than 80%. Cells were cultured for 42 hours and left untreated or treated with the 100 ng/mL of TRAIL for 6 hours, and cells were collected and used either to extract proteins for immunoblot analysis or stain with 100 nM TMRM for 15 minutes at 37°C and subjected to FACS analysis to determine apoptosis.
Immunoblot analysis
Cells were lysed for 30 minutes at 4°C in a lysis buffer (20 mM Tris pH 7.5, 2 mM EDTA, 3 mM EGTA, 2 mM dithiothreitol, 250 mM sucrose, 0.1 mM phenylmethylsulfonyl fluoride, 1% Triton X-100) containing a protease inhibitor cocktail and a phosphatase inhibitor (Sigma-Aldrich). The protein concentration was estimated using the Bradford assay (Biorad). Fifty micrograms of samples was subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis using 5%–15% gel and transferred to nitrocellulose membranes. The membranes were blocked with 5% milk in PBS with 0.05% Tween 20 (PBST) for 1 hour and incubated with the primary (1:1000) and the secondary antibodies (1:10,000) diluted in PBST containing 5% milk. All blots were developed using the ECL plus kit following the manufacturer's protocol (GE Healthcare).
Statistical analysis
All results are expressed as mean±standard error of the mean. The Student's t-test was used to determine p-values using Microsoft Excel (version 2003). p-values <0.05 were considered significant.
Results
TRAIL-induced apoptosis of thyroid cancer cells
The TPC1 cells were highly sensitive to TRAIL-induced apoptosis (>45% cell death) at a lower concentration of TRAIL (6.125 ng/mL), which increased to over 70% at higher concentrations of TRAIL (Fig. 1). Similarly, the BCPAP cells were susceptible and showed 40% apoptosis upon treatment with a dose of 12.5 ng/mL of TRAIL that increased modestly with an increasing dose of TRAIL. Both KTC1 and FTC133 cells were relatively more resistant to TRAIL and showed <20% apoptosis even when treated with the highest concentration of TRAIL (100 ng/mL; Fig. 1).
FIG. 1.
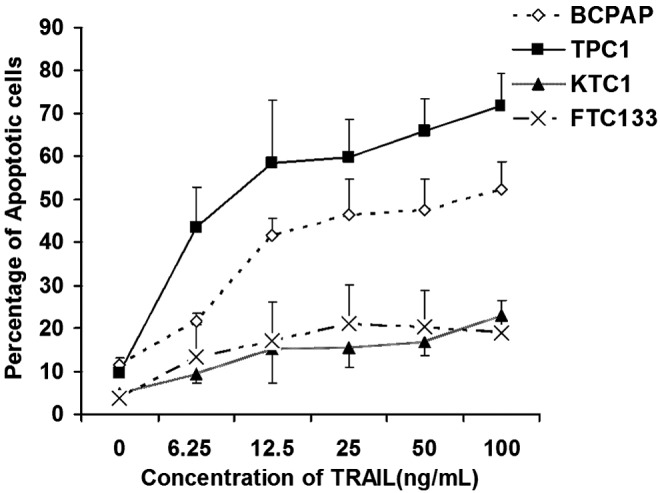
Susceptibility of different thyroid cancer cell lines to tumor necrosis factor–related apoptosis-inducing ligand (TRAIL)–induced apoptosis. Approximately 3×105 of BCPAP, TPC1, KTC1, and FTC133 thyroid cancer cells were cultured in 6-well plates, and treated with different doses of TRAIL. Subsequently, cells were stained with tetramethyl-rhodamine methyl ester (TMRM) and subjected to fluorescence-activated cell sorting (FACS) analysis. Loss of TMRM staining, because of mitochondrial depolarization, was used as a marker of apoptosis. Summarized data are shown from three independent experiments.
Expression of DRs, DcRs, and IG20/MADD in thyroid cancer cell lines
Since the levels of expression of DRs and DcRs can affect TRAIL-induced apoptosis, we evaluated the cell surface expression of these receptors. We found that the DR4 expression was minimal, while DR5 was readily detected in all four cell lines. Examination of DcR1, DcR2, or TRAIL expression revealed little or no expression on any of the cell lines tested (Fig. 2A). Next, we tested for the expression levels of IG20/MADD in normal thyroid tissues and the thyroid cancer cell lines by qRT-PCR. The results showed that the expression level of IG20/MADD RNA was higher in all four thyroid cancer cell lines, but much higher in KTC1 and FTC133 cell lines (Fig. 2B) compared to the levels in normal thyroid tissues.
FIG. 2.
