Abstract
Acute promyelocytic leukemia (APL), a cytogenetically distinct subtype of acute myeloid leukemia (AML), characterized by the t(15;17)-associated PML-RARA fusion, has been successfully treated with therapy utilizing all-trans-retinoic acid (ATRA) to differentiate leukemic blasts. However, among patients with non- APL AML, ATRA-based treatment has not been effective. Here we show that, through epigenetic reprogramming, inhibitors of lysine- specific demethylase 1 (LSD1, also called KDM1A), including tranylcypromine (TCP), unlocked the ATRA-driven therapeutic response in non-APL AML. LSD1 inhibition did not lead to a large-scale increase in histone 3 Lys4 dimethylation (H3K4me2) across the genome, but it did increase H3K4me2 and expression of myeloid-differentiation–associated genes. Notably, treatment with ATRA plus TCP markedly diminished the engraftment of primary human AML cells in vivo in nonobese diabetic (NOD)- severe combined immunodeficient (SCID) mice, suggesting that ATRA in combination with TCP may target leukemia-initiating cells. Furthermore, initiation of ATRA plus TCP treatment 15 d after engraftment of human AML cells in NOD-SCID γ (with interleukin-2 (IL-2) receptor γ chain deficiency) mice also revealed the ATRA plus TCP drug combination to have a potent anti-leukemic effect that was superior to treatment with either drug alone. These data identify LSD1 as a therapeutic target and strongly suggest that it may contribute to AML pathogenesis by inhibiting the normal pro-differentiative function of ATRA, paving the way for new combinatorial therapies for AML.
AML currently accounts for approximately 80% of all adult acute leukemias, with a median age at diagnosis of 67 years (ref. 1). Although clinical advances in AML have been made, treatment failure in non-APL AML remains high, with a particularly poor prognosis commonly seen in the elderly, in patients with certain subtypes of AML and in patients with secondary AML after cancer therapy2. Moreover, given projected improvements in life expectancy in the general population and, concomitantly, an increase in the frequency of AML (with a 38% increase in elderly cases predicted by 2031), the development of new and effective anti-AML therapies is clearly required1. ATRA has held great promise in both cancer treatment and prevention3, and research strategies that seek to extend the efficacy of ATRA-based treatment to non-APL AML are key avenues of investigation4. Evidence points to one of the underlying reasons for ATRA resistance in AML as a failure of ATRA to induce proper transcriptional activation of retinoic acid receptor (RAR) target genes, such as TNFSF10 (ref. 5) and RARA2 (ref. 6). An epigenetic analysis of primary AML samples revealed that relative to normal CD33+ cells, loss of RARα2 expression in AML is associated with a reduction in H3K4me2 on the RARA2 promoter7 (a modification that is associated with transcriptional activation)8. The mono- and di-methyl lysine demethylase LSD1 (KDM1A)9 is highly expressed in patients with AML (http://www.proteinatlas.org/)10, and its overexpression has been implicated in various other tumors11,12. Collectively, these data predicted that the use of small-molecule inhibitors that target LSD1 (LSD1i) could result in epigenetic reprogramming that enhanced or facilitated the execution of the ATRA-induced differentiation program in AML cells. We tested two structurally unrelated compounds, trans-2-phenylcyclopropylamine (tranylcypromine, or TCP)13, which is a time-dependent, mechanism-based irreversible inhibitor of LSD1, and a non-competitive LSD1 inhibitor, 1,15–bis{N5–[3,3–(diphenyl)propyl]–N1–biguanido}–4,12–diazapentadecane (the biguanide polyamine analog 2d)14.
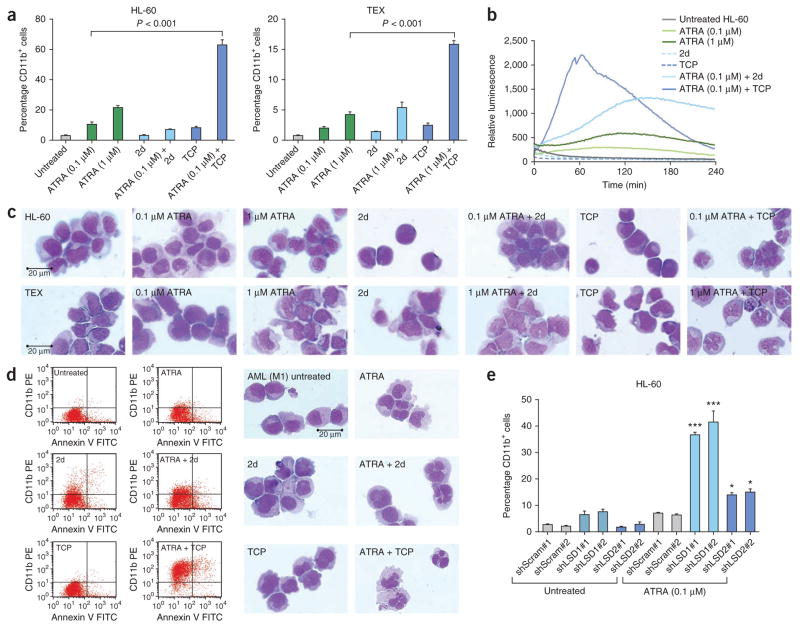
For the in vitro studies, we focused on the ATRA-responsive HL-60 AML M2 (ref. 15) cell line and on ATRA-insensitive TEX cells, which are derived from primitive human cord blood cells immortalized by expression of the FUS (TLS)-ERG oncogene16. TEX cells mimic the features of primary human AML and of leukemia-initiating cells (LIC) and are more than 90% CD34+16. Treatment with ATRA and TCP increased the fraction of cells expressing the myeloid differentiation marker CD11b (which regulates leukocyte adhesion and migration to mediate the inflammatory response) by 21-fold and by 16-fold in HL-60 and TEX cells, respectively (Fig. 1a). We obtained similar results for ATRA-responsive U937 cells and the ATRA-insensitive CD34+ KG-1a (ref. 17) cell line (Supplementary Fig. 1). Although 2 days of treatment with ATRA plus 2d or TCP had little effect on apoptosis in either of the cell lines tested (Supplementary Fig. 2a), after 4 days with the ATRA plus 2d or TCP combinations, we observed early or late apoptosis in 55% of TEX cells, with only a minor increase in apoptosis in p53-null18 HL-60 cells (Supplementary Fig. 2b). These findings together with the gene expression pathway analysis (Supplementary Table 1) are consistent with the onset of post- differentiation cell death5,6, facilitated by the presence of p53 in TEX cells19. Treatment with ATRA and LSD1i led to a marked increase in respiratory burst activity in HL-60 cells (Fig. 1b) and induced the nuclear lobulation that is associated with neutrophilic differentiation in both HL-60 and TEX cells (Fig. 1c).
