Abstract
Prostate carcinoma (CaP) is a heterogeneous multifocal disease where gene expression and regulation are altered not only with disease progression but also between metastatic lesions. The androgen receptor (AR) regulates the growth of metastatic CaPs; however, sensitivity to androgen ablation is short lived, yielding to emergence of castrate-resistant CaP (CRCaP). CRCaP prostate cancers continue to express the AR, a pivotal prostate regulator, but it is not known whether the AR targets similar or different genes in different castrate-resistant cells. In this study, we investigated AR binding and AR-dependent transcription in two related castrate-resistant cell lines derived from androgen-dependent CWR22-relapsed tumors: CWR22Rv1 (Rv1) and CWR-R1 (R1). Expression microarray analysis revealed that R1 and Rv1 cells had significantly different gene expression profiles individually and in response to androgen. In contrast, AR chromatin immunoprecipitation (ChIP) combined with promoter DNA microarrays (ChIP-on-chip) studies showed that they have a similar AR-binding profile. Coupling of the microarray study with ChIP-on-chip analysis identified direct AR targets. The most prominent function of transcripts that were direct AR targets was transcriptional regulation, although only one transcriptional regulator, CCAAT/enhancer binding protein δ, was commonly regulated in both lines. Our results indicate that the AR regulates the expression of different transcripts in the two lines, and demonstrate the versatility of the AR-regulated gene expression program in prostate tumors.
Introduction
Multiple studies have demonstrated heterogeneity in prostate carcinoma (CaP), a multifocal disease where tissue architecture and genetic expression are altered not only with disease progression but also between metastatic lesions of patients with prostate cancer (Nwosu et al. 2001, Beheshti et al. 2002, Liu et al. 2004, Shah et al. 2004). Comparison of metastatic lesions from the same patient as well as different patients showed that metastatic hormone-refractory prostate cancer has a heterogeneous morphology, immunophenotype, and genotype, demonstrating that ‘metastatic disease’ is a group of diseases even within the same patient (Shah et al. 2004). As prostate cancer can progress from an organ-confined, localized state to metastasis, the problem in treating the disease is the identification of therapeutic targets that are common to all the foci within the same patient or in multiple patients. As most CaPs are initially present as androgen-dependent neoplasms, androgen ablation therapy (chemical castration) is an effective treatment, which initially blocks androgen receptor (AR) cell signaling in almost all patients. Although this therapy is initially successful, castration-resistant androgen-independent tumors that are refractory to hormonal therapeutic interventions emerge (Huggins & Hodges 1941, Gittes 1991). Androgen-independent CaPs continue to express the AR and androgen-regulated genes. Thus, a better understanding of the action of AR is a pivotal issue in defining the molecular events that lead to the progression of CaP. However, it is unclear whether requirement for AR function in various foci and metastatic lesions within the same patient or similar groups of patients is the same.
Various studies have indicated that the function of the AR depends on the biochemical environment in which it exists (Ruizeveld de Winter et al. 1994, Li et al. 2002). As a member of the nuclear receptor superfamily that functions as a ligand-dependent transcription factor, AR mediates androgen-regulated gene expression. Androgen-bound AR is stabilized and translocated into the nucleus to regulate the expression of target genes by binding to androgen response elements (AREs) or by interacting with other transcription factors bound to their specific recognition sites. The role of AR in CaP progression is to promote expression of specific target genes. For example, prostate-specific antigen (PSA), the best studied AR target gene, has been reported to contribute to CaP progression through its protease activity and its ability to induce epithelial–mesenchymal transition and cell migration (Borgono & Diamandis 2004, Whitbread et al. 2006). Other AR target genes implicated in CaP progression include FGF8, Cdk1 and Cdk2, PMEPA1, TMRPSS2, and amyloid precursor protein (Gregory et al. 1998, Lin et al. 1999, Gnanapragasam et al. 2002, Xu et al. 2003, Takayama et al. 2009). However, the function of the AR is tightly regulated by the expression of co-factors that are themselves regulated by various transcription factors, including the AR. Studies revealed differential expression of co-regulators with disease progression, which may have led to altered AR function (Li et al. 2002). Hence, an important point of investigation in prostate cancer research is to determine whether the AR functions similarly or differently in various metastatic lesions within the same patient.
Since the last decade, microarray techniques have been applied extensively in searching for genes that are AR regulated specifically in prostate tumors. Although gene expression profiling is a powerful technique for depicting the global function of the AR in a specified model, it does not distinguish whether alteration of gene expression is dependent on a direct or indirect action of AR. Moreover, despite the well-characterized AREs in the promoter and enhancer, little is known about AR cis-regulatory sites across the human genome. Chromatin immunoprecipitation (ChIP-on-chip) technology has been used for the identification of chromosomal-binding sites of transcription factors to identify novel targets (Cawley et al. 2004, Bernstein et al. 2005). Therefore, coupling microarray studies with ChIP-on-chip allows the identification of bona fide AR target genes. Wang et al. (2007) mapped the AR-binding sites on chromosomes 21 and 22 in androgen-dependent LNCaP human prostate cancer cells by combining ChIP with tiled oligonucleotide microarrays. Later, they followed up with comparisons between LNCaP versus a castrate-resistant CaP (CRCaP) variant of LNCaP cells (Wang et al. 2009), in an attempt to identify direct AR-dependent target genes in both androgen-dependent CaP as well as in CRCaP, and determined that the role of the AR in CRCaP is to execute a distinct program resulting in androgen-independent growth. Significantly, targets identified by one group in a CRCaP subline of LNCaP cells (Wang et al. 2009) were not identical to those in another LNCaP subline, C4-2B, shown in a different study (Jia et al. 2008), which mapped AR-occupied regions as well. A third study, which used PC3 cells transfected with wild-type AR (Lin et al. 2009), also identified distinct AR-occupied regions in target genes. The differences in these results may be attributed to differences in the technologies used to study AR-binding sites. An alternative explanation may be that because of the heterogeneity of gene expression in different CaP foci and metastatic lesions, the programs regulated by the AR in each deposit, even within the same patient, may be distinct.
The CWR22 androgen-dependent xenograft model, which mimics human prostate cancer, has been used to study the emergence of CRCaP (Wainstein et al. 1994). In male nude athymic mice, this xenograft exhibits androgen-dependent growth and secretes PSA. After androgen withdrawal, the tumors regress and PSA levels plummet. Importantly, the model simulates the clinical course of prostate cancer, in that PSA levels eventually increase and CRCaP tumors emerge (Nagabhushan et al. 1996). Similar to most CRCaP tumors, CWR22-recurring tumors continue to express the AR (Gregory et al. 1998), which contains a mutation (H847Y) in the ligand-binding domain (LBD) of the molecule (Tan et al. 1997). Since this model recapitulates salient features of human prostate tumors, it has been used extensively to study the emergence of CRCaP.
