Abstract
Gut microbiota is a compilation of microorganisms dwelling in the entire mammalian gastrointestinal tract. They display a symbiotic relationship with the host contributing to its intestinal health and disease. Even a slight fluctuation in this equipoise may be deleterious to the host, leading to many pathological conditions like Clostridium difficile infection or inflammatory bowel disease (IBD). In this review, we focus on the role of microbial dysbiosis in initiation of C. difficile infection and IBD, and we also touch upon the role of specific pathogens, particularly C. difficile, as causative agents of IBD. We also discuss the molecular mechanisms activated by C. difficile that contribute to the development and exacerbation of gastrointestinal disorders.
Keywords: Clostridium difficile, inflammatory bowel disease, microbial dysbiosis
Introduction
Microbial communities colonize all surfaces of the human body, but in the gut, bacterial species are colonized in greater densities known as the microbiota or commensal microflora [Eckburg et al. 2005; Marchesi and Shanahan, 2007]. It is estimated that the adult human gut contains around 1014 bacterial cells and more than 1000 different bacterial species [Eckburg et al. 2005; Savage, 1977]. However, these proportions can vary greatly among individuals, and major shifts in the gut microbiota are based on the host organism’s age, diet, and health status [Hooper et al. 2002]. It is known that these bacterial cells in the intestinal lumen have a continuous communication with the host cells and form long-lasting, interactive associations with their host. These associations play a critical role in conservation of mucosal immune function, epithelial barrier integrity, motility, and nutrient absorption [Ley et al. 2008; Zoetendal et al. 2008; Bäckhed et al. 2005; Mazmanian et al. 2005]. Under normal conditions, commensal microbes and their hosts enjoy a symbiotic relationship. However, even a slight disturbance in normal microbiota of the gut can lead to an imbalance of host–microbe relationships. This state of condition where microbial imbalance exerts adverse effects on the host is known as dysbiosis [Hawrelak and Myers, 2004]. It is known that intestinal microenvironment as a unit provides an important protective, mucosal defense mechanism, but there are ample lines of evidence stating that change in the composition of the commensal microbiota alters the intestinal microenvironment making this niche vulnerable to pathogenic insult [Manichanh et al. 2006; Darfeuille-Michaud et al. 2004; Swidsinski et al. 2002]. Dysbiosis may be detrimental to the host, leading to inflammation and mucosal tissue damage that predisposes them to pathological conditions like Clostridium difficile infection or inflammatory bowel disease (IBD) [Lepage et al. 2008; Tamboli et al. 2004; McFarland, 1998; Jacobs, 1994].
C. difficile is an infectious Gram-positive spore-forming bacillus microorganism of the gastrointestinal tract, and its toxin expression causes gastrointestinal illness with a wide spectrum of severity, ranging from mild diarrhea to pseudomembranous colitis, toxic megacolon, sepsis-like picture and death [Dobson et al. 2003; Mylonakis et al. 2001]. C. difficile is considered a member of the normal gut microflora, however its growth is suppressed by the more dominant anaerobes. Thus, the rate of colonization in human gut for C. difficile is different for different age groups – it is highest in early infancy and decreases with age [Rolfe et al. 1981; Taylor et al. 1981; Testore et al. 1986; Tullus et al. 1989].
Host susceptibility to C. difficile infection and recurrences result partly from inability of the intestinal microbiota to resist C. difficile colonization. Colonization of gut cells by C. difficile is a critical step in their pathogenic process which depends on C. difficile colonization factors, and on the microbiota colonization resistance (barrier effect) [Pechine et al. 2007]. Loss of the commensal microbiota barrier effect and the release of niches previously unavailable following, for example, antimicrobial therapies allow C. difficile to colonize the intestine [Wilson, 1993; Chang et al. 2008; De La Cochetière et al. 2008]. Direct interaction of C. difficile with the intestinal epithelial cells begins a cascade of inflammatory processes that contribute to intestinal diseases such as diarrhea and pseudomembranous colitis.
Thus, on one hand, the composition of the intestinal microbiota could play an important role as a predisposing factor in the onset of the disease, and on the other hand, under specific conditions which alter the intestinal microbiota composition and disrupt barrier effects, allow C. difficile to multiply and colonize the gut.
It is marked from various studies that the microbiota plays an essential role in the pathogenesis of IBD. Moreover, accumulating evidence suggests that composition and function of gut microbiota are abnormal in patients with IBD [Frank et al. 2007]. Crohn’s disease (CD) and ulcerative colitis (UC) are the two major forms of IBD that closely mimic intestinal infections. They occur in areas with the highest luminal bacterial concentrations and many microbial pathogens have been suggested as causes [Lidar et al. 2009; García Rodríguez et al. 2006; Farrell and La Mont, 2002]. Despite the fact that there has been an increasing incidence of C. difficile infection in people with IBD during the past decade, it is still debated whether C. difficile plays a role in the initial onset of IBD [Bossuyt et al. 2009; Ananthakrishnan et al. 2008; Issa et al. 2007; Rodemann et al. 2007].
It is known that C. difficile plays an important role in initiation and perpetuation of intestinal inflammation. However, there are certain questions to be answered: Are gastrointestinal disorders such as IBD associated with C. difficile? What is their reservoir? Are these bacteria sufficient to drive IBD pathogenesis? This article is an overview of the scientific findings about the causal relationship between intestinal microbiota dysbiosis and gastrointestinal diseases, including C. difficile infection and IBD. It also focuses on the role of C. difficile in initiation of the proinflammatory response in human gut. This event is thought to be an early step in the development of mucosal inflammatory responses which characterizes gastrointestinal diseases, such as IBD. Finally we describe the possible relationship between C. difficile and IBD.
Importance of the human intestinal microbiota
The healthy gut microbiota is involved in a dynamic interaction with the host and promotes its health and wellbeing. The combined consortium of the human body and its gut microbiota is known as the ‘superorganism’, which is embraced with human and bacterial genes [Ley et al. 2006]. The totality of microbes and their genetic elements together are signified as the ‘microbiome’. It has been estimated that the microbiome has 100 times more genes than that of human genes present in our body. The microflora of the intestinal microenvironment as an entity provides important protective, immune regulatory and metabolic functions. Intestinal microflora serve a central line of resistance to colonization by exogenous microbes, and thus play a vital role in constraining potential incursion by succeeding pathogens. This defensive mechanism against pathogenic bacteria is known as the barrier effect or colonization resistance. Accruing evidence reveals that the gut microbiota plays a significant role in immune function and energy metabolism of the host. The immune regulatory function consists of priming the mucosal immune system and maintenance of intestinal epithelial homeostasis [Cash et al. 2006; Bry et al. 1996]. The intestinal microflora also makes an important metabolic contribution through breaking down the complex indigestible dietary carbohydrates and proteins, with subsequent generation of fermentation end products like short-chain fatty acids. They also play a major role in production of vitamins, ion absorption and conversion of dietary polyphenolic compounds into their active form [Falony et al. 2006].
Composition of the human gut microbiota
The intestine is an open ecological system that is colonized immediately after birth by a microbial population. It is estimated that the human microbiota contain 1014 bacterial cells. The composition of the intestinal flora changes with age and the gradual changes are observed in early childhood, with a general reduction in the number of aerobes and facultative anaerobes and an increase in the obligate anaerobic species [Hopkins et al. 2005]. A study by Palmer and colleagues showed that, between 1 and 2 years of age, the human gut microbiota starts to resemble that of an adult [Palmer et al. 2007]. In contrast, work by Agans and colleagues showed that the intestinal microbiota of adolescents varied significantly from that of adults [Agans et al. 2011]. However, studies using nucleic-acid-based approaches (targeting 16S rRNA) have shown that normal intestinal flora is conquered by several major bacterial divisions, that is, Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria, Verrucomicrobia, Cyanobacteria and Spirochaeates [Rajilić-Stojanović et al. 2007; Eckburg et al. 2005; Wang et al. 2005]. The same studies described the vast diversity of bacterial species and identified the dominant bacterial groups as Bacteroidetes and Firmicutes. Notably, proportions and compositions of Bacteroidetes were consistently stable within individuals whereas the Firmicutes showed large variation. The gut microbial species composition varies greatly among individuals. Each individual represents a unique collection of bacterial species, which is highly stable over time [Zoetendal et al. 1998]. Moreover, it should be noted that genetic factors play an important role in human gut microbiota development, although the environment also drives species acquisition.
It is known that the adult human intestinal microbiota is dominated by the phylum Firmicutes. Clostridium is a genus of Gram-positive bacteria, belonging to the family of Firmicutes. Clostridium is well known as a gut colonizer, which is found significantly in infants and adults [Eckburg et al. 2005; Wang et al. 2005; Tullus et al. 1989; Bolton et al. 1984].