Figure 1.
LSD1 inhibitors potentiate ATRA-induced differentiation of AML cells. (a) The expression of the myeloid differentiation marker CD11b by fluorescence activated cell sorting (FACS) in HL-60 (left) and TEX (right) cells using phycoerythrin-conjugated human CD11b-specific mouse monoclonal antibody. Results from the FACS analysis performed after the induction of cell differentiation using combinations of ATRA and 2d or TCP compared with untreated control cells. Data were analyzed by one-way analysis of variance (ANOVA) (P < 0.0001) followed by Tukey’s post-hoc test (ATRA compared to ATRA plus TCP, P < 0.0001). Values represent the means and error bars represent the s.d. of three independent experiments. (b) Functional change in myeloid differentiation in HL-60 cells as measured by superoxide anion production performed after treatment with the indicated drug combinations. Data were analyzed by one-way ANOVA (P < 0.0001). (c) Cell morphology of HL-60 (top row) and TEX (bottom row) cells analyzed by May-Grünwald Giemsa staining after treatment with the indicated drug combinations. (d) FACS analysis of primary AML sample #5 (Supplementary Table 2, ICD-O M9873/3) showing CD11b+Annexin-V+ cells (left). Cell morphology of AML cells from primary AML sample #5 (ICD-O M9873/3) analyzed by May-Grünwald Giemsa staining after treatments with combinations of ATRA and 2d or TCP compared with untreated control cells (right). PE, phycoerythrin. (e) Knockdown of LSD1 synergistically enhances ATRA-induced differentiation. Results from HL-60 cells transduced with lentiviral vectors expressing shRNAs targeting LSD1 (shLSD1), LSD2 (shLSD2) and scrambled controls (shScram). Shown is the expression of CD11b, as determined by FACS analysis. Data were analyzed by one-way ANOVA (P < 0.0001) followed by Tukey’s post-hoc test. *P < 0.05, ***P < 0.001. The P values shown for Tukey’s tests represent comparisons between shLSD1#1 or shLSD2#2 compared to both scrambled sequence shRNA controls (shScram#1 and shScram#2). Values represent the means and error bars represent the s.d. of two independent experiments. See Supplementary Statistical Methods for one-way ANOVA values of F and the degrees of freedom.
Mirroring the results in the cell lines, treatment with ATRA and TCP increased the fraction of CD11b+ cells in primary AML samples by a factor of up to 11-fold (Fig. 1d and Supplementary Table 2). Treatment with ATRA plus LSD1i also induced differentiation-associated morphological changes, including the formation of cytoplasmic neutrophil granules (Fig. 1d). In agreement with previously reported findings20, treatment with ATRA alone had, in general, only a limited effect in primary AML samples, and treatment with TCP alone resulted in minimal activity in most samples. Confirming a direct role for LSD1 in myeloid differentiation, shRNA knockdown of LSD1 markedly potentiated the ability of ATRA to induce the expression of CD11b in HL-60 and TEX cells (Fig. 1e and Supplementary Figs. 3 and 4). Given that TCP also inhibits the related H3K4 demethylase LSD2 (ref. 13), which shares homology in the enzymatic domain with LSD1, we also performed knockdown of LSD2 (Fig. 1e and Supplementary Figs. 3 and 4). Although knockdown of LSD2 did increase the effect of ATRA in both HL-60 and TEX cells, it was much less effective at potentiating CD11b expression compared with shRNA knockdown of LSD1.