Two cell lines, R1 and Rv1 (van Bokhoven et al. 2003), were isolated in separate laboratories from CWR22-relapsed tumors. Several lines of evidence indicate that they were derived from a common ancestor. Karyotypes of the two cell lines are very similar; both lines shared the same structural abnormalities, including a reciprocal translocation between chromosomes 6 and 14 (van Bokhoven et al. 2003). Both lines have the same AR (H847Y) mutation that is present in the parental CWR22 cells (van Bokhoven et al. 2003). The Rv1 AR also contains a duplication of exon 3 (that encodes the DNA-binding domain), which results in an insertion of 39 additional amino acids (Tepper et al. 2002). The insertion mutation was present in the relapse tumor, and the subsequent cell line was established from the tumor, but very low levels of this mutated AR could also be detected by RT-PCR in the parent CWR22 tumor (Tepper et al. 2002). Additionally, we and others found that R1 and Rv1 express an ~80 kDa low molecular weight form of AR (LMW-AR) with a deletion of the C-terminal LBD (Gregory et al. 2001, Tepper et al. 2002). However, though the cell lines have significant similarities, they also exhibit differences. In a recent study, we showed that R1 and Rv1 cells were distinct in their AR expression, characterization, and function (Chen et al. 2010). The goals of the current study were to use these two cell lines to determine similarities and differences in AR-regulated programs in two related but distinct systems with a common lineage.
Materials and methods
Cell culture and pharmacological agents
Rv1 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). CWR-R1 cells were provided by Dr Elizabeth Wilson (University of North Carolina). Rv1 and R1 cells were propagated in RPMI 1640 supplemented with 5% fetal bovine serum, 2 mmol/l l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin (Invitrogen Life Science) at 37 °C and 5% CO2. For studies in androgen-depleted conditions, cells were propagated in phenol red-free RPMI 1640 supplemented with 5% charcoal-stripped fetal bovine serum, 2 mmol/l l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin at 37 °C and 5% CO2.
Western immunoblot analysis
Cells were directly placed in a radioimmunoprecipitation assay lysis buffer that contained the Sigma protease inhibitor cocktail (AEBSK, aprotinin, E64, leupeptin, and pepstatin as well as 1 µM calpeptin; Sigma–Aldrich). Thirty micrograms of protein were separated on 8, 10, or 12% SDS-PAGE gels and transferred to 0.22µM nitrocellulose-supported membrane (GE Healthcare, Piscataway, NJ, USA). The membrane was blocked with 5% nonfat dry milk in PBS and 0.1% Tween-20 before the addition of specific antibodies. The following antibodies were used: AR (central) 441 (Ab-1; Lab Vision Corp., Fremont, CA, USA), AR NH2-terminus N-20 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), calpain 2 (Sigma), calpastatin, ERK and phosphoERK (Cell Signaling, Danvers, MA, USA), and focal adhesion kinase (clone 4.47; Millipore, Billerica, MA, USA). Proteins were detected using chemiluminescence (GE Healthcare).
Microarray analysis
Labeling of samples, hybridization to U133A Gene-Chips (Affymetrix, Santa Clara, CA, USA), staining, and scanning were performed as described in the Affymetrix Expression Analysis Technical Manual. Fluorescence intensity values (.CEL files) generated from hybridized, stained GeneChips were analyzed with R statistical software (v.2.01, and ‘affy’ BioConductor package) and BRB Array Tools to identify genes that were differentially expressed. The settings used for Robust Multichip Analysis in R included Microarray Suite 5.0-based background correction, quantile normalization, and Robust Multichip Analysis-based algorithms for calculation of expression values using perfect match only fluorescence intensities. Detection at P≤ 0.05 and a mean fold change of ≥1.5-fold were used as criteria for filtering genes for clustering analyses. Hierarchical clustering and comparative fold change analysis were used to identify and group similar patterns of gene regulation. Assignment of genes to functional categories was done by annotation of gene lists with the program, Database for Annotation, visualization, and Integrated Discovery (http://apps1.niaid.nih.gov/david), and literature-based classification was done by hand. Statistically overrepresented (Fisher’s exact probability score <0.05) biological processes within clusters were identified using Expression Analysis Systematic Explorer v.1.0 analysis software (Hosack et al. 2003).
Quantitative real-time PCR
Total cellular RNA was prepared from Rv1 cells using RNeasy mini kit (Qiagen, Inc.) based on the manufacturer’s protocol. cDNA was synthesized from 1 µg RNA using QuantiTect (Qiagen) reverse transcription kit based on the manufacturer’s protocol. cDNAs were diluted 1:4 in ddH2O, and 2 µl diluted cDNA was added to 5 µl of EXPRESS SYBR GreenERTM qPCR supermix (Invitrogen Life Science) and 200 nM of each primer. GAPDH, HPRT, or RPL13A was used as the endogenous expression standards. PCR conditions were initial denaturation step at 95 °C for 20 s, 40 cycles at 95 °C for 3 s, 60 °C for 30 s, followed by additional 95 °C for 15 s and 60–95 °C over 20 min ramp for melt curve analysis. Primer sequences used in the study are provided in Supplementary Methods (see section on supplementary data at the end of this article). Data were collected by the Mastercycler ep Realplex (Eppendorf AG, Hamburg, Germany). Primer sequences are available upon request.
Ingenuity pathway analysis
The microarray expression data were uploaded into ingenuity pathway analysis (IPA) software using Reference sequence (RefSeq). A total of 2322 genes were mapped using the IPA database. Fold change of 1.5 and P value of ≤0.05 were applied as the cutoff criteria. Gene networks were algorithmically generated based on their connectivity and were assigned a score. A score of 3 or higher indicates a 99.9% confidence level that the network was not generated by chance alone. Canonical pathway analysis identifies the pathways from the IPA library of canonical pathways, which are most significant to the input dataset. The significance of the association between the dataset and the canonical pathway is determined based on two parameters: 1) a ratio of the number of genes from the dataset, which map to the pathway, divided by the total number of genes that map to the canonical pathway and 2) a P value calculated using Fischer’s exact test determining the probability that the association between the genes in the dataset and the canonical pathway is due to chance alone.
ChIP-on-chip analysis
Tiling array analysis was performed with GeneChip Human Promoter 1.0R Arrays (Affymetrix) in order to determine genome-wide analysis of AR recruitment sites. Briefly, AR-associated DNA was enriched by ChIP as described earlier (Louie et al. 2003, Desai et al. 2006). ChIPs using a pre-immune IgG were used as controls. ChIP DNA (10 µl) and input (10 ng) samples were amplified using the GenomePlex Complete Whole Genome Amplification (WGA) kit (Sigma–Aldrich) with a modification to the manufacturer’s protocol to generate product suitable for Affymetrix microarray analysis by including dUTP (80 µM final concentration) in the amplification and re-amplification (if necessary) reactions. WGA products were purified with QIAquick PCR purification kit, eluted in nuclease-free water (Invitrogen), and quantitated with a NanoDrop 2000c spectrophotometer (Thermo Scientific). Target preparation and tiling array-processing procedures were performed according to Affymetrix’s standard protocols. Briefly, 7.5 µg DNA was fragmented through the combined actions of uracil DNA glycosylase and human apurinic endonuclease and then end labeled with biotin using terminal deoxynucleotidyl transferase. Labeled target DNA was hybridized to the arrays at 45 °C for 16 h. Subsequently, the arrays were washed and stained using the Fluidics Station 450 (Affymetrics, Santa Clara, CA, USA) according to the manufacturer’s protocol and then scanned with the GeneChip Scanner 3000 7G. Data analysis was performed with CisGenome software (Ji et al. 2008). AR-binding regions (i.e. ChIP-enriched) were identified by comparing with the nonspecific IgG control using the TileMap peak detection tool (Ji & Wong 2005) with the application of a hidden Markov model. Subsequently, genomic locations of peaks and bound probes were visualized in the CisGenome and UC Santa Cruz genome browsers.