Clostridium is genetically a very heterogeneous group, and is the largest among the anaerobic spore-forming genera. Currently it has more than 120 species which are divided into 19 clusters [Brüggemann and Gottschalk, 2008]. It includes both beneficial species which are involved in metabolic pathways and pathogenic bacteria, responsible for a wide spectrum of diseases. For instance, cluster XIVa (C. coccoides group) dominates the colon of adults, and Roseburia and Eubacterium rectale-C. coccoides group are involved in the production of short-chain fatty acids like butyrate used as fuel for the host’s colonocytes. In addition, butyrate has anti-inflammatory effects that result from inhibition of transcription factor nuclear factor κB (NFκB), leading to a decreased secretion of proinflammatory cytokines [Pryde et al. 2002; Segain et al. 2000]. Cluster IV (C. leptum group) is the second group in the adult colon, and Faecalibacterium prausnitzii is an important member of this cluster [Eckburg et al. 2005; Hold et al. 2002]. Cluster I is the largest of the clostridial groups. Species belonging to this group include pathogens such as C. tetani and C. botulinum, opportunistic pathogens such as C. perfringens, and more harmless members, such as C. butyricum [Collins et al. 1994]. C. perfringens is common in the normal intestinal microbiota and is the most frequently isolated clostridia from clinical specimens.
C. difficile is one of the pathogens belonging to cluster XI. The rate of colonization for C. difficile is different in different age groups; it is highest in infants and decreases with age [Rolfe et al. 1981; Tullus et al. 1989]. During infancy, asymptomatic carriage of C. difficile in the gastrointestinal tract is very common. Many infants are colonized by C. difficile strains during the first 2 years of life but this colonization is rarely associated with C. difficile infection [Collignon et al. 1993], even though high levels of toxins A and B might be present [Tullus et al. 1989; Bolton et al. 1984]. According to previous studies, the prevalence rate of C. difficile in the gut microbiota of healthy human adults was estimated to be 0–17% [Nakamura et al. 1981; Kobayashi et al. 1983; George, 1986]. However, Iizuka and colleagues suggested that toxigenic C. difficile might be present in the gut microbiota of healthy human adults more frequently (53.3%) than previously assumed [Iizuka et al. 2004].
C. difficile does not cause any significant disease when it is present in small numbers. However, disturbance of the normal intestinal flora (dysbiosis) by several potential causative factors may result in unlimited expansion of C. difficile in the microbiota, leading to inflammation and damage of the gut mucosa [Wilson, 1993].
Summarizing, the human gastrointestinal tract is an ecosystem rich in microbial species where the Clostridium genus plays an important role.
Dysbiosis
The gastrointestinal tract is known to be a complex and finely balanced ecosystem. It is one of the largest interfaces between the outside world and the human internal environment. The gut microbiota is highly vulnerable to changes in the gut microenvironment. Under normal conditions, commensal microbes and their hosts enjoy a symbiotic realationship. However, any imbalance in this equilibrium via qualitative and quantitative changes in the intestinal flora itself, changes in their metabolic activities, and changes in their local distribution leads to a condition where microbial imbalance exerts adverse effects on the host known as dysbiosis [Hawrelak and Myers, 2004]. Dysbiosis might have serious health consequences, leading to many chronic and degenerative diseases [Hawrelak and Myers, 2004; Holzapfel et al. 1998]. Many factors like antibiotics, psychological stress, physical stress, modern diet and hygiene can harm the microbial stability, and thus, contribute to intestinal dysbiosis [Hawrelak and Myers, 2004; Bernstein and Shanahan, 2008]. Dysregulation of adaptive and innate immunity and genetic factors might also play a role in contributing to intestinal dysbiosis. Compelling evidence shows a pivotal role of intestinal microbiota dysbiosis in the development of gastrointestinal diseases, such as C. difficile infection and IBD, small intestinal bacterial overgrowth, functional gastrointestinal disorders including irritable bowel syndrome, and colorectal cancer [Lepage et al. 2008; Tamboli et al. 2004; McFarland, 1998; Jacobs, 1994].
Thus, ample evidence in the literature confirmed that dysbiosis is an important clinical entity, and knowledge about the factors which play a causative role in this condition is important.
Dysbiosis and expansion of Clostridium difficile
Composition of the individual’s flora can fluctuate under some circumstances, for example, acute diarrhea, antibiotic treatment, or to lesser extent, dietary intrusions. Any of these factors can influence the composition of the commensal microbiota, which can alter the intestinal environment making the niche susceptible to pathogenic agents. The use of antibiotics is most common and important cause for major change in normal gut microbiota. These antimicrobial agents potentially influence the microbiota by shifting the relative proportion of community members, allowing them the opportunity to establish. Antibiotic therapies eliminate members of the community by destroying them directly or indirectly by breaking necessary mutualistic interactions [Jernberg et al. 2007; Sullivan et al. 2001]. Thus, the loss of microbial equilibrium creates an intestinal environment susceptible to pathogenic agents like C. difficile and subsequent C. difficile-associated disease [Kelly and LaMont, 1998].
Barc and colleagues showed that amoxicillin clavulanic acid treatment did not enumeratively modify the total microbiota; however, the Bacteroidetes and the Enterobacteriaceae groups increased and simultaneously the C. coccoides-Eubacterium rectale group decreased dramatically under the treatment [Barc et al. 2004]. This disequilibrium induced by antibiotics may be responsible for diarrhea and may also facilitate intestinal colonization by C. difficile. But these finding are contrary to the work done by Manges and colleagues, who illustrated that the low levels of Bacteroidetes is significantly associated with development of C. difficile-associated disease (CDAD) [Manges et al. 2010]. In addition, Tvade and Rask-Madsen found that absence of Bacterioides may result in chronic relapsing C. difficile diarrhea [Tvade and Rask-Madsen, 1989]. A study by Rousseau and colleagues showed that in infants a low Firmicutes and Bacteroides ratio and an increase in facultative anaerobes might facilitate colonization by C. difficile without any antibiotic treatment [Rousseau et al. 2011]. Presence of a Firmicutes species such as Ruminococcus gnavus and C. nexile in significant quantities is also associated with C. difficile colonization of the epithelial cells. These two species have been shown to produce a trypsin-dependent antimicrobial substance against C. perfringens but with less activity against C. difficile [Marcille et al. 2002].
Based on epidemiological evidence, the other factors responsible for development of C. difficile pathogenesis are notably H2-receptor antagonists and proton-pump inhibitors (PPIs). PPIs and H2-receptor antagonists are associated with suppression of gastric acid which increases intragastric bacterial counts and small bowel colonization [Dial et al. 2005, 2006; Aldeyab et al. 2009]. These effects may occur by antimicrobial-independent or antimicrobial-dependent mechanisms. PPIs enhance the survival of C. difficile through the stomach [Thomson et al. 2010]. However, a study done by Nerandzic and colleagues presented conflicting evidence about PPIs, stating that their effects in the stomach do not contribute to the pathogenesis of C. difficile infection [Nerandzic et al. 2009].
Another common but less recognized etiological factor which leads to expansion/colonization of the human gut by C. difficile is the prolonged use of elemental diets in critically ill patients with an impaired upper gastrointestinal tract [O’Keefe, 2010; Iizuka et al. 2004]. Experimental studies showed that such diets are absorbed within the small intestine and therefore deprive the colonic microbiota of their source of nutrition, namely dietary fiber, fructose oligosaccharides, and resistant starch [Iizuka et al. 2004]. The absence of fiber and resistant starches not only disturbs microbiotal balance but also deprives the colonic epithelium of its chief energy sources [O’Keefe, 2008]. As a result colonic fermentation is suppressed which leads to a decline in beneficial bacteria, creating a niche for C. difficile expansion/colonization. Once the C. Difficile colonizes the host it results in mucosal inflammation induced by the cytotoxic factors, disrupting the epithelial barrier [Starr, 2005; Pothoulakis and LaMont, 2001]. Toxins A and B enter the colonic cells and kill the cells by multiple mechanisms [O’Keefe, 2010].
Thus, these findings indicate that the intestinal dysbiosis should be considered as a mechanism leading to C. difficile expansion and subsequent infection.
Disbiosis in inflammatory bowel disease
Intestinal balance is sustained by a constant crosstalk between the intestinal microbiota and the host. The coevolution of these two partners leads to an establishment of a mutual beneficial relationship. However, an alteration in the bowel flora composition and its activities leads to many chronic and degenerative diseases. Major modifications of the intestinal microbiota were observed in patients with severe gastrointestinal problems. Despite the fact that the dysbiosis of the gut microbiota is a common feature in patients with IBD, in most cases, it cannot be determined whether these changes are casual or merely consequences of the activated immune and inflammatory response [Peterson et al. 2008]. However, dysbiosis in IBD is considered a symptom when there is an overall decrease in biodiversity, especially a decrease in Firmicutes, in particular clostridia groups IV and XIVa and Bacteroides, with an increase in the number of Enterobacteriaceae and the presence of unusual bacteria [Frank et al. 2007; Manichanh et al. 2006; Swidsinski et al. 2002]. Some characteristics of dysbiosis are not only specific for IBD type (CD and UC) but also for other gastrointestinal inflammatory conditions such as acute self-limited colitis [Sokol et al. 2009]. Any changes associated with the composition of gut microbiota leads to colonization of intestinal epithelial cells by endogenous or exogenous pathogens. The fundamental question of whether specific microorganisms could initiate the onset of IBD was studied by Schumacher and colleagues [Schumacher et al. 1993]. As a result of the microbiological evaluation, 13 out of 61 patients with IBD showed microbial agents, specifically, Yersinia enterocolitica, Salmonella typhi, Campylobacter jejuni, Aeromonas hydrophila, Listeria monocytogenes, Escherichia coli, Myco-bacterium avium paratuberculosis, Chlamydia and C. difficile were suggested as etiologic agents responsible for IBD [Lidar et al. 2009; Darfeuille-Michaud et al. 2004; Schumacher et al. 1993; Burnham et al. 1978]. However, there is no conclusive evidence that a specific pathogen is responsible for onset or relapses of IBD [Frank et al. 2007].