Given that TCP enhanced the effects of ATRA in myeloid differentiation of AML cells more effectively than 2d did, and also taking into consideration that it is already licensed for use as an antidepressant, we focused our subsequent studies on TCP. An in vitro colony formation assay showed that although treatment with either TCP or ATRA alone had a small effect in HL-60 cells, treatment with ATRA and TCP together reduced the clonogenic capacity of the HL-60 cells by 70% compared to treatment with ATRA alone (Fig. 2a). Consistent with this finding, when we treated HL-60 cells in which LSD1 was knocked down with ATRA, they also showed a reduction in clonogenic capacity of 40% compared to treatment with ATRA alone (Fig. 2a). Notably, we confirmed these in vitro data using NOD-SCID mice transplanted intrafemorally with primary AML samples (FAB M1 (ICD-O M9873/3)) treated with ATRA or ATRA plus TCP ex vivo for 16 h prior to transplantation and with the same drug regimens in vivo from day 1 after transplantation. Given that TCP showed only minimal activity as a single agent in the majority of primary AML samples tested (Supplementary Table 2), we did not include it alone in this series of experiments. Here the ATRA plus TCP combination markedly diminished leukemic engraftment in both the injected right femur as well as the non-injected bone marrow sites of the mice (Fig. 2b and Supplementary Figs. 5 and 6). A disseminated 5-week AML graft can only be generated in vivo by LIC21,22, and prior studies have shown that migration and hematopoietic engraftment of non-injected bone marrow sites requires intact stem/progenitor cell function23,24. Thus, the substantially reduced or absent migration and engraftment seen in the distal bone marrow of mice treated with ATRA plus TCP compared to that of untreated mice or mice treated with ATRA alone indicates that the combination regimen eliminated the LICs or severely impaired their function. Furthermore, consistent with the finding that ATRA in combination with TCP did not induce apoptosis in mononuclear cells from the bone marrow of normal healthy human donors in vitro (both total mononuclear cells and CD34+ progenitors; Supplementary Fig. 7), treatment with ATRA plus TCP had no toxic effects on normal cord-blood–derived hematopoietic stem or progenitor cells transplanted into NOD-SCID mice (Fig. 2b and Supplementary Figs. 6 and 8).
Figure 2.
Treatment with ATRA and TCP together diminishes the engraftment of primary AML samples. (a) Results from an in vitro colony forming unit assay performed on HL-60 cells after treatment with drug combinations of ATRA, TCP or ATRA plus TCP (top), or HL-60 cells transduced with LSD1 or control shRNA after treatment with ATRA (bottom). Data were analyzed by one-way ANOVA (P < 0.0001) followed by Tukey’s post-hoc tests (P < 0.001). See Supplementary Statistical Methods for one-way ANOVA values of F and the degrees of freedom. Values represent the means and error bars represent the s.d. of three independent experiments. (b) Levels of human CD45+ cell engraftment in NOD-SCID mice transplanted with AML (FAB M1, ICD-O M9873/3) primary AML samples (n = 5) or lineage-depleted cord blood (CB) cells (n = 5) by intrafemoral injection. The average levels of CD45+ cell engraftment in untreated mice or in mice treated with ATRA or ATRA plus TCP in the injected right femur and non-injected bones as indicated (top). Data were analyzed by robust ANOVA: injected right femur (090240, P < 0.0001; 0840, P < 0.0001; CB, P = 0.1890); non-injected bone marrow (090240, P < 0.0001; 0840, P < 0.0001; CB, P = 0.9600). The ATRA and ATRA plus TCP groups were analyzed by Winsorized t test, with the statistically significant comparisons indicated. For a description of robust the ANOVA statistical analysis performed and values of F and the degrees of freedom, see Supplementary Statistical Methods. Representative flow cytometric analysis of AML or cord blood engraftment in the injected right femur and non-injected pooled bone marrow of untreated mice, mice treated with ATRA alone or mice treated with ATRA plus TCP (bottom). Plots show live cells gated on forward- and side-scatter characteristics, and the percentage of CD45+ human cells is indicated. (c) Levels of CD45+ cell engraftment examined in NSG mice transplanted with AML (M1) primary AML samples (n = 7) and treated with ATRA and ATRA plus TCP and, additionally, TCP alone after transplantation. Data were analyzed by robust ANOVA and Winsorized t test, as above: injected right femur (090240, P < 0.0001; 0840, P < 0.0001; 0858, P = 0.0093); non-injected bone marrow (090240, P < 0.0001; 0840, P < 0.0001; 0858, P = 0.0055). (d) Leukemic engraftment the injected right femur is shown from secondary NSG mouse recipients (n = 5) transplanted with equal numbers of pooled bulk right femur and bone marrow cells from primary mice that received AML sample #0840 untreated control mice and those previously treated with drugs. Data were analyzed using one-way ANOVA: injected right femur (pooled from right femur, P = 0.0961; pooled from bone marrow, P = 0.2646).
To evaluate the ability of the ATRA plus TCP combination treatment to reduce tumor burden, we initiated this treatment 15 d after transplantation in NOD-SCID γ (NSG) mice. The results in the NSG mice mirrored those obtained in the NOD-SCID mice in which treatment was initiated on day 1, with ATRA and TCP showing some activity when used as single agents, but with ATRA plus TCP proving more effective than either alone, in particular where we obtained higher levels of engraftment from untreated mice (primary AML sample #090240) (Fig. 2c and Supplementary Fig. 9). Additionally, we performed secondary transplants with right femur and bone marrow cells from NSG mice after treatment (AML sample #0840). NSG mice transplanted with cells from mice treated with ATRA plus TCP did not engraft in untreated secondary recipients, indicating that this treatment eliminated tumorigenicity (Fig. 2d). However, cells from ATRA-treated mice also did not engraft in secondary mouse recipients (Fig. 2d). It is unclear whether this result was caused by an unexpected efficacy of ATRA toward this particular AML sample or whether higher levels of engraftment would have resulted in a more potent differential effect for treatment with ATRA plus TCP (as observed in Fig. 2b), and further studies will be required to address this issue.