Results
Comparison of the gene expression profiles of R1 and Rv1 cells
We have described earlier the characteristics of R1 and Rv1 cells derived from two different relapsed tumors although both from the same parental CWR22 xenograft (Chen et al. 2010). To further define the differences and similarities between the two CWR22 relapsed lines, we used the Affymatix HG-U133 Plus2.0 Gene Chip microarray to identify differences in gene transcription. The analysis was conducted in duplicate in R1 and Rv1 cells cultured in identical conditions, at the same density in charcoal-stripped serum or 2 h after the addition of 10 nM DHT. The 2 h time point was chosen to identify transcripts that are more likely to be direct AR targets, and other laboratories had previously determined this concentration of DHT to be optimal for AR stimulation (Wang et al. 2007).
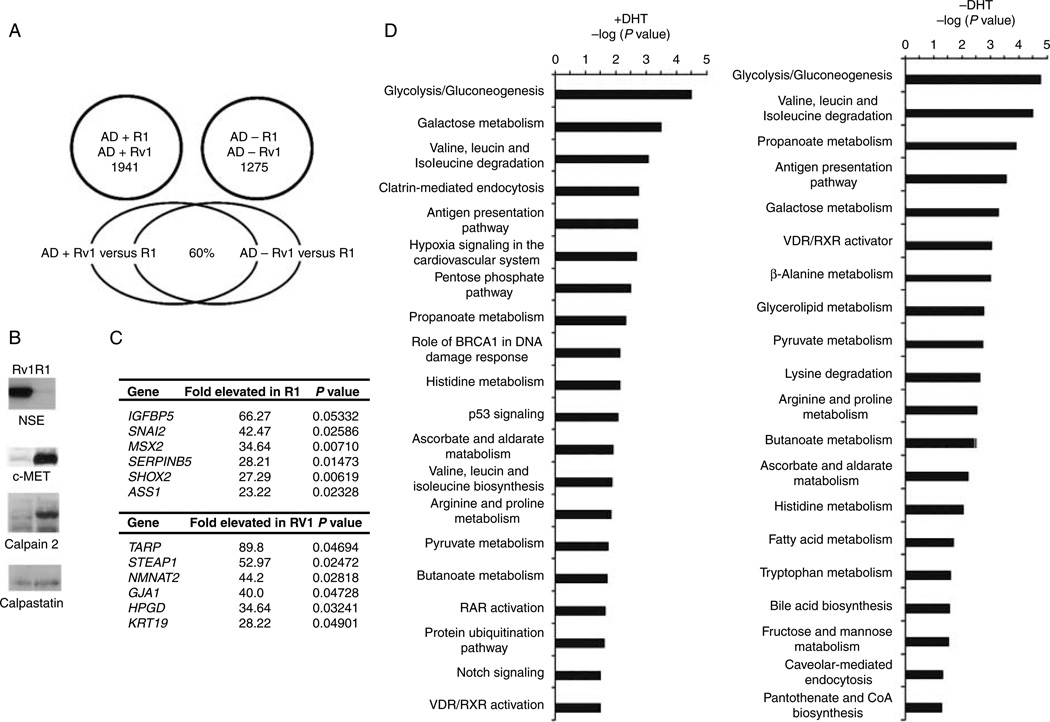
Comparison of R1 and Rv1 gene expression profiles in castrate levels of androgen identified 1275 genes that were differentially expressed (fold change ≥1.5 or ≤−1.5; P≤0.05) in R1 versus Rv1 cells in the absence of androgens and 1941 transcripts that were differentially expressed (fold change ≥1.5 or ≤−1.5; P≤0.05) in R1 versus Rv1 cells treated for 2 h with 10 nM DHT (Fig. 1A; Supplementary Table 1, see section on supplementary data given at the end of this article). Significantly, only 60% of genes differentially expressed in R1 versus Rv1 in the presence of DHT were identical to the transcripts that were differentially expressed in the absence of androgen (Fig. 1A; Supplementary Table 1). These results indicated that the R1 and Rv1 cells were genetically distinct and could serve as models for comparison of two different metastasized CaP lesions derived from the same patient.
Figure 1.
Differences in gene expression of R1 and Rv1 cells in the presence and absence of a DHT. (A) Venn diagram of the number of genes differentially expressed in R1 and Rv1 cells in castrate levels of androgen and after a 2 h treatment with 10 nM DHT. The lower venn diagram shows the cohort of genes differentially expressed in the absence and presence of androgen. (B) Western blot analysis verification of several differentially expressed proteins that were identified by the expression array study. (C) The most differentially expressed transcripts in R1 and Rv1 cells treated with 10 nM DHT. (D) The IPA was used to identify the pathways that differed in the two cell lines in the presence and absence of androgen. The Fisher’s exact test was used to determine the probability that the association between the dataset and a given pathway is due to chance alone. The most significant pathway differences in the presence and absence of androgen involved metabolic pathways.
We verified the specificity and selectivity of the results obtained with the microarray analysis by comparing these results to known differences between the two lines. We had shown earlier that R1 cells expressed increased levels of calpain 2 mRNA compared to Rv1 cells, whereas the levels of the calpain inhibitor calpastatin were similar in both lines (Chen et al. 2010). A similar pattern was seen by the current gene expression studies, thereby authenticating the results (Supplementary Table 1; Fig. 1B). R1 cells also expressed 11.7-fold higher levels of c-MET mRNA (Supplementary Table 1; Fig. 1B). Rv1 cells have more neuroendocrine characteristics than R1 cells because of a greater expression of neuronal-specific enolase (ENO2; 12-fold change; P = 0.02; Supplementary Table 1; Fig. 1B), chromogranin A and B (2.74- and 8.69-fold increase respectively), and synaptophysin (3.34-fold increase; Supplementary Table 1). ENO2 expression was not altered by androgen (data not shown). These results also show the accuracy of the gene expression analysis system in these studies.
Based on the gene expression analysis, the most differentially expressed genes between R1 and Rv1 (expression in the absence of androgen) include TARP, IGFBP5, STEAP1, NMNAT2, and SNAI2 (listed in Fig. 1C). To identify patterns in differential gene expressions, IPA was used to identify the pathways that differed in the two cell lines. The Fisher’s exact test was used to determine the probability that the association between the dataset and a given pathway is due to chance alone. The most significant pathway differences between R1 and Rv1 cells both in the presence and absence of androgen involved metabolic pathways (Fig. 1D). In summary, the gene expression profiles of R1 and Rv1 indicate that although these two lines were derived from the same CWR22 xenograft and have similar morphologies, at the molecular level, they are distinct.