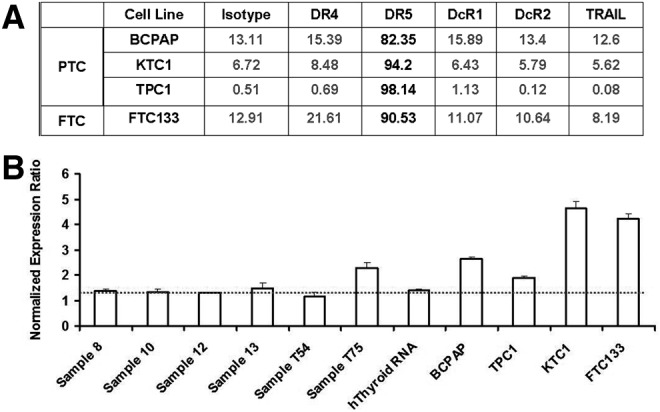
Expression of TRAIL resistance factors in thyroid cancer cell lines. (A) Expression levels of death receptors (DRs), decoy receptors (DcRs), and TRAIL in different thyroid cancer cell lines; (B) expression of endogenous IG20/MADD in thyroid tissues and thyroid cancer cell lines detected by quantitative reverse transcription–polymerase chain reaction (RT-PCR). Sequences of primers and the probe have been published earlier (8), and 18s rRNA primer was used as internal control. The experiment was carried out in triplicate, the dash line indicate the average normal expression level of IG20/MADD gene.
Effects of IG20/MADD knockdown on TRAIL-induced apoptosis
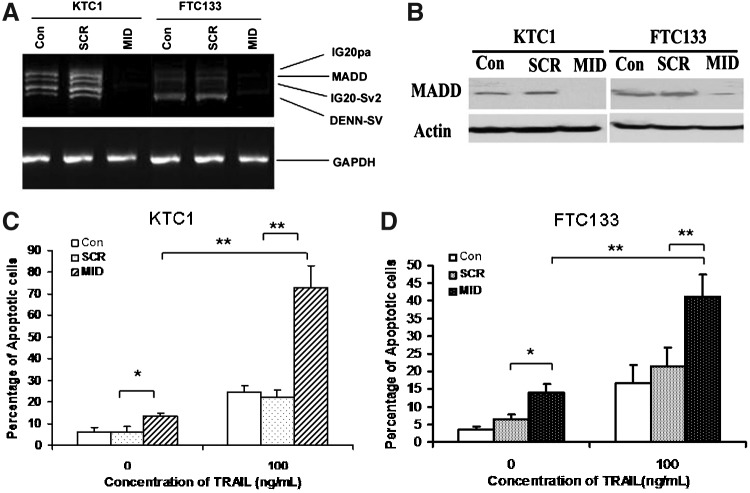
Thyroid cancer cells were transduced with lentiviruses that can express either the MID-shRNA that can target all isoforms of the IG20 gene or the SCR-shRNA, as a control. The transduction efficiency was monitored through coexpression of GFP using flow cytometry. Invariably, greater than 85% of the cells were positive (data not shown). Seventy-two hours post-transduction, cells were harvested and RNA and protein were extracted and subjected to RT-PCR or Western blot, respectively. As shown in Figure 3, the IG20/MADD transcripts (Fig. 3A) and the protein expression (Fig. 3B) were essentially absent by 72 hours post-transduction in cells transduced with MID-shRNA, but not in cells that were left untreated or treated with a SCR-shRNA.
FIG. 3.
IG20/MADD knockdown sensitizes resistant thyroid cancer cells to TRAIL-induced apoptosis. (A, B) Efficiency of IG20 knockdown using MID-shRNA in thyroid cancer cell lines: total of 6×105 of KTC1 and FTC133 cells were cultured in 60-mm dishes and transduced with SCR or MID-shRNA carrying lentivirus for 3 days. (A) One microgram of total RNA was used for RT-PCR. The amplified products were separated using a 2% Tris-Borate-EDTA (TBE) agarose gel. The GAPDH served as a loading control. Western blot (B): fifty micrograms of protein were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel, and transferred to nitrocellulose membranes. Blots were stained using anti-13L antibody. β-actin was used as a loading control. The protein bands were visualized by enhanced chemiluminescence. (C, D) Effect of IG20/MADD knockdown on TRAIL-induced apoptosis in TRAIL-resistant thyroid cancer cell lines: total of 3×105 of KTC1 and FTC133 cells were cultured in six-well plates overnight, and then transduced with SCR or MID-shRNA carrying lentivirus for 72 hours. These cells were incubated with 100 ng/mL of TRAIL for an additional 6 hours. The cells were stained with TMRM and subjected to FACS analysis by gating for green fluorescent protein-positive cells. Summarized data are shown from three independent experiments. *p<0.05; **p<0.01.