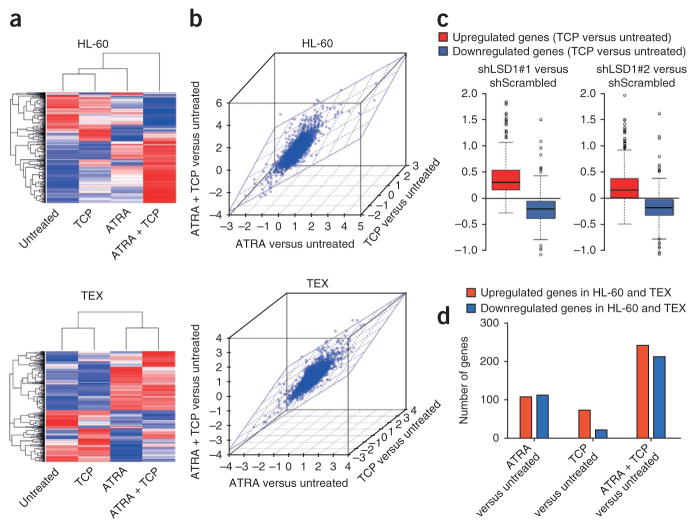
Given that LSD1 is an epigenetic modifier associated primarily with transcriptional silencing, we analyzed the effects of ATRA, TCP or both in combination on gene expression. A hierarchical clustering analysis of the 500 genes that showed the greatest differential response to drug treatments in HL-60 or TEX cells revealed that the majority of these genes were regulated concordantly by ATRA and TCP in the same direction (albeit with TCP alone exerting a lesser effect in TEX cells) and, furthermore, that this regulation was strengthened by the combination of the two drugs (Fig. 3a). Consistent with these findings, the changes in global gene expression induced by treatment with ATRA, TCP or both in combination correlated in HL-60 and TEX cells (Fig. 3b). We also compared the distribution of gene expression intensities between HL-60 cells treated with TCP and those transduced with LSD1 shRNA. This comparison revealed a remarkable association (Fig. 3c) that was consistent with biological data indicating that LSD1 knockdown and TCP treatment had broadly similar effects on ATRA-induced differentiation (Fig. 1). We also analyzed genes regulated in the same direction in both HL-60 and TEX cells and found that similar subsets of genes were altered in both cell lines after drug treatments (Fig. 3d, Supplementary Fig. 10 and Supplementary Data 1). When we further functionally analyzed these subsets, we found that they contained genes associated with the myeloid developmental program or with apoptosis (Supplementary Table 1 and Supplementary Data 2) and that the ATRA plus TCP combination treatment markedly elevated the number of upregulated genes in these pathways. Our analysis focused on upregulated genes, as LSD1 is involved in transcriptional repression; however, it is noteworthy that after treatment with ATRA plus TCP, the downregulated genes included some that are key in AML pathogenesis and treatment response, such as (in HL-60 cells) BCL11A (ref. 25), (downregulated 70%), BCL2 (ref. 26) (downregulated 80%) and MYC (ref. 27) (down-regulated 70%).
Figure 3.
Treatment with ATRA plus TCP enhances the expression of a subset of genes associated with the myeloid differentiation pathway. (a) Heatmap display of unsupervised hierarchical clustering of genes in HL-60 (top) and TEX (bottom) cells differentially expressed in response to treatment with combinations of ATRA, TCP or ATRA plus TCP. A red-blue color scale was used to reflect standardized gene expression, with red indicating higher expression and blue indicating lower expression. (b) Scatter plot analyses showing that global differential gene expression ratios derived from the comparisons of TCP to untreated, ATRA to untreated and ATRA plus TCP to untreated are highly comparable for HL-60 (top) and TEX (bottom) cells. The Spearman’s correlation coefficients were calculated as: ATRA compared to untreated and TCP compared to untreated: HL-60, r = 0.416, P < 0.001; TEX, r = 0.308, P < 0.001. ATRA compared to untreated and ATRA + TCP compared to untreated: HL-60, r = 0.502, P < 0.001; TEX, r = 0.452, P < 0.001. TCP compared to untreated and ATRA + TCP compared to untreated: HL-60, r = 0.454, P < 0.001; TEX, r = 0.420, P < 0.001. (c) Box plots of the distribution of gene expression intensities after knockdown of LSD1 in genes regulated by TCP (400 upregulated and 315 downregulated genes). (d) The bars represent the number of genes upregulated and downregulated in both HL-60 and TEX cells by ATRA, TCP and ATRA plus TCP. A cutoff value of a 1.4-fold change relative to the untreated groups was selected. The identities of these genes are listed in Supplementary Data 1.