Analysis of genes differentially regulated in R1 versus Rv1 cell lines in response to androgen treatment
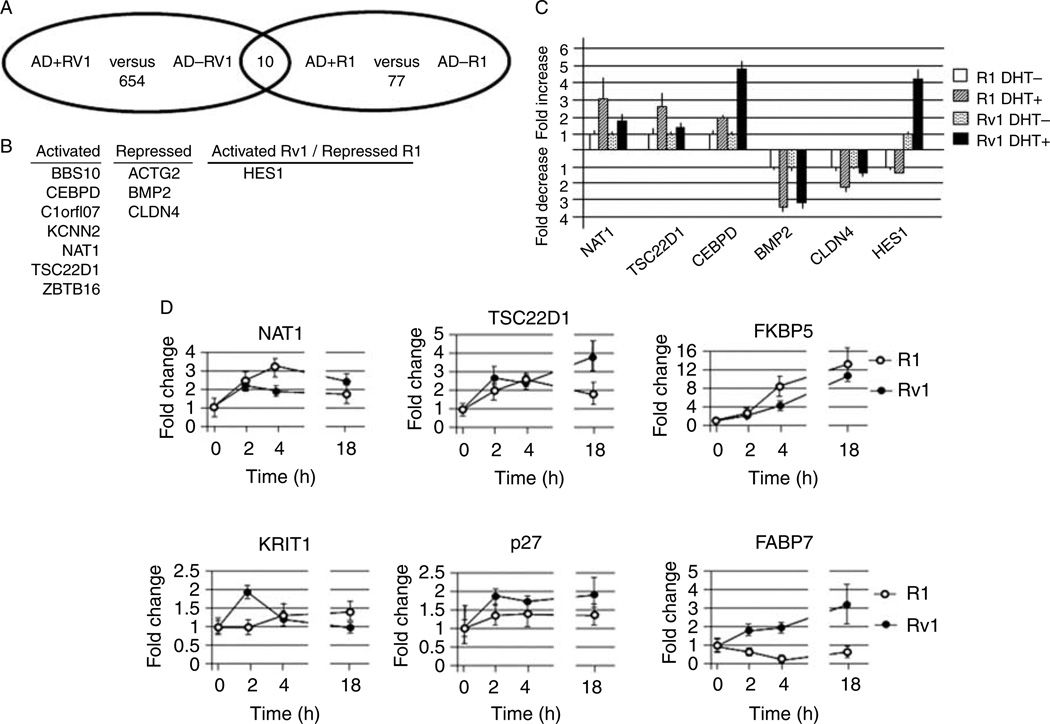
Since the gene expression profile of R1 versus Rv1 cells was vastly different, we investigated whether these genes behaved similarly in response to DHT treatment. Using the same gene expression data that were used in Fig. 1, we analyzed genes that were differentially regulated in the two cell lines in response to a 2 h androgen treatment. Using a cutoff value of fold change ≥1.5 or ≤−1.5 and P≤0.05, we found that in Rv1 cells, the expression of 854 transcripts was altered by a 2 h DHT treatment, whereas in R1 cells, the expression of only 77 transcripts changed after the addition of DHT for 2 h (Fig. 2A; Supplementary Table 2, see section on supplementary data given at the end of this article). Therefore, the transcriptional response to DHT was greater in Rv1 cells than in R1 cells. A comparison of the DHT-responsive R1 and Rv1 transcripts identified only ten genes that were commonly regulated in both cell lines (Fig. 2B), again indicating the large differences between these two lines. This included seven genes that were upregulated by DHT in both R1 and Rv1 cells, including CEBPD and N-acetyltransferase type I (NAT1), and three that genes were repressed in both cells, including CLDN4. Interestingly, the expression of HES1, a component of the Notch signaling pathway (Fischer & Gessler 2007), was DHT regulated in both cell lines, but expression was repressed in R1 cells and activated in Rv1 cells (Fig. 2B). Androgen-dependent regulation of six of these transcripts was validated by real-time PCR, which verified the accuracy of these results (compare Fig. 2C with Supplementary Table 2). The expression of two well-known androgen-responsive genes KLK3 (PSA) and TMPRSS2 was not significantly altered by DHT in either cell line, thus confirming previous reports that the transcripts are not androgen regulated in these cell lines (Riegman et al. 1991, Lin et al. 1999, Tomlins et al. 2005).
Figure 2.
Differences in AR-dependent gene expression of R1 and Rv1 cells. (A) Venn diagram of AR-regulated transcripts in R1 and Rv1 cells. (B) Transcripts that are commonly regulated in the two cell lines. Note that although HES1 is androgen regulated in both cell lines, HES1 expression is elevated in Rv1 cells, but repressed in R1 cells. (C) Real-time PCR verification of several AR-regulated transcripts. (D) Time course of DHT-inducible gene expression in R1 and Rv1 cells.
It is possible that the discrepancy in the number of DHT-regulated transcripts in R1 and Rv1 cells is due to a delay in the DHT response in R1 cells. To address this possibility, we looked at the expression of transcripts that were DHT regulated in R1 and Rv1 cells (NAT1 and TSC22D1), only in R1 cells (FKBP5), or only in Rv1 cells (KRIT1, p27, and FABP7) at 0, 2, 4, and 18 h after DHT addition (Fig. 2D). The time course for transcripts that were regulated in both lines was similar but not identical in the two cell lines. Interestingly, the induction of FKBP5 was more robust in R1 cells than in Rv1 cells; therefore, in the array, study expression in Rv1 cells 2 h after DHT addition was below our cutoff value. In concordance with the array analysis, KRIT1 and p27 were not DHT transactivated in R1 cells. The expression of FABP7 was elevated in Rv1 cells in a time-dependent manner, but in R1 cells, the expression was repressed in a time-dependent manner. This analysis argues that the smaller number of DHT-regulated transcripts in R1 cells is not due to a general delay in response to hormone stimulation.