In summary, it is more likely that microbial dysbiosis and lack of beneficial bacteria may lead to the inflammatory response characteristic of IBD.
Evidence for role of Clostridium difficile in inflammatory bowel disease
It is stated in the literature that C. difficile infection should be included in the diagnosis of patients with IBD symptoms as it is well known to induce or mimic a flare of IBD [Issa et al. 2007; Rodemann et al. 2007]. The similarity in symptoms (diarrhea, abdominal pain, low-grade fever) between C. difficile infection and IBD makes it difficult to distinguish between them. However, the past decade has seen an alarming increase in the burden of disease associated with C. difficile and its role in IBD [Musa et al. 2010; Ananthakrishnan et al. 2008; Jarvis et al. 2009; Issa et al. 2007; Rodemann et al. 2007; Kuijper et al. 2006; Pepin et al. 2004]. The association between IBD and C. difficile may be due to a variety of factors, including drugs used in medical treatment, which might alter the intestinal flora and promote colonization, altered immune status possibly related to therapeutic agents, nutritional status and frequent hospitalizations [Freeman, 2008; McFarland et al. 2007].
The role of C. difficile infection in IBD has been noted; however, the fundamental question of whether C. difficile infection is a cause of IBD or a consequence of the inflammatory state of the intestinal environment still needs to be answered. C. difficile may cause an infectious colitis superimposed on IBD, or in some patients, may precipitate an IBD flare leading to two separate but simultaneous inflammatory processes. The other possibility is that C. difficile may be just a colonizer and that IBD flare probably occurs independently. Given the uncertainty in the pathogenesis of IBD and its multifactorial character [Danese and Fiocchi, 2006] we can conclude that disturbance of the intestinal flora by a broad spectrum of causative factors gives C. difficile the chance to expand and produce toxins that cause inflammation and damage to gut epithelial cells. The adhesion of bacteria and the increased permeability of intestinal mucosa in gastroenteritis are held to be important in the pathogenesis of IBD. In this context, C. difficile is known to have a vital role in the etiology of IBD [Stallmach and Carstens, 2002]. Moreover, it appears that the events during C. difficile infection are responsible for initiation of the cascade of inflammatory processes which could play an important role in the clinical initiation of IBD.
Several epidemiological studies have documented more cases of C. difficile infection in patients with UC and CD (adult and pediatric patients) than controls and the worse outcome in was found in patients with IBD [Musa et al. 2010; Wultanska et al. 2010; Bossuyt et al. 2009; Ananthakrishnan et al. 2008; Issa et al. 2007; Rodemann et al. 2007]. Patients with IBD and colonic involvement exhibited a significant association with development of C. difficile infection, both those with CD and UC. More recent data from a retrospective observational study by Issa and colleagues found that the rate of C. difficile infection in patients with IBD increased from 1.8% in 2004 to 4.6% in 2005. The majority of cases reported in 2005 were colonic IBD (91%) and outpatient-acquired infections (76%) [Issa et al. 2007]. The study by Powell and colleagues suggested that the relative increase in C. difficile infection in UC compared with CD was due to the extent of colonic involvement in UC rather than the difference in nature of the two diseases [Powell et al. 2008].
However, there is an ongoing debate as to whether C. difficile infection represents a risk factor for the development of IBD, or whether inflammation present during the course of IBD predisposes people to C. difficile infection.
Role of Clostridium difficile toxin in inflammatory bowel disease
Almost three decades ago, LaMont and Trnka postulated that C. difficile toxin complicates chronic IBD and contributes to relapse in some patients [LaMont and Trnka, 1980]. C. difficile toxin was found in the stool of patients with IBD, suggesting that C. difficile can cause worsening of disease activity in patients with IBD [Trnka and LaMont, 1981]. C. difficile toxin was also suggested as a contributory factor to mucosal injury in IBD by Meyers and colleagues [Meyers et al. 1981]. However, the authors showed that C. difficile toxin appeared only in patients exposed to antimicrobials and was unlikely to be a significant contributory factor in IBD, and the presence of toxin was not correlated with disease severity. Keighley and colleagues concluded that C. difficile cytotoxin was rare in patients with IBD, unless the patient had been exposed to antibiotics, and the isolation of this organism (without toxin) was therefore of doubtful pathological significance [Keighley et al. 1982]. The authors showed that the absence of toxin detection might have been due to insufficient numbers of clostridia to produce a measurable level of cytotoxin or production of only small amounts of toxin by the clostridia. However, some studies showed that C. difficile and its toxins have been implicated as risk factors for exacerbation of the inflammatory process in up to 5% of patients with UC or CD. A severe clinical course may result from C. difficile infection superimposed on IBD, including toxic megacolon and toxic colitis [Bolton and Read, 1982].
C. difficile infection can cause acute enteritis in patients with IBD and ileostomies, while colitis in the rectal stump of a patient with ileostomy and colectomy for UC has been reported [Tsironi et al. 2006]. Moreover, C. difficile toxin was detected in ileostomy fluid with symptomatically increased ileostomy output [LaMont and Trnka, 1980]. This finding suggested that C. difficile, under special circumstances, might be able to cause small bowel and colonic disease.
Typical evidence of colonic changes with C. difficile infection, including pseudomembranous exudate, are often not present in patients with underlying CD or UC [Nomura et al. 2009; Skaros et al. 2007]. However, a severe clinical course may result, including precipitation of toxic colitis and toxic megacolon. Why pseudomembranes are infrequently seen in IBD is unclear. One possible explanation is that the weakened intestinal epithelial environment of a patient with chronically active IBD is unable to mount an adequate inflammatory response to form a pseudomembrane. The absence of pseudomembranes can be associated with immunosuppression as studies suggest that patients with bone marrow or stem cell transplants on immunosuppression, and those with UC on steroids, did not develop pseudomembranes with C. difficile infection [Nomura et al. 2009].
In summary, at present there is no clear epidemiological evidence that C. difficile infection precedes IBD. In the next section we describe the mechanisms by which C. difficile activates the proinflammatory response that contribute to intestinal diseases such as diarrhea and pseudomembranous colitis. We hypothesize that these responses may contribute towards the development or exacerbation of IBD.
Pathogenesis of Clostridium difficile
The clinical appearance of C. difficile infection is highly variable, ranging from mild self-limited diarrhea to severe pseudomembraneous colitis.
Epithelial expansion/colonization by bacteria and toxins, and the induction of cytokines enhance mucosal permeability. If the mucosal barrier is broken, an influx of luminal antigens may result in the perpetuation of intestinal inflammation by chronically stimulating resident and recruited immune cells of the lamina propria [Stallmach and Carstens, 2002].
The first step in C. difficile pathogenesis is that initially it must be entrenched in the gut and attach to epithelial cells. Well known virulence factors which play a role in adherence and intestinal colonization of C. difficile have been identified and these include proteolytic enzymes (i.e. cysteine protease) and adhesins (cell-wall protein Cwp66, the GroEL heat-shock protein, a 68 kDa fibronectin-binding protein), and the flagella components FliC (flagellin) and FliD (flagellar cap protein) [Hennequin et al. 2001, 2003; Waligora et al. 2001; Tasteyre et al. 2001].
The second phase of the pathogenic process is the production of toxins. C. difficile toxins (toxins A and B) target the colonic epithelium directly. This is one of the most important events as it disrupts tight junctions to breach the intestinal epithelial barrier. As a result of toxin-induced changes, filamentous actin comprising the perijunctional actomyosin ring, known to be important in regulating tight junction permeability, is condensed into discrete plaques, resulting in a loss of electrical resistance and increased paracellular permeability from the luminal domain [Moore et al. 1990; Hecht et al. 1992]
The loss of tight junctions and enhanced mucosal permeability leads to an array of consequences, including penetration, translocation of bacteria and their products between epithelial cells, and also promotes access of neutrophils to the intestinal lumen. An intense inflammatory response and neutrophil infiltration are essential for the pathophysiology of CDAD. Neutrophilic infiltration can be pronounced in patients with CDAD in that the neutrophils, together with fibrin, form plaques known as pseudomembranes on the colonic wall [Gerding et al. 1995]. Release of reactive oxygen species and enzymes from activated neutrophils is likely to act synergetically with both toxins to produce tissue destruction. The mechanism by which neutrophils are recruited to sites of inflammation is complex and involves the expression of leukocyte and endothelial cell adhesion molecules, followed by neutrophil attachment and adhesion to the endothelium, and finally transmigration of neutrophils into the intestine mucosa [Springer, 1994]. These events are driven by production of a wide range of chemoattractants, and by activating cytokines that establish a chemotactic gradient and induce the expression of adhesion molecules in both endothelial cells and neutrophils. IL-8 is the principle cytokine involved in migration and activation of neutrophils. Release of IL-8 from immune cells and epithelial cells appears to be a critical event in inflammatory cascade after C. difficile infection. The molecular mechanism by which C. difficile induces the inflammatory process involves activation of NFκB, which appears to be required for transcription of the IL-8 gene [Kim et al. 2006; Jefferson et al. 1999].