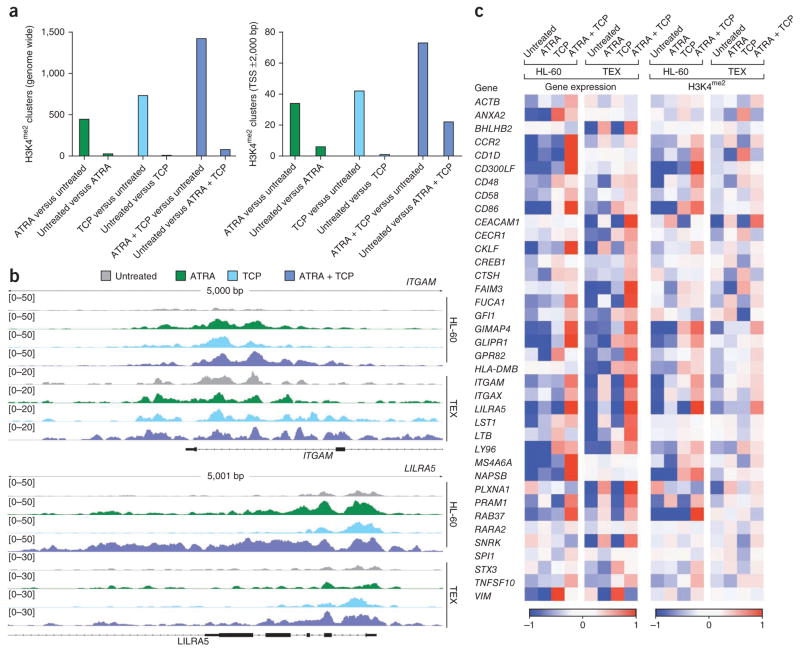
We next examined the effects of treatment with ATRA, TCP and ATRA plus TCP on H3K4me2 genome wide. Although we found no genome-wide increase in H3K4me2 in response to LSD1 inhibition, we found unique H3K4me2 clusters after the drug treatments (Fig. 4a). Although treatment with ATRA or TCP alone led to an increase in H3K4me2, the combined treatment had an even greater effect both genome wide and within the gene promoter and 5′ transcribed regions (−2,000 to +2,000 bp from transcriptional start sites). These data are consistent with the results previously obtained after deletion of Kdm1a (also known as Aof2) in mouse embryonic stem cells in which the total amounts of H3K4me2 remained unchanged but in which 4% of promoters were found to gain dimethyl H3K4 (ref. 28). We found a positive correlation between upregulation of gene expression and H3K4me2, with genes that gained clusters in promoter regions after treatments being more highly expressed in treated samples than in untreated samples (P < 0.001) and vice versa (that is, genes that did not gain H3K4me2 clusters were more highly expressed in untreated samples (P = 0.043)). Furthermore, in genes associated with the myeloid differentiation program that are upregulated by treatment with ATRA plus TCP, our chromatin immunoprecipitation sequencing (ChIP-Seq) analysis revealed a strong correlation with increased amounts of H3K4me2 at –2,000 or +2,000 bp from transcriptional start sites (Fig. 4b, c, Supplementary Table 3 and Supplementary Fig. 11).
Figure 4.
Gene-specific increases in H3K4me2 induced by treatment with ATRA plus TCP correlate with the upregulation of myeloid-differentiation–associated gene expression. (a) Comparison of unique peaks and clusters genome wide (left) and -2,000 or +2,000 bp from transcriptional start sites (TSS, right) that are present in samples treated with an indicated drug but not the untreated controls (for example, ATRA compared to untreated) or in the untreated control but not the treatment sample (for example, untreated compared to ATRA). (b) Chip-Seq profiles of H3K4me2 modifications associated with ATRA, TCP or ATRA plus TCP treatment for the ITGAM (CD11b, top) and LILRA5 (CD85f, bottom) genes in HL-60 and TEX cells. (c) The heatmap shown represents the standardized expression (left) and standardized H3K4me2 (right) of the respective genes between untreated and samples treated with ATRA, TCP or ATRA plus TCP.
Drugs targeting aberrant but reversible epigenetic modifications (so called ‘epi drugs’) have therapeutic potential, and epigenetic enzymes such as DNA methyltransferases, histone deacetylases or histone methyltransferases or demethylases are bona fide targets for anti-AML drug development4,29. Recently, several groups have developed new small-molecule LSD1is based on monoamine oxidase inhibitors (of which TCP is an example), biguanide polyamine analogs or oligoamine analogs30–34, and these efforts may yield results that lead to future treatments with greater specificity and potency than TCP. At present, however, TCP has a major advantage over other LSD1i in development in that it is well tolerated by patients and has been used as an antidepressant and anxiolytic agent since 1960 (sold under the brand names Parnate and Jatrosom)35. Furthermore, the TCP concentration used in this study is representative of the range of peak plasma concentrations reported in patients treated with this drug35. In summary, these data show the existence of therapeutically relevant crosstalk between the ATRA-induced differentiation pathway and histone H3K4 methylation and that targeting LSD1 in combination with ATRA may be a promising treatment for AML.