To test whether IG20/MADD knockdown can render TRAIL-resistant thyroid cancer cells susceptible, we transduced KTC1 and FTC133 cells with lentivirus encoding either SCR-shRNA or MID-shRNA. Starting at 72 hours post-transduction, these cells were treated with 100 ng/mL of TRAIL for 6 hours. Upon IG20/MADD knockdown, the TRAIL-sensitive cell lines, BCPAP and TPC1, remained sensitive to TRAIL-induced apoptosis (data not shown). Interestingly, as shown in Figure 3C and D, knockdown of IG20/MADD caused a modest spontaneous apoptosis in both KTC1 (6.04±2.04 vs. 13.66±0.83; p<0.05) and FTC133 cell (3.53±0.95 vs. 14.65±2.37; p<0.05). The KTC1 cells showed ∼24.63%±2.9% apoptosis when treated with 100 ng/mL of TRAIL; however, upon IG20/MADD knockdown, the percentage of KTC1 cells undergoing TRAIL-induced apoptosis increased to over 72.80%±10.25% (p<0.05). Similarly, the FTC133 cells showed further dramatically increased apoptosis (∼45.04%±6.7% apoptosis, p<0.01) upon IG20/MADD knockdown followed by TRAIL treatment (Fig. 3D). These results indicate that IG20/MADD knockdown can help overcome resistance to TRAIL-induced apoptosis in certain TRAIL-resistant thyroid cancer cell lines.
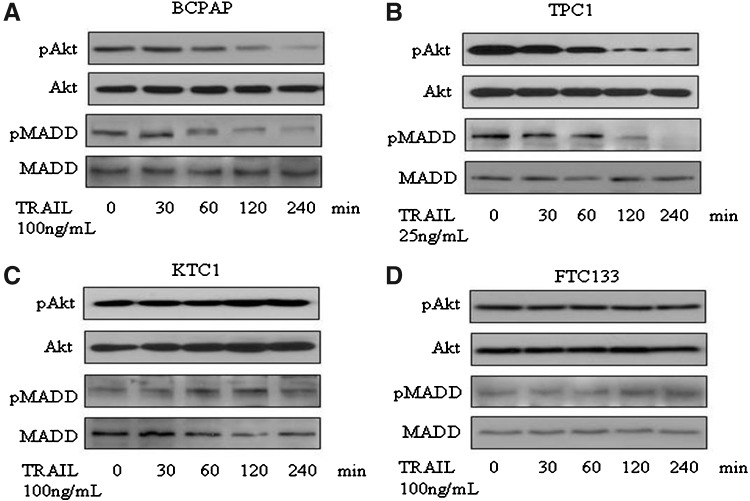
Effect of TRAIL treatment on pAkt and pMADD levels in thyroid cancer cells
We treated thyroid cancer cells with optimal doses of TRAIL for different durations (0, 30, 60, 120, and 240 minutes). Cell lysates were prepared and separated on a 5%–15% gradient gel and subjected to immunoblotting to detect pAkt and pMADD. Total Akt and MADD served as loading controls. As expected, in TRAIL-sensitive cell lines (BCPAP and TPC1) (Fig. 4A, B), the levels of pAkt were significantly reduced beginning at 1 hour after TRAIL treatment, while in the resistant cell lines (KTC1 and FTC133), the levels of pAkt were not reduced upon TRAIL treatment (Fig. 4C, D). In BCPAP and TPC1 cell lines, there was a concomitant decline in the levels of pMADD with very little or no detectable pMADD by 4 hours (Fig. 4A, B), while the levels of pMADD remained unchanged in the KTC1 and FTC133 cells (Fig. 4C, D). The importance of pAkt/pMADD in TRAIL-induced apoptosis in thyroid cancer cells became more apparent when expression of constitutively active Akt (ca-Akt) in TPC1 and BCPAP cells could confer resistance to TRAIL-induced apoptosis in those cells (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/thy).
FIG. 4.
Expression levels of phospho-MADD (pMADD) and phospho-Akt (pAkt) upon TRAIL treatment in thyroid cancer cell lines: total of 6×105 of (A) BCPAP, (B) TPC1, (C) KTC1, and (D) FTC133 thyroid cancer cells were cultured in 60-mm dishes overnight and treated with TRAIL at the concentration and for the duration indicated. Cell lysates containing 50 μg of protein were subjected to SDS-PAGE and transferred to nitrocellulose membranes. Blots were stained using specific antibodies against Akt, pAkt, MADD, and pMADD. The protein bands were visualized by enhanced chemiluminescence.