NFκB was originally identified as the transcription factor that activates the promoter of the κ region of the immunoglobulin light chain locus in B lymphocytes [Barnes, 1997]. Since its initial discovery, investigators have demonstrated that NFκB is a key transcriptional factor controlling the expression of genes mediating the inflammatory response. Moreover, it is clear from various studies that NFκB is very likely to play a key role and it seems that by analyzing transcriptional regulatory mechanisms in intestinal inflammation we can gain fundamental insights into the pathogenesis, prognosis and treatment of intestinal diseases, including C. difficile infection and IBD.
The excessive immune reaction in the mucosa in response to C. difficile could be responsible for the persisting and destructive nature of the inflammation that is a common feature in gastrointestinal disorders, such as IBD.
Clostridium difficile toxicity
C. difficile plays a crucial role in the pathogenesis of pseudomembranous colitis and diarrhea by releasing two toxins, A and B, with potent cytotoxic and proinflammatory properties [Savidge et al. 2003; Pothoulakis and LaMont, 2001]. These toxins are glucosyltransferases, which catalyze the inactivation of Rho proteins that control cellular functions, including the organization of the actin cytoskeleton and epithelial barrier function [Jank et al. 2007; Voth and Ballard, 2005; Schirmer and Aktories, 2004]. Both toxins act via translocation into target cells, and do their damage through autocatalytic processes by inactivating guanosine triphosphate (GTP)-binding proteins of the Rho GTPase family, such as Rho, Rac, Cdc42, involved in cellular signaling [Jank et al. 2007; Henriques et al. 1987; Just et al. 1995]. Thus, glucosylation of Rho GTPases leads to actin cytoskeleton disaggregation, increased membrane permeability, loss of barrier function, cell rounding, cytotoxicity, and ultimately cell death [Voth and Ballard, 2005]. In addition, the C. difficile toxins are involved in tissue-damaging inflammatory processes by initiating massive cellular immune responses; that is, neutrophilic infiltration with upregulation and release of cytokines, such as IL-8, IL-6, IL-1β, leukotrienes B4 and interferon γ [Voth and Ballard, 2005].
C. difficile toxins also induce both apoptosis and necrosis of epithelial cells. Moreover, the apoptogenic properties of toxin A and B are well known, and they may contribute to the inflammatory process induced by these toxins [Mahida et al. 1996; Fiorentini et al. 1998; Brito et al. 2002].
Binding of C. difficile toxins to specific surface receptors appears to be an important step in the expression of the biological actions of C. difficile toxins. It was previously shown that toxin A binds to trisaccharide α-Gal-(1,3)-β-Gal-(1,4)-β-GlcNac [Krivan et al. 1986]. However, because of a lack of the functional α-galactosyltransferase required for assembly of this trisaccharide in humans, this structure cannot be part of intestinal receptors in humans [Jank et al. 2007; Koike et al. 2000]. This implies that toxin A binds to human enterocytes via different oligosaccharides or possibly by protein–protein interaction. Thus, the disaccharide Galβ1-4GlcNAc, which is present in humans, has been suggested to be part of a possible receptor. Our understanding of the toxin B receptor is even more limited.
Toxin A and proinflammatory signaling
The mechanism of toxin A-mediated inflammation involves activation of immune and inflammatory cells, leading to the release of proinflammatory mediators, acute neutrophil infiltration, and intestinal mucosal injury [Warny et al. 2000; Jefferson et al. 1999; Linevsky et al. 1997]. NFκB appears to be the most important transcription factor of various inflammatory mediators and the main regulator of inducible expression of the IL-8 gene in cells exposed to C. difficile toxin A. In addition to NFκB, expression of the IL-8 gene in response to toxin A is dependent on activation of a second transcription factor, activator protein 1 (AP-1). Toxin A induced IL-8 expression at the level of gene transcription and this effect occurs through a mechanism requiring intracellular Ca2+, calmodulin and calcium/calmodulin-dependent kinase (CaMK) [Kim et al. 2006; Jefferson et al. 1999]. Generally, transcriptional activation of cytokine genes occurs when the stimulus interacts with the cell in such a way that its message is relayed to the transcription factor directly responsible for transcriptional activation of the gene. This signal transduction pathway involves activation of a cascade of intracellular protein kinases. Although phosphatidylinositol 3 kinase (PI3 kinase) plays an important role in a variety of intracellular signaling pathways, Jefferson and colleagues showed that the PI3 kinase inhibitor wortmannin did not block the upregulation of IL-8 by toxin A [Jefferson et al. 1999]. However, toxin A-induced release of IL-8 decreased in the presence of protein tyrosine kinase (PTK) inhibitor, indicating a presence of PTK-dependent pathways implicated in upregulation of IL-8. Internalization of toxin A through an endosome-like pathway is required to effectively induce IL-8 secretion from monocytes, while cell surface binding is not sufficient to induce IL-8 secretion. In summary, toxin A-induced secretion of IL-8 from human monocytes results from an activation pathway that involves Ca2+/calmodulin, CaMK, PTK, NFκB, and AP-1 activation (Figure 1(a)) [Kim et al. 2006; Jefferson et al. 1999]. NFκB may also be involved in the transcriptional activity of other cytokines known to be induced by C. difficile toxin A, including IL-1, IL-6, and tumor necrosis factor α (TNFα) [Flegel et al. 1991].
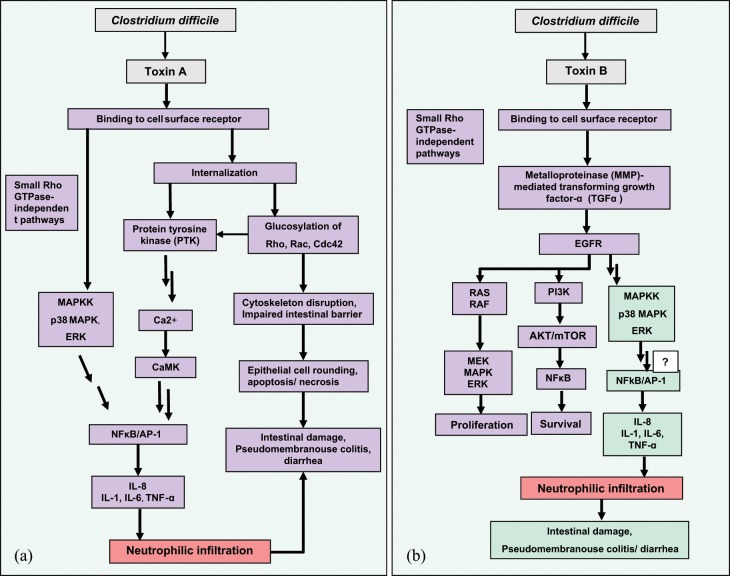
Figure 1.
(a) Mechanism of Clostridium difficile toxin A-mediated inflammation. After receptor binding, toxin A is internalized into target cells (Rho-dependent signaling) and induced interleukin (IL)-8 expression at the level of gene transcription; this effect occurred through a mechanism requiring intracellular Ca2+, calmodulin, calcium/calmodulin-dependent kinase (CaMK), protein tyrosine kinase (PTK), and nuclear factor κB (NFκB), and activator protein 1 (AP-1) activation. The Rho-independent signaling pathway induced by toxin A in human monocytic cells occurred through a mechanism requiring interaction with surface receptor, p38 mitogen-activated protein (MAP) kinase and extracellular signal-regulated kinase (ERK) activation. Kinase activation leads to NFκB translocation and IL-8 gene expression in target cells. (b). Mechanism of C. difficile toxin B-mediated inflammation. Toxin B through an unidentified membrane receptor activates the ERK-MAP kinase pathway via signaling dependent on epidermal growth factor receptor (EGFR). Metalloproteinase (MMP)-mediated transforming growth factor α (TGFα) is needed for phosphorylation of EGFR. Toxin B-mediated EGFR and ERK-MAP kinase activation and IL-8 gene expression are independent of Rho glucosylation. NFκB activation is not linked to toxin B-mediated IL-8 expression gene, but other unidentified signaling pathways may be involved in this process. EGFR activation of the MAP kinase-associated pathway has also been associated with cell proliferation and tissue repair. This process recruits the RAS-RAF-MAP kinase pathway, resulting in increased cellular proliferation. EFGR signaling also activates the phosphatidylinositol 3 kinase (PI3K)-serine threonine kinase Akt (AKT)-mammalian target of rapamycin (mTOR) pathway, which activates the major cellular survival and antiapoptosis signals via activating NFκB. Color legend: bacterial cell (gray); epithelial cell (green/violet); immune cells (red).