DHT-regulated pathways in R1 and Rv1 cells
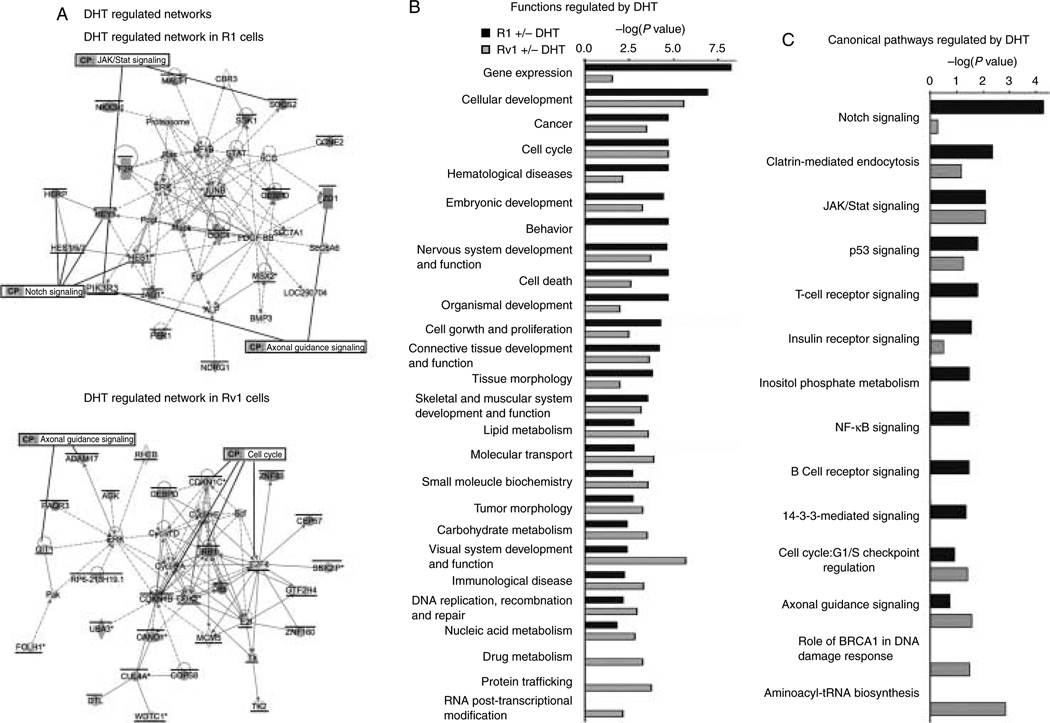
Since the genes regulated by androgens in the two cell lines are different, we asked whether the pathways they regulated were also different, or whether androgen was regulating the same programs in both cells but through different mechanisms. The differentially expressed genes in response to DHT for 2 h were analyzed by IPA to identify most significantly associated biological networks and canonical pathways (metabolic and cell signaling) altered in the two cell lines. The Fisher’s exact test was used to determine the probability that the association between the dataset and a given pathway is due to chance alone. IPA identified two significant biological networks associated with the differentially expressed genes in R1 cells (the major one is shown in Fig. 3A, and the other one is shown in Supplementary Figure 1, see section on supplementary data given at the end of this article), whereas in Rv1 cells, a total of 18 biological networks were identified, which are significantly associated with the differentially expressed genes (the major network is shown in Fig. 3A, and the others are shown in Supplementary Figure 2, see section on supplementary data given at the end of this article). In R1 cells, the significantly associated functions affected by DHT treatment include gene expression, cellular development, cell cycle, and embryonic development (Fig. 3B). The canonical pathways most significantly associated with DHT treatment are notch signaling, clathrin-mediated endocytosis, JAK/Stat signaling, and p53 signaling (Fig. 3C). The significantly associated functions in Rv1 cells include cellular development, visual system development and function, cancer, cell cycle, molecular transport, and protein trafficking (Fig. 3B), whereas the most associated canonical pathways include aminoacyl-tRNA biosynthesis, axonal guidance signaling, DNA damage response, cell cycle, p53 signaling, and clatrin-mediated endocytosis (Fig. 3C). These results indicate a greater biological role of AR in Rv1 cells compared to R1 cells. However, a number of cellular functions and canonical pathways regulated by DHT treatment in the two cells lines are similar, suggesting that in the two cell lines, the AR plays a similar role, but employs different mechanisms.
Figure 3.
Comparison of biological networks, pathways, and function of R1 and Rv1 DHT-regulated transcripts. (A) The most prominent DHT-regulated network in R1 and Rv1 cells. A bar above the gene denotes transcripts that are DHT transactivated, and an underscore denotes transcripts that are DHT repressed. Several components of the Notch signaling pathway are DHT regulated in R1 cells, whereas components of cell cycle are DHT regulated in Rv1 cells. (B) The most common functions of transcripts regulated by DHT in R1 and Rv1 cells. (C) The most common DHT-regulated canonical pathways in R1 and Rv1 cells.
The activity of the AR is affected by multiple co-regulators that serve as co-activators or co-repressors of AR-dependent transcription (Devlin & Mudryj 2009, Heemers et al. 2009). Since the differences in DHT-inducible gene expression could be due to the expression of a different cohort of AR co-regulators, we compared the expression of these proteins in the two cell lines in the presence and absence of androgen (Table 1). The number of co-regulators that were expressed at higher levels was greater in R1 than in Rv1 cells. R1 cells had higher levels of 22 co-activators, whereas Rv1 cells had higher levels of 13 co-activators. R1 cells had higher levels of eight co-repressors, and Rv1 cells had higher levels of seven co-repressors. However, it is notable that there were differences in co-regulator levels in the presence or absence of androgen. Most of these differences were due to the DHT-dependent regulation of co-regulator expression in Rv1 cells. The only AR co-regulator that was differentially expressed after DHT addition in R1 cells was HEY1.
Table 1.
Androgen receptor (AR) co-regulators differentially expressed in R1 and Rv1 cells
| Elevated in R1 | Elevated in Rv1 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Symbol | Act./Rep. | −DHT | +DHT | DHT reg. | Symbol | Act./Rep. | −DHT | +DHT | DHT reg. |
| BRCA2 | Act. | ✓ | Repressed in Rv1* | ARID1A | Act. | ✓ | ✓ | ||
| CARM1 | Act. | ✓ | Induced in Rv1* | CREBBP | Act. | ✓ | Induced in Rv1* | ||
| CDK7 | Act. | ✓ | ✓ | DDC | Act. | ✓ | ✓ | ||
| COPS5 | Act. | ✓ | Induced in Rv1 | HMGB2 | Act. | ✓ | ✓ | ||
| FHL2 | Act. | ✓ | Repressed in Rv1 | HTATIP2 | Act. | ✓ | ✓ | ||
| GSN | Act. | ✓ | ✓ | IDE | Act. | ✓ | ✓ | ||
| HIPK3 | Act. | ✓ | Repressed in Rv1 | IQWD1 | Act. | ✓ | Induced in Rv1 | ||
| HRMT1L2 | Act. | ✓ | PLAGL1 | Act. | ✓ | Induced in Rv1 | |||
| HTATIP | Act. | ✓ | Repressed in Rv1 | PRKDC | Act. | ✓ | Induced in Rv1 | ||
| NCOA1 | Act. | ✓ | ✓ | RB1 | Act. | ✓ | ✓ | Induced in Rv1 | |
| NCOA2 | Act. | ✓ | ✓ | Repressed in Rv1 | RNF14 | Act. | ✓ | Induced in Rv1 | |
| NONO | Act. | ✓ | ✓ | TMF1 | Act. | ✓ | Induced in Rv1 | ||
| PIAS2 | Act. | ✓ | Repressed in Rv1 | TRIM24 | Act. | ✓ | Induced in Rv1 | ||
| PNRC1 | Act. | ✓ | ✓ | APPL | Rep. | ✓ | ✓ | ||
| PXN | Act. | ✓ | ✓ | Repressed in Rv1 | DNAJA1 | Rep. | ✓ | Induced in Rv1 | |
| RAN | Act. | ✓ | ✓ | HDAC1 | Rep. | ✓ | Induced in Rv1 | ||
| SMARCA2 | Act. | ✓ | ✓ | NCOR1 | Rep. | ✓ | Induced in Rv1 | ||
| SMARCC1 | Act. | ✓ | Repressed in Rv1 | PA2G4 | Rep. | ✓ | Induced in Rv1 | ||
| STAT3 | Act. | ✓ | PAK6 | Rep. | ✓ | ✓ | |||
| TSC2 | Act. | ✓* | ✓ | ZNF278/PATZ1 | Rep. | ✓ | ✓ | ||
| ZMIZ1 | Act. | ✓ | ✓* | ||||||
| AES | Rep. | ✓ | ✓ | ||||||
| APPBP2 | Rep. | ✓ | Repressed in Rv1 | ||||||
| CALR | Rep. | ✓ | Repressed in Rv1 | ||||||
| FLNA | Rep. | ✓ | ✓ | ||||||
| HEY1 | Rep. | ✓ | ✓ | Induced in R1 | |||||
| PTEN | Rep. | ✓ | ✓ | ||||||
| TGIF | Rep. | ✓ | ✓ | ||||||
| TLE1 | Rep. | ✓ | ✓ | ||||||
| FKBP5 | Act. | Elevated in R1 in the presence of DHT, elevated in Rv1 in absence of DHT; DHT induced in R1 cells |
|||||||
Act., activator; Rep., repressor; −DHT, castrate levels of androgen; +DHT, following addition of 10 nM DHT for 2 h; DHT reg., DHT regulated.
indicates that the P value is slightly above 0.1 (up to o.12).