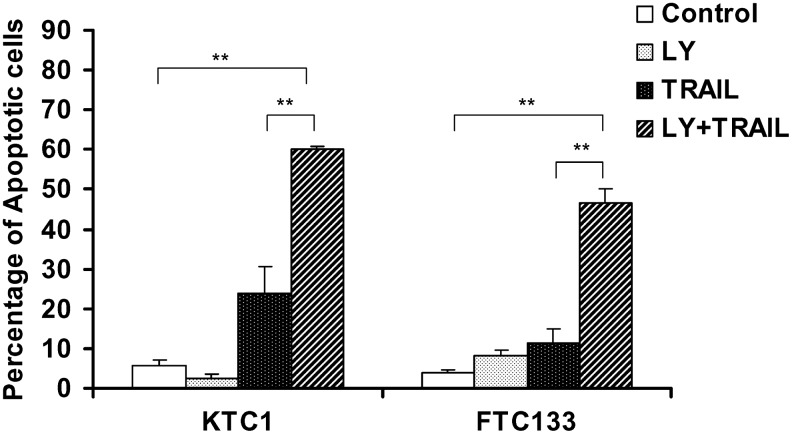
LY294002 treatment can render KTC1 and FTC133 cells TRAIL susceptible
To determine if Akt activation that is required for MADD phosphorylation is essential for TRAIL resistance, we treated the cells with LY294002, a PI3K inhibitor. We found that pretreatment of the KTC1 and FTC133 cells, which were resistant to TRAIL-induced apoptosis, with LY294002 (10 μM) for 1 hour, sensitized them to TRAIL-induced apoptosis. TRAIL-induced apoptosis of KTC1 cells increased from ∼25% to nearly 60%, while that of FTC133 increased from ∼12% to nearly 50% (Fig. 5). These data indicate the potential importance of Akt activation in TRAIL resistance observed in KTC1 and FTC133 cells.
FIG. 5.
Pretreatment with the PI3K inhibitor LY 294002 (LY) sensitizes KTC1 and FTC133 cells to TRAIL-induced apoptosis. Total of 1.6×105 of KTC1 and FTC133 cells were cultured in 6-well plates. The cells were serum starved overnight, and treated with 10 μM of LY for 1 hour followed by treatment with TRAIL (100 μg/mL) for 6 hours. The cells were harvested and stained with PE-labeled anti-caspase-3 antibody and subjected to FACS analysis. Data are summarized from three experiments. **p<0.01.
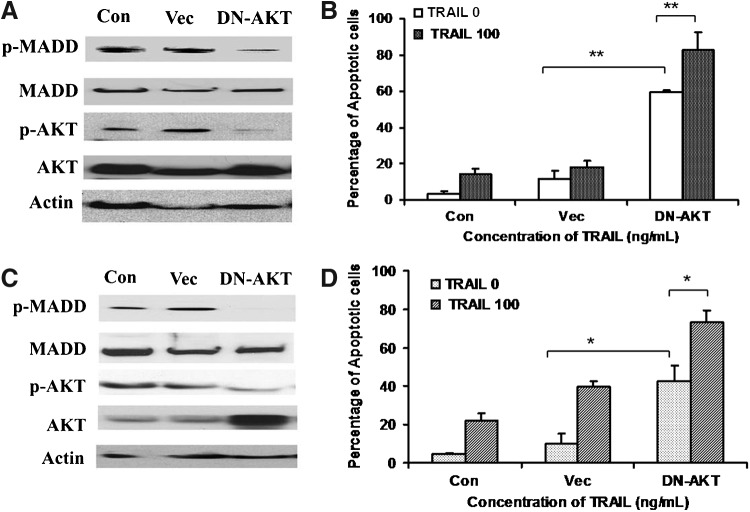
Expression of a DN-Akt sensitizes KTC1 and FTC133 to TRAIL-induced apoptosis
To more specifically demonstrate the contribution of Akt pMADD in TRAIL resistance of KTC1 and FTC133 cells, we transfected these cells with a control vector or a DN-Akt. Forty-two hours post-transfection, the cells were left untreated or treated with 100 ng/mL of TRAIL for 6 hours and analyzed for apoptosis. As shown in Figure 6, overexpression of DN-Akt resulted in reduced levels of pAkt and pMADD in both KTC1 (Fig. 6A) and FTC133 (Fig. 6C) cells. This treatment also caused spontaneous apoptosis in both KTC1 (control vector: 11.76±4.3 vs. DN-Akt vector: 59.68±1.06; p<0.01, Fig. 6B) and FTC133 (control vector: 10.14±5.1 vs. DN-Akt: 42.69±8.13; p<0.05, Fig. 6D) cells. The apoptosis was further increased upon TRAIL treatment in both KTC1 (59.68±1.06 vs. 82.78±9.87; p<0.05, Fig. 6B) and FTC133 (42.69±8.13 vs. 73.0±6.3; p<0.01, Fig. 6D). These results indicate that DN-Akt reduced the levels of pAkt and, as a consequence, it also reduces the levels of pMADD and renders these cells more susceptible to spontaneous as well as TRAIL-induced apoptosis.