A study by Warny and colleagues showed that, in human monocytic cells, p38 mitogen-activated protein (MAP) kinase and extracellular signal-regulated kinase (ERK) are activated by C. difficile toxin A and are responsible for IL-8 gene expression (Figure 1(a)). Interestingly, toxin A interaction with cell-surface receptors may trigger p38 MAP kinase and ERK activation before Rho protein glucosylation, indicating the presence of a Rho-independent pathway. Rho glucosylation is detected later because this event is required for toxin internalization and translocation into the cytoplasm. MAP kinase activation in a Rho-independent manner gave rise to the hypothesis that toxin A-induced IL-8 production might be independent of enzymatic activity of toxin A. This hypothesis could be supported by the fact that the active forms of the GTP-binding proteins have been implicated in the activation of inflammatory cytokine gene transcription. The Rho family GTPases, including Cdc42 and Rac, have been shown to regulate p38 MAP kinase and c-Jun N-terminal kinase (JNK), whereas Rho, Cdc42, and Rac have been associated with NFκB activation. However, Rho inactivation by C. difficile toxins blocked the activation of p38 and JNK kinases as well as activation of transcriptional factors, NFκB, and AP-1 [Montaner et al. 1998; Naumann et al. 1998; Pan et al. 1998; Wesselborg et al. 1998; Perona et al. 1997]. Because Rho proteins appear to coordinate cytokine gene transcription in response to inflammatory stimuli, their inactivation by toxin A would not be expected to trigger an inflammatory response. Activation of p38 MAP kinase has been reported to be a glucosyltransferase-independent effect of toxin A, possibly mediated by reactive oxygen species [Kim et al. 2005]. However, a role for Rho glucosylation in IL-8 gene expression cannot be excluded. A study by Beltman and colleagues showed that inactivation of RhoA protein by C. botulinum C3 toxin might induce MAP kinase activation [Beltman et al. 1999].
Toxin B and proinflammatory signaling
In human intestinal cells, toxin B, like toxin A, has been shown to induce intestinal epithelial damage, increase mucosal permeability, stimulate IL-8 synthesis, and elicit an acute inflammatory response characterized by neutrophil recruitment [Lyras et al. 2009; Na et al. 2005; Savidge et al. 2003]. C. difficile toxin B mediates the inflammatory response, which is linked to increased IL-8 gene expression by activation of epidermal growth factor receptor (EGFR) and ERK-MAP kinase activation [Na et al. 2005]. Activation of EGFR has been shown to mediate ERK-MAP phosphorylation in response to extracellular stimuli, including bacteria, bacterial products, and ligands for G protein-coupled receptors [Daub et al. 1996]. Although both toxins of C. difficile are able to phosphorylate ERK1/2 in human colonic epithelial cells, only toxin B has been shown to activate EGFR. Toxin A-mediated ERK1/2 activation could not be blocked by EGFR inhibitor, suggesting that these toxins probably bind to separate cell surface receptors and trigger the different signaling cascade to activate MAP kinase signaling. Toxin B activates ERK-MAP kinase by binding to an as yet undefined colonocyte membrane receptor, and via a pathway dependent on EGFR phosphorylation induced by metalloproteinase (MMP)-dependent transforming growth factor α (TGFα) secretion (Figure 1(b)). Release of TGFα from colonocytes was detected within 5 min of toxin B exposure, but Rho glucosylation during 30–60 min because this process needs toxin internalization and translocation. Rapid release of TGFα suggested that toxin B-induced EGFR-MAP kinase activation and secretion of IL-8 are independent of Rho glucosylation [Na et al. 2005]. IL-8 gene expression is often linked to the NFκB pathway, but showed that the toxin B-mediated NFκB signaling response, that is, IκB phosphorylation, degradation, and NFκB phosphorylation, was not detected, indicating that EGRF-ERK activation but not NFκB are important for toxin B-mediated IL-8 expression [Na et al. 2005].
Although, the ERK-MAP-EGFR signaling pathway is critical in toxin B-mediated IL-8 expression, inhibitors of ERK and EGFR only decreased but did not abolish the effect of toxin B on IL-8 activity. Thus, it is possible that other unidentified signaling pathways dependent on NFκB and AP-1 may be involved in IL-8 gene expression. Toxin B may also be involved in induction of other cytokines, including IL-1β, TNFα, and IL-6 [Flegel et al. 1991].
EGFR activation of the MAP kinase-associated pathway is usually linked to cell proliferation and tissue repair. This process recruits the RAS-RAF-MAP kinase pathway, resulting in increased cellular proliferation. RAS-RAF-MAP are all serine/threonine-selective protein kinases. Moreover, EGFR signaling also activates PI3 kinase, resulting in activation of the downstream effector molecules, such as serine-threonine kinase Akt (AKT) and mammalian target of rapamycin (mTOR) kinase. These molecules induce proliferation and block apoptosis, resulting in cell survival via activating nuclear transcription factors such as NFκB [Suzuki et al. 2010]. However in C. difficile infection activation of the EGFR pathway may have dual roles in both tissue repair and proinflammatory response.
Conclusion
Alterations in the bowel flora and its activities are now believed to be contributing factors to many chronic and degenerative diseases, and any change in the composition of the microbiota alters the intestinal environment making this niche vulnerable to pathogenic bacteria. Thus, disturbance of the intestinal microbiota equilibrium may promote inflammation and mucosal tissue damage that predisposes people to pathological conditions of the gastrointestinal tract, such as C. difficile infection and IBD. Current data do not convincingly support a persistent, specific pathogen as a cause of IBD; however, specific microbiological virulence factors could initiate intestinal inflammation and injury. Whether C. difficile belongs to the group of pathogens that have a role in the initial onset of IBD is subject to debate. C. difficile infection has a broad spectrum of manifestations, including microscopic and gross lesions, diarrhea, and intestinal inflammation. Most of these distinguishing symptoms may be attributed to inflammatory events mediated by two large clostridial toxins, A and B. Both toxins induce intestinal injury and inflammation through disruption of the intestinal epithelial barrier, induction of proinflammatory mediators and causing cell apoptosis or necrosis. Moreover, toxins A and B are glucosyltransferases that irreversibly inactivate small Rho GTPases, leading to disruption of cytoskeleton and tight junctions and subsequent cell rounding, detachment, and cell death. The mechanisms of toxin A- and B-mediated inflammation, involving activation of MAP kinase, NFκB and AP-1, and stimulation of IL-8, occurred via two different Rho-dependent and -independent pathways.
At present there is no clear evidence that C. difficile infection precedes IBD, however the mechanisms by which C. difficile activate the proinflammatory response and which contribute to intestinal diseases such as diarrhea and pseudomembranous colitis may also be responsible for the development or exacerbation of IBD. A better understanding of all events associated with development of gastrointestinal disorders will hopefully provide new insights into the mechanism of intestinal responses to microorganisms.
Footnotes
Funding: This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of interest statement: The authors declare no conflicts of interest in preparing this article.
Contributor Information
Justyna Bien, Witold Stefanski Institute of Parasitology of the Polish Academy of Sciences, Warsaw, Poland.
Vindhya Palagani, Department of Internal Medicine I, Faculty of Medicine, Tübingen University, Tübingen, Germany.
Przemyslaw Bozko, Department of Internal Medicine I, Faculty of Medicine, Tübingen University, Otfried-Müller-Straße 10, 72076 Tübingen, Germany.