✓ indicates overexpression.
AR chromosomal-binding sites in R1 and Rv1 cells in response to DHT
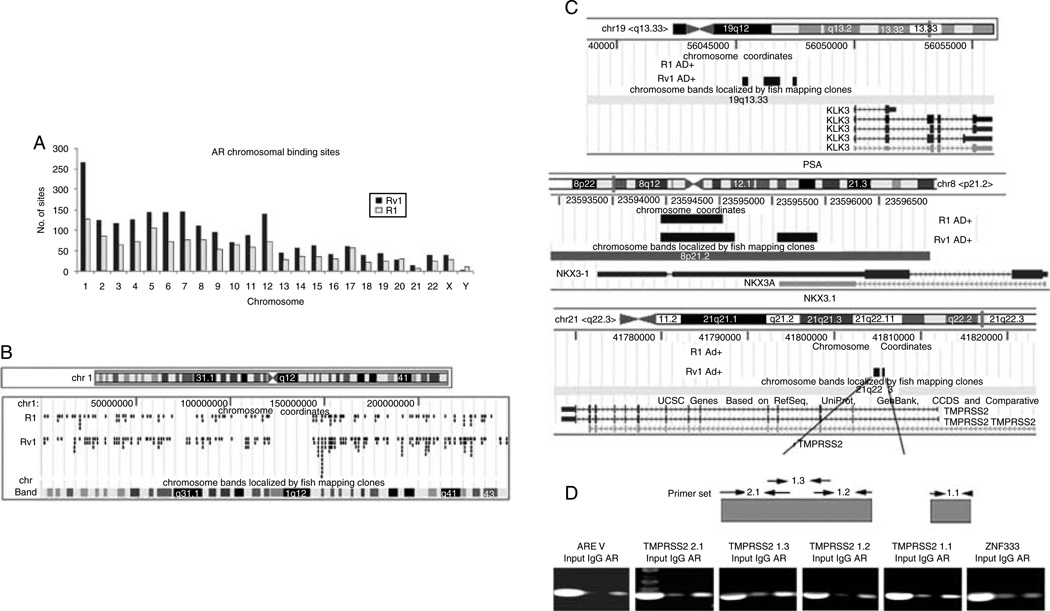
Next, we asked why the cohort of androgen-regulated transcripts differed in R1 and Rv1 cells. Hence, we determined whether AR binding to regulatory regions differed significantly between the two cell lines. The Human Promoter 1.0R Array (Affymetrix) oligonucleotide (25-mer)-based, high-density tiling array that covers 25 500 promoters with probe sets spanning at least 10 kb of genomic content per gene (~7.5 kb upstream and ~2.45 kb downstream of the transcriptional start site (TSS)) and at a resolution of 35 bp was used to identify AR-binding sites in the entire genome in both cell lines. A total of 1225 and 2021 AR-binding sites (FDR≤0.05) were identified in R1 and Rv1 cells respectively when treated with DHT for 2 h. Figure 4A shows the distribution of the binding sites along chromosomes in two cell lines. A comparison of AR binding across chromosomes in R1 and Rv1 cells treated with DHT showed that AR-binding pattern was similar, but not identical (Fig. 4B). Certain sites were AR bound only in Rv1 cells, whereas others were AR bound only in R1 cells. This analysis indicated that AR binding after the addition of DHT was more extensive in Rv1 than in R1 cells, and most of the R1 AR-bound sites were also AR bound in Rv1 cells. Therefore, although the androgen-regulated gene expression profile of the two cell lines is different, the AR-binding pattern is similar.
Figure 4.
Distribution of AR-binding sites in R1 and Rv1 cells. (A) The number of AR-binding sites detected on individual chromosomes after a 2 h DHT treatment is lower in R1 than in Rv1 cells. (B) More detailed mapping of AR binding on chromosome 1 in R1 and Rv1 cells. Few AR-binding sites are unique in R1 cells. (C) Precise location of AR binding to PSA, NKX3.1, and TMPRSS2 genes in R1 and Rv1 cells. AR bound to common sequences of the NKX3.1 gene, but AR binding to the PSA and TMPRSS2 genes was detected only in Rv1 cells. (D) ChIP analysis of AR binding to sites in the TMPRSS2 gene in Rv1 cells. The upper panel notes the location of the promoter sequences. ARE V contains an AR-binding site ~14 kb upstream of the TMPRSS2 TSS. Sequences in the ZNF333 promoter served as a negative control.
To validate our results, we focused on the binding pattern for three well-known androgen-responsive genes KLK3 (PSA; Riegman et al. 1991), NKX3.1 (He et al. 1997), and TMPRSS2 (Tomlins et al. 2005) in R1 and Rv1 cells. Gene expression studies had shown that neither KLK3 nor TMPRSS2 was androgen regulated in either cell line; however, both genes bound AR. In Rv1 cells, sequences near the KLK3 (PSA) gene bound AR (−4603, −3484, and −2499 upstream of its TSS), whereas there was no AR binding near or in the KLK3 gene in R1 cells (Fig. 4C). AR binding to NKX3.1 chromosomal region was identified in both R1 and Rv1 cells. In R1 cells, AR bound in the 3′-UTR (2149 downstream of TSS) of the NKX3.1 gene, whereas in Rv1 cells, AR bound not only in the 3′-UTR (2059 downstream of TSS) but also in the intron (1164 downstream of TSS) of NKX3.1 (Fig. 4C). This result is consistent with a recent study that identified androgen-responsive elements in the 3′-UTR of the NKX3.1 gene (Thomas et al. 2010).
AR binding in the 5′-UTR (two sites: 6382 and 7179 downstream of TSS) of the androgen-regulated gene TMPRSS2 was detected in Rv1 cells, but no binding near or in the TMPRSS2 gene in R1 cells (Fig. 4B). Previous studies conducted in LNCaP cells detected AR binding to sequences ~14 kb upstream of the TMPRSS2 TSS, but this sequence was not present in our promoter array. Hence, even if the AR bound to this section of TMPRSS2 gene in R1 or Rv1 cells, we would not detect AR binding. Therefore, we further analyzed AR binding to the four sites identified in our study using ChIP analysis (Fig. 4D). After the addition of 10 nM DHT for 2 h in Rv1 and LNCaP, AR binding was detected in Rv1 cells, but not in LNCaP cells, further confirming our results.