FIG. 6.
Dominant-negative Akt (DN-Akt) sensitizes thyroid cancer cells to TRAIL-induced apoptosis. (A, C) Expression of DN-Akt reduces the levels of pAkt and pMADD in KTC1 and FTC133 cells. Total of 8×104 of KTC1 and FTC133 cells were cultured in six-well plates and transfected with either an empty vector (Vec) or a DN-Akt and cultured for 42 hours. The cells were treated with 100 ng/mL of TRAIL for an additional 6 hours, washed in cold PBS, and then proteins were subjected to Western blotting to detect pAkt and pMADD; β-actin was used as a loading control. (B, D) Overexpression of DN-Akt leads to spontaneous as well as TRAIL-induced apoptosis in KTC1 and FTC133 cells. The same set of KTC1 and FTC133 cells as shown in (A) and (C) were stained with TMRM and subjected to FACS analysis. Summarized data are shown from three independent experiments, *p<0.05; **p<0.01.
Discussion
Thyroid cancer is the most common endocrine malignancy with a rapidly increasing incidence in recent decades. Although surgical and RAI treatments are generally effective, about 10% of the patients with well-differentiated thyroid cancer (both PTC and FTC) lose their ability to take up RAI, leading to recurrent disease and death (1). Therefore, more effective molecular-targeted therapeutic strategies need to be developed.
TRAIL is a promising anti-cancer drug, which induces apoptosis in many human cancer cells with little or no effect on normal cells (12,18–20). Therefore, it is an attractive potential therapeutic agent for thyroid cancer. Earlier, we had shown that the IG20 gene was expressed at higher levels in thyroid cancer tissues and WRO and FRO cell lines relative to normal thyroid, and the MADD and DENN-SV were the predominant isoforms. Additionally, we had shown that MADD knockdown can render WRO, not FRO, cells susceptible to spontaneous apoptosis. Similarly, WRO cells were susceptible, while FRO cells were resistant to TRAIL-induced apoptosis because of the lack of caspase-8 expression in FRO cells (10). From this limited study, we had concluded that MADD can act as a resistance factor against TRAIL-induced apoptosis.
In the current study, we tested the sensitivity of four different thyroid cancer cell lines to TRAIL-induced apoptosis and observed that among the PTC-derived cell lines, BCPAP cells and TPC1 cells were highly sensitive to TRAIL-induced apoptosis even when TRAIL was used at a lower concentration. In contrast, the PTC-derived cell line, the KTC1 and the FTC-derived cell line FTC133 were only marginally susceptible even when a high concentration of TRAIL was used. These results are consistent with a previous publication (7) that demonstrated that TRAIL resistance factors may oppose TRAIL-induced apoptosis.
The differential TRAIL susceptibility of these thyroid cancer cells led us to wonder whether differential expression of various TRAIL resistance factors, including the DR1 and DR2, DcR1 and DcR2 could account for the noted differential susceptibility. However, our analysis showed little or no relationship between the levels of expression of these molecules (Fig. 2A) and susceptibility to TRAIL, and suggest that factors other than the levels of expression of these molecules are most likely responsible for the differential susceptibility to TRAIL. Since other studies in our laboratory had shown that the MADD splice variant was responsible for the resistance to spontaneous as well as TRAIL-induced apoptosis (8–14), we sought to test for MADD protein expression in various cell lines used in the current study. Interestingly, thyroid cancer cell lines expressed higher levels of IG20/MADD relative to normal thyroid tissues. The levels of expression were higher in TRAIL-resistant KTC1 and FTC 133 cells relative to TRAIL-susceptible TPC1 and BCPAP cells and suggest that it may be contributing to TRAIL resistance.
To determine whether IG20/MADD was contributing to the observed resistance of KTC1 and FTC133 cells to TRAIL-induced apoptosis, we knocked down the IG20/MADD gene expression, and found that it could render KTC1 and FTC133 cells susceptible to TRAIL-induced apoptosis (Fig. 3). These findings are consistent with our earlier findings of increased TRAIL susceptibility, upon MADD knockdown, of different cancer cell lines, including the WRO thyroid cancer cells (8–14).
In a more recent study, we had shown that the ability of MADD to confer resistance to either spontaneous or TRAIL-induced apoptosis of HeLa cells was related to its phosphorylation by Akt. Only pMADD could bind to DRs and prevent FADD recruitment and DISC formation. However, the non-pMADD was unable to bind to DRs and allowed FADD recruitment to DRs followed by DISC formation that resulted in apoptosis (9). Moreover, we had noted that in TRAIL-susceptible cells, the levels of pAkt as well as pMADD were significantly lower upon TRAIL treatment, but they remained unchanged in TRAIL-resistant cells (9). Therefore, we wondered whether the susceptibility or resistance of thyroid cancer cells to TRAIL-induced apoptosis was also related to the levels of pMADD. Upon TRAIL treatment, the levels of both pAkt and pMADD were significantly reduced in the TRAIL-sensitive BCPAP and TPC1 cells, while they remained essentially unchanged in the TRAIL-resistant KTC1 and FTC133 cells (Fig. 4). These data are consistent with our earlier observation in other cancer cell lines that showed decline in the levels of both pAkt and pMADD in TRAIL-susceptible cells, while they remained unchanged in resistant cells upon TRAIL treatment (9).