References
- Agans R., Rigsbee L., Kenche H., Michail S., Khamis H., Paliy O. (2011) Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol Ecol 77: 404–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldeyab M., Harbarth S., Vernaz N., et al. (2009) Quasiexperimental study of the effects of antibiotic use, gastric acid-suppressive agents, and infection control practices on the incidence of Clostridium difficile-associated diarrhea in hospitalized patients. Antimicrob Agents Chemother 53: 2082–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthakrishnan A., McGinley E., Binion D. (2008) Excess hospitalisation burden associated with Clostridium difficile in patients with inflammatory bowel disease. Gut 57: 205–210 [DOI] [PubMed] [Google Scholar]
- Bäckhed F., Ley R., Sonnenburg J., Peterson D., Gordon J. (2005) Host–bacterial mutualism in the human intestine. Science 307: 1915–1920 [DOI] [PubMed] [Google Scholar]
- Barc M., Bourlioux F., Janoir C., Rigothier-Gois L., Boureau H, Doré J., et al. (2004) Effect of amoxicillin clavulanate on human fecal flora in a gnotobiotic mouse model measured with group specific 16S rRNA targeted oligonucleotide probes in combination with flow cytometry. Antimicrob Agents Chemother 48: 1365–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes P. (1997) Nuclear factor-κB. Int J Biochem 29: 867. [DOI] [PubMed] [Google Scholar]
- Beltman J., Erickson J., Martin G., Lyons J., Cook S. (1999) C3 toxin activates the stress signaling pathways, JNK and p38, but antagonizes the activation of AP-1 in rat-1 cells. J Biol Chem 274: 3772–3780 [DOI] [PubMed] [Google Scholar]
- Bernstein C., Shanahan F. (2008) Disorders of a modern lifestyle: reconciling the epidemiology of inflammatory bowel diseases. Gut 57: 1185–1191 [DOI] [PubMed] [Google Scholar]
- Bolton R., Read A. (1982) Clostridium difficile in toxic megacolon complicating acute inflammatory bowel disease. Br Med J (Clin Res Ed) 285: 475–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton R., Tait S., Dear P. (1984) Asymptomatic neonatal colonization by Clostridium difficile. Arch Dis Child 59: 466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossuyt P., Verhaegen J., Van Assche G., Rutgeerts P., Vermeire S. (2009) Increasing incidence of Clostridium difficile-associated diarrhea in inflammatory bowel disease. J Crohns Colitis 3: 4–7 [DOI] [PubMed] [Google Scholar]
- Brito G., Fujji J., Carneiro-Filho B., Lima A., Obrig T., Guerrant R. (2002) Mechanism of Clostridium difficile toxin A-induced apoptosis in T84 cells. J Infect Dis 186: 1438–1447 [DOI] [PubMed] [Google Scholar]
- Bry L., Falk P., Midtvedt T., Gordon J. (1996) A model of host microbial interactions in an open mammalian ecosystem. Science 273: 1380–1383 [DOI] [PubMed] [Google Scholar]
- Burnham W., Lennard-Jones J., Stanford J., Bird R.(1978) Mycobacteria as a possible cause of inflammatory bowel disease. Lancet 2: 693–696 [DOI] [PubMed] [Google Scholar]
- Brüggemann H., Gottschalk G. (2008) Comparative genomics of clostridia link between the ecological niche and cell surface properties. Ann N Y Acad Sci 1125: 73–81 [DOI] [PubMed] [Google Scholar]
- Cash H., Whitham C., Behrendt C., Hooper L. (2006) Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313: 1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Antonopoulos D., Kalra A., Tonelli A., Khalife W., Schmidt T., et al. (2008) Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 197: 435–438 [DOI] [PubMed] [Google Scholar]
- Collignon A., Ticchi L., Depitre C., Gaudelus J., Delmée M., Corthier G., et al. (1993) Heterogeneity of Clostridium difficile isolates from infants. Eur J Pediatr 152: 319–322 [DOI] [PubMed] [Google Scholar]
- Collins M., Lawson P., Willems A., Cordoba J., Fernandez-Garayzabal J., Garcia P., et al. (1994) The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 44: 812–826 [DOI] [PubMed] [Google Scholar]
- Danese S., Fiocchi C. (2006) Etiopathogenesis of inflammatory bowel diseases. World J Gastroenterol 12: 4807–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A., Boudeau J., Bulois P., Neut C., Glasser A., Barnich N., et al. (2004) High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127: 412–421 [DOI] [PubMed] [Google Scholar]
- Daub H., Weiss F., Wallasch C., Ullrich A. (1996) Role of transactivation of the EGF receptor in signalling by G-protein-coupled receptors. Nature 379: 557–560 [DOI] [PubMed] [Google Scholar]
- De La Cochetière M., Durand T., Lalande V., Petit J., Potel G., Beaugerie L. 2008. Effect of antibiotic therapy on human fecal microbiota and the relation to the development of Clostridium difficile. Microb Ecol 56:395–402 [DOI] [PubMed] [Google Scholar]
- Dial S., Delaney J., Barkun A., Suissa S. (2005) Use of gastric acid-suppressive agents and the risk of community-acquired Clostridium difficile-associated disease. JAMA 294: 2989–2995 [DOI] [PubMed] [Google Scholar]
- Dial S., Delaney J., Schneider V., Suissa S. (2006) Proton pump inhibitor use and risk of community-acquired Clostridium difficile-associated disease defined by prescription for oral vancomycin therapy. CMAJ 175: 745–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson G., Hickey C., Trinder J. (2003) Clostridium difficile colitis causing toxic megacolon, severe sepsis and multiple organ dysfunction syndrome. Intensive Care Med 29: 1030. [DOI] [PubMed] [Google Scholar]
- Eckburg P., Bik E., Bernstein C., Purdom E., Dethlefsen L., Sargent M., et al. (2005) Diversity of the human intestinal microbial flora. Science 308: 1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falony G., Vlachou A., Verbrugghe K., De Vuyst L. (2006) Crossfeeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl Environ Microbiol 72: 7835–7841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell R., La Mont J. (2002) Microbial factors in inflammatory bowel disease. Gastroenterol Clin North Am 31: 41–62 [DOI] [PubMed] [Google Scholar]
- Fiorentini C., Fabbri A., Falzano L., Fattorossi A., Matarrese P., Rivabene R., et al. (1998) Clostridium difficile toxin B induces apoptosis in intestinal cultured cells. Infect Immun 66: 2660–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flegel W., Müller F., Daubener W., Fischer H., Hadding U., Northoff H. (1991) Cytokine response by human monocytes to Clostridium difficile toxin A and toxin B. Infec Immun 59: 3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D., St Amand A., Feldman R., Boedeker E., Harpaz N., Pace N. (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 104: 13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman H. (2008) Recent developments on the role of Clostridium difficile in inflammatory bowel disease. World J Gastroenterol 14: 2794–2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García Rodríguez L., Ruigómez A., Panés J. (2006) Acute gastroenteritis is followed by an increased risk of inflammatory bowel disease. Gastroenterology 130: 1588–1594 [DOI] [PubMed] [Google Scholar]
- George R. (1986) The carrier state: Clostridium difficile. Antimicrob Chemother 18(Suppl. A): 47–58 [DOI] [PubMed] [Google Scholar]
- Gerding D., Johnson B., Peterson L., Mulligan M., Silva J., Jr. (1995) Clostridium difficile-associated diarrhea and colitis. Infect Control Hosp Epidemiol 16: 459. [DOI] [PubMed] [Google Scholar]
- Hawrelak J., Myers S. (2004) The causes of intestinal dysbiosis: a review. Altern Med Rev 9: 180–197 [PubMed] [Google Scholar]
- Hecht G., Koutsouris A., Pothoulakis C., LaMont J., Madara J. (1992) Clostridium difficile toxin B disrupts the barrier function of T84 monolayers. Gastroenterology 102: 416–423 [DOI] [PubMed] [Google Scholar]
- Hennequin C., Janoir C., Barc M., Collignon A., Karjalainen T. (2003) Identification and characterization of a fibronectin-binding protein from Clostridium difficile. Microbiology 149: 2779–2787 [DOI] [PubMed] [Google Scholar]
- Hennequin C., Porcheray F., Waligora-Dupriet A., Collignon A., Barc M., Bourlioux P., et al. (2001) GroEL (Hsp60) of Clostridium difficile is involved in cell adherence. Microbiology 147: 87–96 [DOI] [PubMed] [Google Scholar]
- Henriques B., Florin I., Thelestam M. (1987) Cellular internalisation of Clostridium difficile toxin A. Microb Pathog 2: 455–463 [DOI] [PubMed] [Google Scholar]
- Hopkins M., Macfarlane G., Furrie E., Fite A., Macfarlane S. (2005) Characterisation of intestinal bacteria in infant stools using real-time PCR and northern hybridisation analyses. FEMS Microbiol Ecol 54: 77–85 [DOI] [PubMed] [Google Scholar]
- Hold G., Pryde S., Russell V., Furrie E., Flint H. (2002) Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol Ecol 39: 33–39 [DOI] [PubMed] [Google Scholar]
- Holzapfel W., Haberer P., Snel J., Schillinger U., Huis in’t Veld J. (1998) Overview of gut flora and probiotics. Int J Food Microbiol 41: 85–101 [DOI] [PubMed] [Google Scholar]