ONLINE METHODS
Cell lines, primary samples and cell culture
We obtained HL-60 cells from the German Collection of Microorganisms and Cell Cultures (DSMZ) and maintained in them RPMI 1640 supplemented with 10% FBS (Sigma), 100 μml−1 penicillin and 100 μg ml−1 streptomycin. We maintained TEX cells in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 15% FBS, 20 ng ml−1 stem cell factor (SCF; PeproTech) and 2 ng ml−1 interleukin-3 (IL-3) (PeproTech). We grew normal bone marrow mononuclear cells (AllCells, ABM011F) in IMDM supplemented with 20 ng ml−1 SCF, 10 ng ml−1 each of IL-3, IL-6 and fms-related tyrosine kinase 3 (FLT3) and 10% FBS. Primary AML samples collected in Germany were provided by the University Hospital of Münster and were obtained at the time of diagnosis. Informed consent was obtained from all patients in accordance with the Declaration of Helsinki, and the study was approved by the Ethics committee of the University of Münster. In Canada, peripheral blood cells from patients diagnosed with AML, and umbilical cord blood cells were collected after obtaining informed consent according to procedures approved by the Research Ethics Boards of the University Health Network and Trillium Health Centre. Low-density mononuclear cells were isolated from AML samples and viably frozen. We obtained lineage-depleted cord blood cells by negative selection using StemSep columns according to the manufacturer’s protocol (STEMCELL Technologies, Canada) and frozen viably.
Analysis of myeloid differentiation
We treated cells with ATRA (Sigma), and the LSD1 inhibitors 2d13 or TCP (Sigma) for 4 d before analysis. We performed FACS analysis of CD11b expression on 5 × 105 cells using a phycoerythrin-conjugated human CD11b-specific mouse monoclonal antibody at a 1:5 dilution (BD Pharmingen, #555388) on a BD LSRII FACS machine (Becton Dickinson) with CellQuest software. We performed FACS analysis on live cells, which were sorted using a LIVE/DEAD violet fluorescent stain (Invitrogen). We analyzed the level of superoxide anion in HL-60 cells after differentiation using the Lumimax Superoxide Anion Detection Kit (Stratagene) following the manufacturer’s instructions. Briefly, we treated 5 × 105 cells with the above described ATRA plus LSD1i combinations for 4 d. We then resuspended the cells in 200 μl reagent mix, to which we added phorbol-12-myristate-13-acetate (PMA) to a final concentration of 200 ng ml−1. We read luminescence on a Mithras LB940 multimode plate reader (Berthold Technologies). For the morphological analysis of myeloid cell differentiation, we prepared cytospins by centrifugation in 150 μl PBS at a speed of 300 rpm for 5 min using Superfrost Plus (VWR) positively charged glass slides. We stained cytospun slides at room temperature with May-Grünwald-Giemsa (Sigma) and examined cellular morphology using an Axiokop 2 plus microscope with an AxioCam MRc camera (Carl Zeiss).
Analysis of apoptosis
We treated cells with the ATRA plus LSD1i combinations as described above for 2 or 4 d before analysis. We performed FACS analyses on 5 × 105 cells using the Annexin-V FITC apoptosis detection kit II (BD Pharmingen) according to the manufacturer’s instructions, and we used propidium Iodide (Invitrogen) at 1 μg ml−1. We carried out the FACS analysis as described above. We isolated CD34+ normal mononuclear cells by FACS using FITC-conjugated human CD34-specific mouse monoclonal antibody at a 1:5 dilution (BD Pharmingen, #555821).
In vivo treatment of AML in NOD-SCID and NSG mice
We cultured thawed primary AML and cord blood cells in X-VIVO 10 medium (Lonza) supplemented with 15% BIT (STEMCELL Technologies) and a cytokine cocktail containing SCF (100 ng ml−1), Flt3 ligand (Flt3L) (100 ng ml−1), IL-6 (20 ng ml−1) and thrombopoietin (TPO) (50 ng ml−1). We cultured the cells in vitro for 16 h before transplantation into mice, which were either untreated or were treated with ATRA alone (1 μM) or with ATRA (1 μM) plus TCP (10 μM).
NOD-SCID and NSG mice were bred and housed at the University Health Network, Princess Margaret Hospital. All animal experimental protocols were approved by the institutional Animal Care Committee of the University Health Network, Princess Margaret Hospital. We performed the mouse repopulation assay as previously described36. Briefly, we sublethally irradiated 10-week-old female NOD-SCID mice with 225 cGy from a 137 Cs γ-irradiator and then injected them intraperitoneally with 200 μg purified CD122-specific antibody 24 h before intrafemoral transplantation of AML or cord blood cells. All mice in the same experiment received equal numbers of cells (3–4 × 106 AML cells or 5.3 × 104 cord blood cells per mouse). Immediately after cell transplantation, mice injected with cells treated with ATRA or with ATRA plus TCP received a 21-d– release 10-mg ATRA pellet (Innovative Research of America) implanted subcutaneously at the lateral side of the neck, whereas mice injected with untreated cells received a placebo pellet (five mice per treatment arm). Starting the next day, we injected mice intraperitoneally with PBS (for the cells treated with ATRA alone or the untreated cells) or TCP (10 mg kg−1, for the cells treated with ATRA plus TCP) daily for 21 consecutive days. To evaluate the ability of the ATRA plus TCP combination treatment to reduce tumor burden, we injected AML cells into irradiated NSG mice (seven mice per treatment arm), and we initiated treatment as described above 15 d after transplantation. We killed the mice 5 weeks after cell transplantation and human engraftment in the injected right femur and evaluated the non-injected bones for human CD45+ chimerism by flow cytometry.