Interestingly, sustaining pAkt/pMADD levels through the expression of a ca-Akt rendered BCPAP and TPC1 cells resistant to TRAILL-induced apoptosis (Supplementary Fig. S1). In contrast, treatment of TRAIL-resistant KTC1 and FTC1 cells with LY294002, a PI3K inhibitor, rendered them susceptible to TRAIL-induced apoptosis (Fig. 5). Since the effects of LY is not limited to inhibiting the PI3K activity, and as a consequence Akt activity, we transfected KTC1 and FTC1 cells with either an empty vector or a vector capable of expressing DN-Akt to specifically inhibit the Akt activity and treated the cells with TRAIL. Overexpression of DN-Akt caused a significant decrease in pAkt and pMADD in KTC1 and FTC133 cells, and rendered these cells more susceptible to spontaneous as well as TRAIL-induced apoptosis (Fig. 6B, D). A reduction in pMADD levels upon TRAIL treatment could result from either upregulation of PTEN or activation of a protein phosphatase; either of which can reduce the levels of pAkt with a resultant reduction in the levels of pMADD. However, the precise mechanism by which the levels of pAkt and pMADD are reduced in TRAIL-sensitive thyroid cancer cell lines is unknown and is under active investigation.
For the first time, we have shown that Akt pMADD can contribute to TRAIL resistance in thyroid cancer. This is profoundly interesting, particularly, in light of the importance of activating mutations in Ras, PI3K, and Akt, and inactivating mutation in PTEN often found in thyroid cancer (21–32). These mutations eventually result in constitutive enhanced activation of Akt. Since MADD is an Akt substrate, it is likely to result in a concomitant increase in the levels of pMADD resulting in an increased resistance to TRAIL-induced apoptosis. Moreover, our data show that cells that are TRAIL susceptible show a significant decrease in pAkt and pMADD levels, while their levels remain unaltered in resistant cells. More importantly, if the levels of pMADD can be reduced (e.g., with LY or DN-Akt), then even the resistant cells can be rendered susceptible to TRAIL-induced apoptosis. This latter observation has profound implications for co-treating thyroid cancer cells with TRAIL and PI3K/Akt inhibitors.
In summary, our data show that TRAIL alone might be sufficient to induce apoptosis in certain thyroid cancer cells. However, in TRAIL-resistant thyroid cancer cells, TRAIL can still be used in conjunction with down-modulation of MADD expression or dephosphorylation to effectively induce apoptosis. Since TRAIL treatment and down-modulation of MADD expression or dephosphorylation can selectively induce apoptosis primarily in cancer cells but not in normal cells, combining these modalities of apoptosis induction might be a highly effective strategy to treat thyroid cancer.
Supplementary Material
Footnotes
The splice isoform is indicated by regular font, while the gene is referred to in italics.
Acknowledgments
We thank Dr. Bryan Haugen for providing the cell lines. This work was supported, in part, by the grant RO1 CA107506 to B.S.P. from the National Institutes of Health (Bethesda, MD), and by grants from Elsa U. Pardee Foundation to L.-C.L. and the American Thyroid Association to T.P.
Disclosure Statement
The authors declare that there is no conflict of interest that would prejudice the impartiality of this scientific work.