- Hooper L., Midwedt T., Gordon J. (2002) How host microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 22: 283–307 [DOI] [PubMed] [Google Scholar]
- Iizuka M., Itou H., Konno S., Chihara J., Tobita M., Oyamada H., et al. (2004) Elemental diet modulates the growth of Clostridium difficile in the gut flora. Aliment Pharmacol Ther 20: 151–157 [DOI] [PubMed] [Google Scholar]
- Issa M., Vijayapal A., Graham M., Beaulieu D., Otterson M., Lundeen S., et al. (2007) Impact of Clostridium difficile on inflammatory bowel disease. Clin Gastroenterol Hepat 5: 345–351 [DOI] [PubMed] [Google Scholar]
- Jacobs N., Jr. (1994) Antibiotic-induced diarrhea and pseudomembranous colitis. Postgrad Med 95: 111–120 [DOI] [PubMed] [Google Scholar]
- Jank T., Giesemann T., Aktories K. (2007) Rho-glucosylating Clostridium difficile toxins A and B: new insights into structure and function. Glycobiology 17: 15–22 [DOI] [PubMed] [Google Scholar]
- Jarvis W., Schlosser J., Jarvis A., Chinn R. (2009) National point prevalence of Clostridium difficile in US health care facility inpatients, 2008. Am J Infect Control 37: 263–270 [DOI] [PubMed] [Google Scholar]
- Jefferson K., Smith M., Bobak D., Jr (1999) Roles of intracellular calcium and NF-κB in the Clostridium difficile toxin A-induced up-regulation and secretion of IL-8 from human monocytes. J Immunol 163: 5183–5191 [PubMed] [Google Scholar]
- Jernberg C., Lofmark S., Edlund C., Jansson J. (2007) Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J 1: 56–66 [DOI] [PubMed] [Google Scholar]
- Just I., Selzer J., Wilm M., von Eichel-Streiber C., Mann M., Aktories K. (1995) Glucosylation of Rho proteins by Clostridium difficile toxin B. Nature 375: 500–503 [DOI] [PubMed] [Google Scholar]
- Keighley M., Youngs D., Johnson M., Allan R., Burdon D. (1982) Clostridium difficile toxin in acute diarrhoea complicating inflammatory bowel disease. Gut 23: 410–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly C., LaMont J. (1998) Clostridium difficile infection. Annu Rev Med 49: 375–390 [DOI] [PubMed] [Google Scholar]
- Kim J., Lee J., Yoon Y., Ohz Y., Youny J., Kim Y. (2006) NF-κB activation pathway is essential for the chemokine expression in intestinal epithelial cells stimulated with Clostridium difficile toxin A. Scan J Immunol 63: 453–446 [DOI] [PubMed] [Google Scholar]
- Kim H., Rhee S., Kokkotou E., Na X., Savidge T., Moyer M., et al. (2005) Clostridium difficile toxin A regulates inducible cyclooxygenase-2 and prostaglandin E2 synthesis in colonocytes via reactive oxygen species and activation of p38 MAPK. J Biol Chem 280: 21237–21245 [DOI] [PubMed] [Google Scholar]
- Kobayashi T. (1983) Studies on Clostridium difficile and antimicrobial associated diarrhea or colitis. Jpn J Antibiot 36: 464–476 [PubMed] [Google Scholar]
- Koike T., Kuzuya M., Asai T., Kanda S., Cheng X., Watanabe K., et al. (2000) Activation of MMP-2 by Clostridium difficile toxin B in bovine smooth muscle cells. Biochem Biophys Res Commun 277: 43–46 [DOI] [PubMed] [Google Scholar]
- Krivan H., Clark G., Smith D., Wilkins T. (1986) Cell surface binding site for Clostridium difficile enterotoxin: evidence for a glycoconjugate containing the sequence Gal alpha 1–3Gal beta 1–4GlcNAc. Infect Immun 53: 573–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijper E., Coignard B., Tull P. (2006) Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect 12: 2–18 [DOI] [PubMed] [Google Scholar]
- LaMont J., Trnka Y. (1980) Therapeutic implications of Clostridium difficile toxin during relapse of chronic inflammatory bowel disease. Lancet 1: 381–383 [DOI] [PubMed] [Google Scholar]
- Lepage P., Colombet J., Marteau P., Sime-Ngando T., Dore J., Leclerc M. (2008) Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut 57: 424–425 [DOI] [PubMed] [Google Scholar]
- Ley R., Hamady M., Lozupone C., Turnbaugh P., Ramey R., Bircher J., et al. (2008) Evolution of mammals and their gut microbes. Science 320: 1647–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R., Peterson D., Gordon J. (2006) Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 124: 837–848 [DOI] [PubMed] [Google Scholar]
- Lidar M., Langevitz P., Shoenfeld Y. (2009) The role of infection in inflammatory bowel disease: initiation, exacerbation and protection. IMAJ 11: 558–563 [PubMed] [Google Scholar]
- Linevsky J., Pothoulakis C., Keates S., Warny M., Keates A., Lamont J., et al. (1997) IL-8 release and neutrophil activation by Clostridium difficile toxin-exposed human monocytes. Am J Physiol 273: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Lyras D., O’Connor J., Howarth P., Sambol S., Carter G., Phumoonna T., et al. (2009) Toxin B is essential for virulence of Clostridium difficile. Nature 458: 1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahida Y., Makh S., Hyde S., Gray T., Borriello S. (1996) Effect of Clostridium difficile toxin A on human intestinal epithelial cells: induction of interleukin 8 production and apoptosis after cell detachment. Gut 38: 337–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manges A., Labbe A., Loo V., Atherton J., Behr M., Masson L., et al. (2010) Comparative metagenomic study of alterations to the intestinal microbiota and risk of nosocomial Clostridum difficile-associated disease. J Infect Dis 202: 1877–1884 [DOI] [PubMed] [Google Scholar]
- Manichanh C., Rigottier-Gois L., Bonnaud E., Gloux K., Pelletier E., Frangeul L., et al. (2006) Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 55: 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchesi J., Shanahan F. (2007) The normal intestinal microbiota. Curr Opin Infect Dis 20: 508–513 [DOI] [PubMed] [Google Scholar]
- Marcille F., Gomez A., Joubert P., Ladiré M., Veau G., Clara A., et al. (2002) Distribution of genes encoding the trypsin-dependent antibiotic ruminococcin A among bacteria isolated from human fecal microbiota. Appl Environ Microbiol 68: 3424–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazmanian S., Liu C., Tzianabos A., Kasper D. (2005) An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122: 107–118 [DOI] [PubMed] [Google Scholar]
- McFarland L. (1998) Epidemiology, risk factors and treatments for antibiotic-associated diarrhea. Dig Dis 16: 292–307 [DOI] [PubMed] [Google Scholar]
- McFarland L., Beneda H., Clarridge J., Raugi G. (2007) Implications of the changing face of Clostridium difficile disease for health care practitioners. Am J Infect Control 35: 237–253 [DOI] [PubMed] [Google Scholar]
- Meyers S., Mayer L., Bottone E., Desmond E., Janowitz H. (1981) Occurrence of Clostridium difficile toxin during the course of inflammatory bowel disease. Gastroenterology 80: 697–670 [PubMed] [Google Scholar]
- Montaner S., Perona R., Saniger L., Lacal J. (1998) Multiple signalling pathways lead to the activation of the nuclear factor κB by the Rho family of GTPases. J Biol Chem 273: 12779–12785 [DOI] [PubMed] [Google Scholar]
- Moore R., Pothoulakis C., LaMont J., Carlson S., Madara J. (1990) C. difficile toxin A increases intestinal permeability and induces Cl– secretion. Am J Physiol 259: G165–G172 [DOI] [PubMed] [Google Scholar]
- Musa S., Thomson S., Cowan M., Rahman T. (2010) Clostridium difficile infection and inflammatory bowel disease. Scand J Gastroenterol 45: 261–272 [DOI] [PubMed] [Google Scholar]
- Mylonakis E., Ryan E., Calderwood S. (2001) Clostridium difficile-associated diarrhea. Arch Intern Med 161: 525–533 [DOI] [PubMed] [Google Scholar]
- Na X., Zhao D., Koon H., Kim H., Husmark J., Moyer M., et al. (2005) Clostridium difficile toxin B activates the EGF receptor and the ERK/MAP kinase pathway in human colonocytes. Gastroenterology 128: 1002–1011 [DOI] [PubMed] [Google Scholar]
- Nakamura S., Mikawa M., Nakashio S., Takabatake M., Okado I., Yamakawa K., et al. (1981) Isolation of Clostridium difficile from the feces and the antibody in sera of young and elderly adults. Microbiol Immunol 25: 345–351 [DOI] [PubMed] [Google Scholar]
- Naumann M., Rudel T., Wieland B., Bartsch C., Meyer T. (1998) Coordinate activation of activator protein 1 and inflammatory cytokines in response to neisseria gonorrhoeae epithelial cell contact involves stress response kinases. J Exp Med 188: 1277–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerandzic M., Pultz M., Donskey C. (2009) Examination of potential mechanisms to explain the association between proton pump inhibitors and Clostridium difficile infection. Antimicrob Agents Chemother 53: 4133–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Fujimoto Y., Yamashita M., Morimoto Y., Ohshiro M., Sato K., et al. (2009) Absence of pseudomembranes in Clostridium difficile-associated diarrhea in patients using immunosuppression agents. Scand J Gastroenterol 44: 74–78 [DOI] [PubMed] [Google Scholar]
- O’Keefe S. (2008) Nutrition and colonic health: the critical role of the microbiota. Curr Opin Gastroenterol 24: 51–58 [DOI] [PubMed] [Google Scholar]