Motif analysis of AR-binding sites
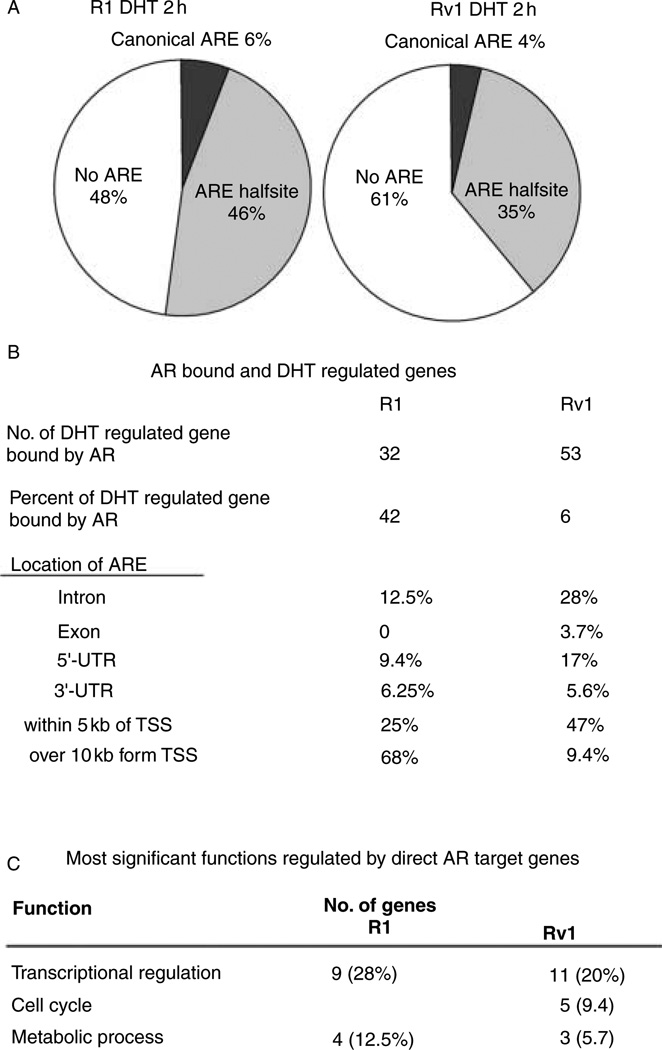
A motif analysis of the AR-binding sites was conducted to determine whether AR binds to the established consensus ARE in these target genes. Previous studies conducted in LNCaP, LNCaP-derived cells, or AR-transfected PC3 cells (Wang et al. 2007, Jia et al. 2008, Lin et al. 2009) reported that only 10% or less of the AR-binding regions had a canonical class 1 ARE (AGAACAnnnTGTTCT)-binding motif when two positions were allowed to vary from the palindromic consensus with three nucleotides spacing. Previous studies also found that between 7.8 and 8.4% of the binding regions contained the AR-binding half-site motif (AGAACA). In this study, we found that in Rv1 cells, only 4% (86/2021) of the sites had the canonical ARE and 35% (700/2021) had the AR half-site motif. Likewise, in R1 cells, 6% (76/1225) of the sites had the canonical ARE and 46% (568/1225) had the AR half-site motif (Fig. 5A). These studies indicate that the canonical ARE is not required for AR binding in the majority of the genes examined, and that the half-site is sufficient for AR function.
Figure 5.
Characteristics of AR-binding sites and direct AR transcriptional target genes. (A) The half ARE is present in many AR-binding sites, whereas the canonical ARE is not. (B) AR binding was more prevalent in intronic sequences that are present in the 5′-UTR. (C) The most significant function of transcripts that are near an AR-binding site and are androgen regulated in R1 and Rv1 is transcriptional regulation.
The expression profile of genes closest to the AR-binding sites in R1 and Rv1 cells in response to DHT
Next, we investigated whether the AR directly regulated the same cohort of genes in the two cell lines. The AR-binding sites identified in R1 and Rv1 cells were closest to 965 and 1518 genes respectively (data not shown). Notably, although some closest genes only contained one AR-binding site, many others had more than one AR-binding sites.
By combining the ChIP-on-chip with microarray expression data, we identified that, of the 854 differentially regulated genes in Rv1 cells in response to DHT for 2 h (Fig. 2A), AR bound to nearby chromosomal sites (FDR≤0.05) of only 53 genes (6%). The location of the AR-binding sites includes intron (15 genes), exon (2 genes), 5′-UTR (9 genes), 3′-UTR (3 genes), and within 5 kb upstream from the TSS (25 genes) (Fig. 5B; Supplementary Table 3A, see section on supplementary data given at the end of this article). Additionally, two genes had AR-binding sites that were more than 10 kb upstream of TSS, whereas three genes had AR-binding sites that were more than 10 kb downstream of the transcriptional end site (TES). The same analysis was performed in R1 cells. Of the 77 differentially regulated genes after adding DHT for 2 h, AR bound to the nearby chromosomal regions (FDR≤0.05) of 32 genes (42%). The identified AR-binding sites include intron (4 genes), 5′-UTR (3 genes), 3′-UTR (2 genes), within 5 kb upstream of the TSS (8 genes), more than 10 kb upstream of the TSS (14 genes), and more than 10 kb downstream of the TES (8 genes) (Fig. 5B; Supplementary Table 3). The AR-binding site that is far upstream or downstream of the androgen-responsive genes resides within or downstream of other annotated genes (Supplementary Table 3). However, the nearest genes are not androgen regulated. The ability of the AR to bind to sequences within one gene, but regulate transcription of a more distant gene has been reported earlier (Wang et al. 2009). Taken together, our results indicate that a much higher number of genes are androgen regulated in Rv1 cells compared to R1 cells. However, of the androgen-regulated genes in RV1 cells, only a few are regulated directly by AR, whereas in R1 cells, almost 50% of the genes that are androgen regulated resulted from direct AR transcriptional activity.
IPA analysis showed that the biological functions most prominently associated with these 53 genes in RV1 cells were transcriptional regulation, cell cycle, and metabolic process (Fig. 5C), whereas the major biological functions associated with the 32 genes identified in R1 cells are transcriptional regulation and metabolic process (Fig. 5C); hence, transcriptional regulation and metabolic process are biological functions, which are AR regulated in both cell lines, whereas cell cycle regulation is apparent in Rv1 cells. Therefore, these results support our earlier assertion that the AR regulated multiple common pathways in R1 and Rv1 cells, but that the AR has additional roles in Rv1 cells. The AR effect on gene expression in Rv1 cells is more extensive; however, the effect of the AR on majority of androgen-regulated genes in R1 is through direct transcriptional activity, whereas the effect of the AR on the majority of androgen-regulated genes in Rv1 cells is indirect.
Identification of common genes that are androgen regulated by direct AR transcriptional activity in both R1 and Rv1 cells
A comparison of R1 and Rv1 cells revealed that the majority of the AR-bound sites near the differentially regulated genes were common. However, only two closest genes (CCAAT/enhancer binding protein δ (CEBPD) and arylamine NAT1) adjacent to the common AR-binding sites in both R1 and Rv1 cells showed correlated transcriptional regulation (fold change 1.5 and P<0.05) in both lines. Claudin 4 (CLDN4) was androgen regulated in both cell lines, and in both lines, AR binding was detected, but the AR sites were distinct. This argues that only a subset of AR chromosomal-binding sites exhibit transcriptional regulation. Of these three common AR direct targets, CEBPD and NAT1 have been reported to be androgen-responsive genes (Yang et al. 2001, Butcher et al. 2007), and CLDN4 has been reported to be deregulated in both primary and metastatic prostate cancer (Landers et al. 2008).
Previous studies from other laboratories showed that other transcription factors collaborate with the AR to induce gene regulation (Wang et al. 2007, 2009, Jia et al. 2008, Lin et al. 2009). Considering that other transcription factors might play a collaborative role in AR function, we used a transcription element search system (TESS) to screen for motifs that most frequently co-existing with AR-binding motifs present in the above differentially regulated genes. TESS identifies transcription factor motifs using site or consensus strings and positional weight matrices from the TRANSFAC, JASPAR, IMD, and the CBIL-GibbsMat database (www.cbil.upenn.edu/tess). Using these databases, we found that the transcription factor motifs that most frequently co-exist with AR-binding motifs included GRE, GATA-binding protein (GATA), Sp-1, and forkhead box J2 (FoxJ2) in both R1 and Rv1 cells (not shown). Significantly, previous studies identified GATA- and FoxA1-binding site near AR-binding site (Wang et al. 2007, Lin et al. 2009).