References
- 1.Kim CS. Zhu X. Lessons from mouse models of thyroid cancer. Thyroid. 2009;19:1317–1331. doi: 10.1089/thy.2009.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlumberger M. Sherman SI. Clinical trials for progressive differentiated thyroid cancer: patient selection, study design, and recent advances. Thyroid. 2009;19:1393–1400. doi: 10.1089/thy.2009.1603. [DOI] [PubMed] [Google Scholar]
- 3.Nagane M. Huang HJ. Cavenee WK. The potential of TRAIL for cancer chemotherapy. Apoptosis. 2001;6:191–197. doi: 10.1023/a:1011336726649. [DOI] [PubMed] [Google Scholar]
- 4.Bodmer JL. Holler N. Reynard S. Vinciguerra P. Schneider P. Juo P. Blenis J. Tschopp J. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat Cell Biol. 2000;2:241–243. doi: 10.1038/35008667. [DOI] [PubMed] [Google Scholar]
- 5.Wang SH. Mezosi E. Wolf JM. Cao Z. Utsugi S. Gauger PG. Doherty GM. Baker JR., Jr IFNgamma sensitization to TRAIL-induced apoptosis in human thyroid carcinoma cells by upregulating Bak expression. Oncogene. 2004;23:928–935. doi: 10.1038/sj.onc.1207213. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L. Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther. 2005;12:228–237. doi: 10.1038/sj.cgt.7700792. [DOI] [PubMed] [Google Scholar]
- 7.Park JW. Wong MG. Lobo M. Hyun WC. Duh QY. Clark OH. Modulation of tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis by chemotherapy in thyroid cancer cell lines. Thyroid. 2003;13:1103–1110. doi: 10.1089/10507250360731497. [DOI] [PubMed] [Google Scholar]
- 8.Li LC. Jayaram S. Ganesh L. Qian L. Rotmensch J. Maker AV. Prabhakar BS. Knockdown of MADD and c-FLIP overcomes resistance to TRAIL-induced apoptosis in ovarian cancer cells. Am J Obstet Gynecol. 2011;205:362.e12–362.e25. doi: 10.1016/j.ajog.2011.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li P. Jayarama S. Ganesh L. Mordi D. Carr R. Kanteti P. Hay N. Prabhakar BS. Akt-phosphorylated mitogen-activated kinase-activating death domain protein (MADD) inhibits TRAIL-induced apoptosis by blocking Fas-associated death domain (FADD) association with death receptor 4. J Biol Chem. 2010;285:22713–22722. doi: 10.1074/jbc.M110.105692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subramanian M. Pilli T. Bhattacharya P. Pacini F. Nikiforov YE. Kanteti PV. Prabhakar BS. Knockdown of IG20 gene expression renders thyroid cancer cells susceptible to apoptosis. J Clin Endocrinol Metab. 2009;94:1467–1471. doi: 10.1210/jc.2008-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kurada BR. Li LC. Mulherkar N. Subramanian M. Prasad KV. Prabhakar BS. MADD, a splice variant of IG20, is indispensable for MAPK activation and protection against apoptosis upon tumor necrosis factor-alpha treatment. J Biol Chem. 2009;284:13533–13541. doi: 10.1074/jbc.M808554200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prabhakar BS. Mulherkar N. Prasad KV. Role of IG20 splice variants in TRAIL resistance. Clin Cancer Res. 2008;14:347–351. doi: 10.1158/1078-0432.CCR-07-0493. [DOI] [PubMed] [Google Scholar]
- 13.Li LC. Sheng JR. Mulherkar N. Prabhakar BS. Meriggioli MN. Regulation of apoptosis and caspase-8 expression in neuroblastoma cells by isoforms of the IG20 gene. Cancer Res. 2008;68:7352–7361. doi: 10.1158/0008-5472.CAN-07-6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mulherkar N. Ramaswamy M. Mordi DC. Prabhakar BS. MADD/DENN splice variant of the IG20 gene is necessary and sufficient for cancer cell survival. Oncogene. 2006;25:6252–6261. doi: 10.1038/sj.onc.1209650. [DOI] [PubMed] [Google Scholar]
- 15.Al-Zoubi AM. Efimova EV. Kaithamana S. Martinez O. El-Idrissi Mel A. Dogan RE. Prabhakar BS. Contrasting effects of IG20 and its splice isoforms, MADD and DENN-SV, on tumor necrosis factor alpha-induced apoptosis and activation of caspase-8 and -3. J Biol Chem. 2001;276:47202–47211. doi: 10.1074/jbc.M104835200. [DOI] [PubMed] [Google Scholar]
- 16.Schweppe RE. Klopper JP. Korch C. Pugazhenthi U. Benezra M. Knauf JA. Fagin JA. Marlow LA. Copland JA. Smallridge RC. Haugen BR. Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J Clin Endocrinol Metab. 2008;93:4331–4341. doi: 10.1210/jc.2008-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Simon P. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 2003;19:1439–1440. doi: 10.1093/bioinformatics/btg157. [DOI] [PubMed] [Google Scholar]