- O’Keefe S. (2010) Tube feeding, the microbiota, and Clostridium difficile infection. World J Gastroenterol 16: 139–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer C., Bik E., DiGiulio D., Relman D., Brown P. (2007) Development of the human infant intestinal microbiota. PLoS Biol 5: e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z., Ye R., Christiansen S., Jagels M., Bokoch G., Zuraw B. (1998) Role of Rho GTPase in bradykinin-stimulated nuclear factor-kB activation and IL-1β gene expression in cultured human epithelial cells. J Immunol 160: 3038–3045 [PubMed] [Google Scholar]
- Pechine S., Janoir C., Boureau H., Gleizes A., Tsapis N., Hoys S., et al. (2007) Diminished intestinal colonization by Clostridium difficile and immune response in mice after mucosal immunization with surface proteins of Clostridium difficile. Vaccine 25: 3946–3954 [DOI] [PubMed] [Google Scholar]
- Pepin J., Valiquette L., Alary M., Villemure P., Pelletier A., Forget K., et al. (2004) Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAJ 171:466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona R., Montaner S., Saniger L., Sanchez-Perez I., Bravo R., Lacal J. (1997) Activation of the nuclear factor-κB by Rho, CDC42 and Rac-1 proteins. Genes Dev 11: 463–475 [DOI] [PubMed] [Google Scholar]
- Peterson D., Frank D., Pace N., Gordon J. (2008) Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell Host Microbe 3: 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothoulakis C., LaMont J. (2001) Microbes and microbial toxins: paradigms for microbial-mucosal interactions II. The integrated response of the intestine to Clostridium difficile toxins. Am J Physiol Gastrointest Liver Physiol 280: 178–183 [DOI] [PubMed] [Google Scholar]
- Powell N., Jung S., Krishnan B. (2008) Clostridium difficile infection and inflammatory bowel disease: a marker for disease extent? Gut 57: 1183–1184 [PubMed] [Google Scholar]
- Pryde S., Duncan S., Hold G., Stewart C., Flint H. (2002) The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett 217: 133–139 [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M., Smidt H., de Vos W. (2007) Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol 9: 2125–2136 [DOI] [PubMed] [Google Scholar]
- Rodemann J., Dubberke E., Reske K., da Seo H., Stone C. (2007) Incidence of Clostridium difficile infection in inflammatory bowel disease. Clin Gastroenterol Hepatol 5: 339–344 [DOI] [PubMed] [Google Scholar]
- Rolfe R., Helebian S., Finegold S. (1981) Bacterial interference between Clostridium difficile and normal fecal flora. J Infect Dis 143: 470–475 [DOI] [PubMed] [Google Scholar]
- Rousseau C., Levenez F., Fouqueray C., Dore J., Collignon A., Lepage P. (2011) Clostridium difficile colonization in early infancy is accompanied by changes in intestinal microbiota composition. J Clin Microbiol 49: 858–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage D. (1977) Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol 31: 107–133 [DOI] [PubMed] [Google Scholar]
- Savidge T., Pan W., Newman P., O’Brien M., Anton P., Pothoulakis C. (2003) Clostridium difficile toxin B is an inflammatory enterotoxin in human intestine. Gastroenterology 125: 413–420 [DOI] [PubMed] [Google Scholar]
- Schirmer J., Aktories K. (2004) Large clostridial cytotoxins: cellular biology of Rho/Ras-glucosylating toxins. Biochim Biophys Acta 1673: 66–74 [DOI] [PubMed] [Google Scholar]
- Schumacher G., Kollberg B., Sandstedt B., Jorup C., Grillner L., Ljungh A., et al. (1993) A prospective study of first attacks of inflammatory bowel disease and non-relapsing colitis. Scand J Gastroenterol 28: 1077–1085 [DOI] [PubMed] [Google Scholar]
- Segain J., de la Raingeard B., Bourreille A., Leray V., Gervois N., Rosales C., et al. (2000) Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut 47: 397–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaros S., Weber L., Komorowski R., Knox J., Emmons J., Bajaj J., et al. (2007) Impact of Clostridium difficile on inflammatory bowel disease. Clin Gastroenterol Hepatol 5: 345–351 [DOI] [PubMed] [Google Scholar]
- Sokol H., Seksik P., Furet J., Firmesse O., NionLarmurier I., Beauqerie G., et al. (2009) Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 15: 1183–1189 [DOI] [PubMed] [Google Scholar]
- Springer T. (1994) Traffic signals for lymphocyte recirculation and leukocyte emigration: the multistep paradigm. Cell 76: 301–314 [DOI] [PubMed] [Google Scholar]
- Stallmach A., Carstens O. (2002) Role of infections in the manifestation or reactivation of inflammatory bowel diseases. Inflamm Bowel Dis 8: 213–218 [DOI] [PubMed] [Google Scholar]
- Starr J. (2005) Clostridium difficile associated diarrhoea: diagnosis and treatment. BMJ 331: 498–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan A., Edlund C., Nord C. (2001) Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect Dis 1: 101–114 [DOI] [PubMed] [Google Scholar]
- Suzuki A., Sekiya S., Gunshima E., Fujii S., Taniguchi H. (2010) EGF signaling activates proliferation and blocks apoptosis of mouse and human intestinal stem/progenitor cells in long-term monolayer cell culture. Lab Invest 90:1425–1436 [DOI] [PubMed] [Google Scholar]
- Swidsinski S., Ladhoff A., Pernthaler A., Swidsinski S., Loening-Baucke V., Ortner M., et al. (2002) Mucosal flora in inflammatory bowel disease. Gastroenterology 122: 44–54 [DOI] [PubMed] [Google Scholar]
- Tamboli C., Neut C., Desreumaux P., Colombel J. (2004) Dysbiosis in inflammatory bowel disease. Gut 53: 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasteyre A., Barc M., Collignon A., Boureau H., Karjalainen T. (2001) Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect Immun 69: 7937–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R., Borriello S., Taylor A. (1981) Isolation of Clostridium difficile from the small bowel. Br Med J (Clin Res Ed) 283: 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testore G., Nardi F., Babudieri S., Giuliano M., Di Rosa R., Panichi G. (1986) Isolation of Clostridium difficile from human jejunum: identification of a reservoir for disease? J Clin Pathol 39: 861–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson A., Sauve M., Kassam N., Kamitakahara H. (2010) Safety of the long-term use of proton pump inhibitors. World J Gastroenterol 16: 2323–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trnka Y., LaMont J. (1981) Association of Clostridium difficile toxin with symptomatic relapse of chronic inflammatory bowel disease. Gastroenterology 80: 693–696 [PubMed] [Google Scholar]
- Tsironi E., Irving P., Feakins R., Rampton D. (2006) ‘Diversion’ colitis caused by Clostridium difficile infection: report of a case. Dis Colon Rectum 49: 1074–1077 [DOI] [PubMed] [Google Scholar]
- Tullus K., Aronsson B., Marcus S., Mollby R. (1989) Intestinal colonization with Clostridium difficile in infants up to 18 months of age. Eur J Clin Microbiol Infect Dis 8: 390–393 [DOI] [PubMed] [Google Scholar]
- Tvade M., Rask-Madsen J. (1989) Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet 1: 1156–1160 [DOI] [PubMed] [Google Scholar]
- Voth D., Ballard J. (2005) Clostridium difficile toxins: mechanism of action and role in disease. Clin Microbiol Rev 18: 247–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waligora A., Hennequin C., Mullany P., Bourlioux P., Collignon A., Karjalainen T. (2001) Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect Immun 69: 2144–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Ahrne S., Jeppsson B., Molin G. (2005) Comparison of bacterial diversity along the human intestinal tract by direct cloning and sequencing of 16S rRNA genes. FEMS Microbiol Ecol 54: 219–231 [DOI] [PubMed] [Google Scholar]
- Warny M., Keates A., Keates S., Castagliuolo I., Zacks J., Aboudola S., et al. (2000) p38 MAP kinase activation by Clostridium difficile toxin A mediates monocyte necrosis, IL-8 production, and enteritis. J Clin Invest 105: 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesselborg S., Bauer M., Vogt M., Schmitz M., Schulze-Osthoff K. (1998) Activation of transcription factor NF-kB and p38 mitogen-activated protein kinase is mediated by distinct and separate stress effector pathways. J Biol Chem 272: 12422–12429 [DOI] [PubMed] [Google Scholar]
- Wilson K. (1993) The microecology of Clostridium difficile. Clin Infect Dis 16: 214–218 [DOI] [PubMed] [Google Scholar]
- Wultańska D., Banaszkiewicz A., Radzikowski A., Obuch-Woszczatyński P., Młynarczyk G., Brazier J., et al. (2010) Clostridium difficile infection in Polish pediatric outpatients with inflammatory bowel disease. Eur J Clin Microbiol Infect Dis 29: 1265–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal E., Akkermans A., de Vos W. (1998) Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol 64: 3854–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal E., Rajilic-Stojanovic M., de Vos W. (2008) High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57: 1605–1615 [DOI] [PubMed] [Google Scholar]