Secondary transplants of AML-engrafted mice
We treated human AML-engrafted female NSG mice (five mice per treatment arm) with ATRA, TCP or ATRA plus TCP as described above, with treatment starting 15 d after cell transplantation. We transplanted a total of 1.5 × 106 pooled right femur or bone marrow cells harvested from primary untreated control or drug-treated mice into sublethally irradiated female NSG adult mice. We assessed AML engraftment of secondary recipients in the right femur 9 weeks later, as described above.
Supplementary Material
Acknowledgments
T.S., L.H., K.P. and A.Z. were supported by a Specialist Programme Grant from Leukaemia and Lymphoma Research. T.S., L.H., K.P. and A.Z. were also supported in part by the Samuel Waxman Cancer Research Foundation. S.G. and C.M.-T. were supported by the Interdisziplinäres Zentrum für Klinische Forschung (IZKF) and the Deutsche Forschungsgemeinschaft (1328/6-1, 8-1 and 9-1). W.C.C., L.J., J.C.Y.W. and J.E.D. were supported by the Cancer Stem Cell Consortium with funding from the Government of Canada through Genome Canada and the Ontario Genomics Institute (OGI-047) and through the Canadian Institute of Health Research (CSC-105367), as well as with funding from Canadian Institutes for Health Research (CIHR), the Canadian Cancer Society Research Institute, the Terry Fox Foundation; Genome Canada through the Ontario Genomics Institute; Ontario Institute for Cancer Research with funds from the province of Ontario; and a Canada Research Chair. W.C.C., L.J., J.C.Y.W. and J.E.D. were also funded in part by the Ontario Ministry of Health and Long Term Care (OMOHLTC). The views expressed here do not necessarily reflect those of the OMOHLTC. R.A.C. Jr. was supported by a National Cancer Institute Grant (CA51085). The authors would like to thank D. Leongamornlert, L. Jasnos and S. Bashir for valuable discussions on the interpretation of the ChIP-Seq data and the statistical analyses. The authors would also like to thank M. Greaves for support and critical reading of the manuscript.
Footnotes
Additional methods. Detailed methodology is described in the Supplementary Methods.
Accession codes. Raw and normalized expression data are deposited in the Gene Expression Omnibus (GEO) with the accession number GSE34672. ChIP-Seq data are deposited in the GEO with the accession number GSE34726. Data are also deposited in the Leukemia Gene Atlas (http://www.leukemia-gene-atlas.org/).
Note: Supplementary information is available on the Nature Medicine website.
AUTHOR CONTRIBUTIONS
T.S. performed cell drug treatments, ChIP-Seq, quantitative PCR, RNAi and western blotting; L.H. performed cell drug treatments, FACS, the superoxide assay, immunohistochemical staining and light microscopy; S.G. and C.M.-T. performed ChIP-Seq and primary AML sample drug treatments; W.C.C., L.J., A.C.P., J.C.Y.W. and J.E.D. performed in vivo treatments of AML in NOD-SCID and NSG mice; K.H., H.-U.K. and M.D. performed bioinformatic analyses of expression microarray and ChIP-Seq data; A.B., K.M. and M.D.M. isolated primary AML samples; P.W., L.M. and R.A.C. Jr. collaborated on the 2d LSD1 inhibitor; T.S., K.P. and A.Z. designed the study and analyzed the data; K.P. and A.Z. wrote the paper. All authors discussed the results and commented on the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturemedicine/.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Pollyea DA, Kohrt HE, Medeiros BC. Acute myeloid leukaemia in the elderly: a review. Br J Haematol. 2011;152:524–542. doi: 10.1111/j.1365-2141.2010.08470.x. [DOI] [PubMed] [Google Scholar]
- 2.Krug U, et al. Complete remission and early death after intensive chemotherapy in patients aged 60 years or older with acute myeloid leukaemia: a web-based application for prediction of outcomes. Lancet. 2010;376:2000–2008. doi: 10.1016/S0140-6736(10)62105-8. [DOI] [PubMed] [Google Scholar]
- 3.Altucci L, Gronemeyer H. The promise of retinoids to fight against cancer. Nat Rev Cancer. 2001;1:181–193. doi: 10.1038/35106036. [DOI] [PubMed] [Google Scholar]
- 4.Petrie K, Zelent A, Waxman S. Differentiation therapy of acute myeloid leukemia: past, present and future. Curr Opin Hematol. 2009;16:84–91. doi: 10.1097/MOH.0b013e3283257aee. [DOI] [PubMed] [Google Scholar]
- 5.Altucci L, et al. Retinoic acid–induced apoptosis in leukemia cells is mediated by paracrine action of tumor-selective death ligand TRAIL. Nat Med. 2001;7:680–686. doi: 10.1038/89050. [DOI] [PubMed] [Google Scholar]
- 6.Glasow A, Prodromou N, Xu K, von Lindern M, Zelent A. Retinoids and myelomonocytic growth factors cooperatively activate RARA and induce human myeloid leukemia cell differentiation via MAP kinase pathways. Blood. 2005;105:341–349. doi: 10.1182/blood-2004-03-1074. [DOI] [PubMed] [Google Scholar]