- 18.Ashkenazi A. Pai RC. Fong S. Leung S. Lawrence DA. Marsters SA. Blackie C. Chang L. McMurtrey AE. Hebert A. DeForge L. Koumenis IL. Lewis D. Harris L. Bussiere J. Koeppen H. Shahrokh Z. Schwall RH. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ashkenazi A. Dixit VM. Apoptosis control by death and decoy receptors. Curr Opin Cell Biol. 1999;11:255–260. doi: 10.1016/s0955-0674(99)80034-9. [DOI] [PubMed] [Google Scholar]
- 20.Ramaswamy M. Efimova EV. Martinez O. Mulherkar NU. Singh SP. Prabhakar BS. IG20 (MADD splice variant-5), a proapoptotic protein, interacts with DR4/DR5 and enhances TRAIL-induced apoptosis by increasing recruitment of FADD and caspase-8 to the DISC. Oncogene. 2004;23:6083–6094. doi: 10.1038/sj.onc.1207804. [DOI] [PubMed] [Google Scholar]
- 21.Karga H. Lee JK. Vickery AL., Jr. Thor A. Gaz RD. Jameson JL. Ras oncogene mutations in benign and malignant thyroid neoplasms. J Clin Endocrinol Metab. 1991;73:832–836. doi: 10.1210/jcem-73-4-832. [DOI] [PubMed] [Google Scholar]
- 22.Manenti G. Pilotti S. Re FC. Della Porta G. Pierotti MA. Selective activation of ras oncogenes in follicular and undifferentiated thyroid carcinomas. Eur J Cancer. 1994;30A:987–993. doi: 10.1016/0959-8049(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 23.Basolo F. Pisaturo F. Pollina LE. Fontanini G. Elisei R. Molinaro E. Iacconi P. Miccoli P. Pacini F. N-ras mutation in poorly differentiated thyroid carcinomas: correlation with bone metastases and inverse correlation to thyroglobulin expression. Thyroid. 2000;10:19–23. doi: 10.1089/thy.2000.10.19. [DOI] [PubMed] [Google Scholar]
- 24.Fagin JA. Minireview: branded from the start-distinct oncogenic initiating events may determine tumor fate in the thyroid. Mol Endocrinol. 2002;16:903–911. doi: 10.1210/mend.16.5.0838. [DOI] [PubMed] [Google Scholar]
- 25.Saavedra HI. Knauf JA. Shirokawa JM. Wang J. Ouyang B. Elisei R. Stambrook PJ. Fagin JA. The RAS oncogene induces genomic instability in thyroid PCCL3 cells via the MAPK pathway. Oncogene. 2000;19:3948–3954. doi: 10.1038/sj.onc.1203723. [DOI] [PubMed] [Google Scholar]
- 26.Ricarte-Filho JC. Ryder M. Chitale DA. Rivera M. Heguy A. Ladanyi M. Janakiraman M. Solit D. Knauf JA. Tuttle RM. Ghossein RA. Fagin JA. Mutational profile of advanced primary and metastatic radioactive iodine-refractory thyroid cancers reveals distinct pathogenetic roles for BRAF, PIK3CA, and AKT1. Cancer Res. 2009;69:4885–4893. doi: 10.1158/0008-5472.CAN-09-0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carpten JD. Faber AL. Horn C. Donoho GP. Briggs SL. Robbins CM. Hostetter G. Boguslawski S. Moses TY. Savage S. Uhlik M. Lin A. Du J. Qian YW. Zeckner DJ. Tucker-Kellogg G. Touchman J. Patel K. Mousses S. Bittner M. Schevitz R. Lai MH. Blanchard KL. Thomas JE. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature. 2007;448:439–444. doi: 10.1038/nature05933. [DOI] [PubMed] [Google Scholar]
- 28.Kim CS. Vasko VV. Kato Y. Kruhlak M. Saji M. Cheng SY. Ringel MD. AKT activation promotes metastasis in a mouse model of follicular thyroid carcinoma. Endocrinology. 2005;146:4456–4463. doi: 10.1210/en.2005-0172. [DOI] [PubMed] [Google Scholar]
- 29.Vasko V. Saji M. Hardy E. Kruhlak M. Larin A. Savchenko V. Miyakawa M. Isozaki O. Murakami H. Tsushima T. Burman KD. De Micco C. Ringel MD. Akt activation and localisation correlate with tumour invasion and oncogene expression in thyroid cancer. J Med Genet. 2004;41:161–170. doi: 10.1136/jmg.2003.015339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paes JE. Ringel MD. Dysregulation of the phosphatidylinositol 3-kinase pathway in thyroid neoplasia. Endocrinol Metab Clin North Am. 2008;37:375–387. doi: 10.1016/j.ecl.2008.01.001. , viii-ix. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shinohara M. Chung YJ. Saji M. Ringel MD. AKT in thyroid tumorigenesis and progression. Endocrinology. 2007;148:942–947. doi: 10.1210/en.2006-0937. [DOI] [PubMed] [Google Scholar]
- 32.Ngeow J. Mester J. Rybicki LA. Ni Y. Milas M. Eng C. Incidence and clinical characteristics of thyroid cancer in prospective series of individuals with Cowden and Cowden-like syndrome characterized by germline PTEN, SDH, or KLLN alterations. J Clin Endocrinol Metab. 2011;96:E2063–E2071. doi: 10.1210/jc.2011-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.