- 7.Glasow A, et al. DNA methylation-independent loss of RARA gene expression in acute myeloid leukemia. Blood. 2008;111:2374–2377. doi: 10.1182/blood-2007-05-088344. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Berglund L, et al. A genecentric Human Protein Atlas for expression profiles based on antibodies. Mol Cell Proteomics. 2008;7:2019–2027. doi: 10.1074/mcp.R800013-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Lim S, et al. Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis. 2010;31:512–520. doi: 10.1093/carcin/bgp324. [DOI] [PubMed] [Google Scholar]
- 12.Hayami S, et al. Overexpression of LSD1 contributes to human carcinogenesis through chromatin regulation in various cancers. Int J Cancer. 2011;128:574–586. doi: 10.1002/ijc.25349. [DOI] [PubMed] [Google Scholar]
- 13.Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, et al. Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci USA. 2007;104:8023–8028. doi: 10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalton WT, Jr, et al. HL-60 cell line was derived from a patient with FAB-M2 and not FAB-M3. Blood. 1988;71:242–247. [PubMed] [Google Scholar]
- 16.Warner JK, et al. Direct evidence for cooperating genetic events in the leukemic transformation of normal human hematopoietic cells. Leukemia. 2005;19:1794–1805. doi: 10.1038/sj.leu.2403917. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, Zheng X, Behm FG, Ratnam M. Differentiation-independent retinoid induction of folate receptor type β, a potential tumor target in myeloid leukemia. Blood. 2000;96:3529–3536. [PubMed] [Google Scholar]
- 18.Wolf D, Rotter V. Major deletions in the gene encoding the p53 tumor antigen cause lack of p53 expression in HL-60 cells. Proc Natl Acad Sci USA. 1985;82:790–794. doi: 10.1073/pnas.82.3.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDermott SP, et al. A small molecule screening strategy with validation on human leukemia stem cells uncovers the therapeutic efficacy of kinetin riboside. Blood. 2012;119:1200–1207. doi: 10.1182/blood-2011-01-330019. [DOI] [PubMed] [Google Scholar]
- 20.Howell AL, Stukel TA, Bloomfield CD, Davey FR, Ball ED. Induction of differentiation in blast cells and leukemia colony-forming cells from patients with acute myeloid leukemia. Blood. 1990;75:721–729. [PubMed] [Google Scholar]
- 21.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 22.Lapidot T, et al. Cytokine stimulation of multilineage hematopoiesis from immature human cells engrafted in SCID mice. Science. 1992;255:1137–1141. doi: 10.1126/science.1372131. [DOI] [PubMed] [Google Scholar]
- 23.Doulatov S, et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 24.McKenzie JL, Gan OI, Doedens M, Wang JC, Dick JE. Individual stem cells with highly variable proliferation and self-renewal properties comprise the human hematopoietic stem cell compartment. Nat Immunol. 2006;7:1225–1233. doi: 10.1038/ni1393. [DOI] [PubMed] [Google Scholar]
- 25.Yin B, et al. A retroviral mutagenesis screen reveals strong cooperation between Bcl11a overexpression and loss of the Nf1 tumor suppressor gene. Blood. 2009;113:1075–1085. doi: 10.1182/blood-2008-03-144436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milojkovic D, et al. Antiapoptotic microenvironment of acute myeloid leukemia. J Immunol. 2004;173:6745–6752. doi: 10.4049/jimmunol.173.11.6745. [DOI] [PubMed] [Google Scholar]
- 27.Zuber J, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–129. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 29.Florean C, Schnekenburger M, Grandjenette C, Dicato M, Diederich M. Epigenomics of leukemia: from mechanisms to therapeutic applications. Epigenomics. 2011;3:581–609. doi: 10.2217/epi.11.73. [DOI] [PubMed] [Google Scholar]
- 30.Binda C, et al. Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. J Am Chem Soc. 2010;132:6827–6833. doi: 10.1021/ja101557k. [DOI] [PubMed] [Google Scholar]
- 31.Ueda R, et al. Identification of cell-active lysine specific demethylase 1-selective inhibitors. J Am Chem Soc. 2009;131:17536–17537. doi: 10.1021/ja907055q. [DOI] [PubMed] [Google Scholar]
- 32.Culhane JC, Wang D, Yen PM, Cole PA. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors. J Am Chem Soc. 2010;132:3164–3176. doi: 10.1021/ja909996p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, et al. Novel oligoamine analogues inhibit lysine-specific demethylase 1 and induce reexpression of epigenetically silenced genes. Clin Cancer Res. 2009;15:7217–7228. doi: 10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma SK, et al. (Bis)urea and (bis)thiourea inhibitors of lysine-specific demethylase 1 as epigenetic modulators. J Med Chem. 2010;53:5197–5212. doi: 10.1021/jm100217a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whyte IM. Monoamineoxidase inhibitors. In: Dart RC, editor. Medical Toxicology. Lippincott Williams and Wilkins; Philadelphia, Pennsylvania, USA: 2003. pp. 823–833. [Google Scholar]
- 36.McKenzie JL, Gan OI, Doedens M, Dick JE. Reversible cell surface expression of CD38 on CD34-positive human hematopoietic repopulating cells. Exp Hematol. 2007;35:1429–1436. doi: 10.1016/j.exphem.2007.05.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.