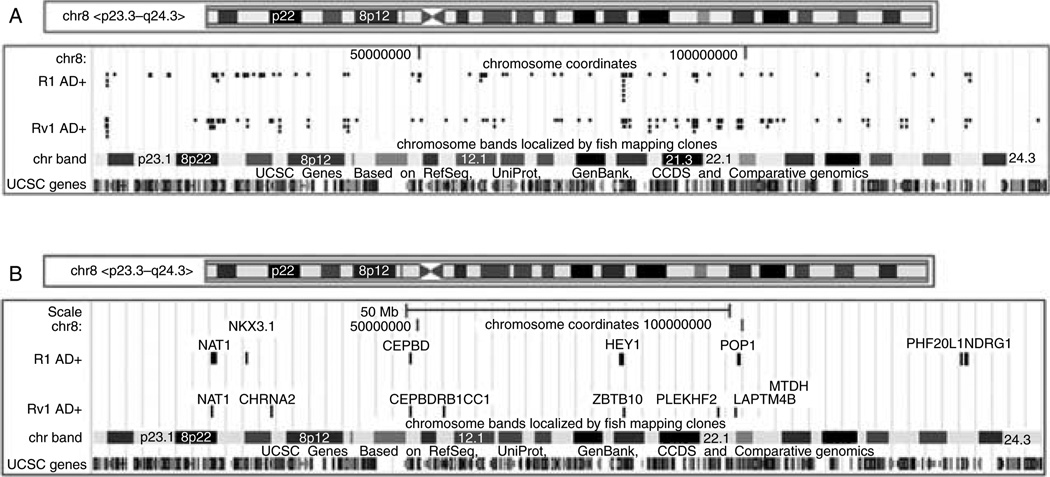
Our analysis of direct AR target genes in R1 cells revealed that ~25% of genes (seven genes – NAT1, NKX3.1, CEBPD, HEY, POP1, PHF20L1, and NDRG1) mapped to chromosome 8 (Fig. 6A), and all were positively regulated by androgen. Hey is one of the primary targets of the Delta-Notch signaling pathway and functions primarily as transcriptional repressors (Fischer & Gessler 2007). Furthermore, one of the AR sites has a single half ARE (POP1), whereas all of the other sites have two or three half ARE sites separated by between 40 and several thousand nucleotides. In Rv1 cells, 15% of the genes (eight genes – NAT1, CHRNA2, CEBPD, RB1CC1, ZBTB10, PLEKHF2,LAPTM4B, andMTDH) mapped to chromosome 8. Seven were positively regulated by androgen, whereas one was repressed (CHRNA2). Five of the eight AR-binding sites (NAT1, CHRNA2, CEBPD, RB1CC1, and PLEKHF2) contained at least one ARE half-site. Two of the genes were commonly regulated in both cell lines – NAT1 and CEBDP, whereas the others were not. The high percentage of direct AR target genes on one chromosome indicates that chromosome 8 is exceptionally rich in AR-regulated genes.
Figure 6.
AR-binding pattern on chromosome 8 in R1 and Rv1 cells. (A) AR-binding sites in R1 and Rv1 cells detected after DHT addition. (B) AR sites associated with transcripts that are AR regulated in R1 and Rv1 cells. Most androgen-responsive genes are transactivated, since only the expression of CHRNA2 in Rv1 cells is repressed.
Discussion
Outwardly, the morphology and phenotypical characteristics of the R1 and Rv1 cells appear very similar. Both are derived from CWR22-relapsed tumors, are not responsive to the anti-androgen Casodex, and express a LMW-AR that is ligand independent. Both cell lines have the same chromosomal translocations and harbor the same AR and p53 mutations (van Bokhoven et al. 2003), indicating they were derived from a common progenitor. However, the extensive difference in gene expression of R1 and Rv1 cells strongly argues that although they are derived from a common progenitor, at the molecular levels, they are very different. This progenitor cell must have had enough plasticity to give rise to cells with distinct molecular phenotypes. We hypothesize that the initial parent xenograft is composed of several distinct cell types and that the CRCaP cells constitute a small component of morphologically indistinct cells that expanded after the selective pressure of androgen ablation. The current study indicated that even tumors that appear to be homogenous may be composed of several cell types, thus further complicating gene expression analysis.
Most CRCaP tumors and cells continue to express the AR, and the AR continues to regulate gene expression. It was, however, unclear whether in these cells the AR is regulating a distinct set of genes. Previous studies have characterized the gene expression profile of LNCaP-derived CRCaP cells. One study found that the expression of several genes associated with mitosis was AR regulated in the LNCaP-abl cells (Wang et al. 2009), whereas an analysis of C4-2B cells identified a different cohort of DHT-regulated transcripts (Jia et al. 2008). These studies revealed that AR gene regulation patterns change with disease progression; however, the results from the two studies revealed significant differences in AR-mediated gene regulation in the two LNCaP-derived cell lines. This difference could have arisen because of differences in experimental conditions, or could have been due to inherent alterations in AR-driven programs in the two cell lines. To distinguish between these two possibilities, we analyzed AR-driven gene expression in two related but distinctive cell lines arising from a common progenitor.
The current analysis of androgen-regulated gene transcription in R1 and Rv1 cells revealed that AR-regulated gene expression patterns are very different between these two related cell lines. In R1 cells, the Notch signaling pathway is the most identifiable DHT-regulated pathway. In Rv1 cells, the Notch pathway is not DHT regulated, and the most prominent DHT-regulated pathway includes aminoacyl-tRNA biogenesis, DNA damage response, axonal guidance signaling, and JAK/Stat signaling. However, an analysis of direct AR target genes in R1 (9 mappable genes by IPA analysis) and Rv1 (11 mappable genes by IPA analysis) cells revealed that the most common function by far involved regulation of transcription, whereas the second most common functions were regulation of the cell cycle or metabolism. Therefore, even though the AR uses different tools for gene regulation in the two cell lines, it performs the same function in both, pointing to the versatility in the regulation of cell function. Our results indicate that in the two cell lines, the AR achieves the same goal using different pathways. This may be the reason why so many cancer drugs fail – each drug targets a single pathway, but as soon as one lesion is affected by one drug, other pathways that can bypass the drug target arise.
The most common function of DHT-regulated transcripts involves regulation of transcription. In R1, this includes DHT-mediated regulation of two downstream effectors of Notch signaling, the Hes-1 and Hey-1 transcriptional repressors. However, the DHT-mediated regulation of the two repressors is different. Hes1 expression is DHT repressed, whereas Hey1 levels are higher in R1 cells in the absence of androgen. Hey1 levels are even further elevated after DHT treatment. Moreover, Hey1 has been shown to be an AR co-repressor (Belandia et al. 2005); therefore, transactivation of Hey1 may serve as a negative feedback loop to limit AR-regulated transcription. HES1 expression is DHT transactivated in Rv1 and in C4-2B cells (Jia et al. 2008). HES1, but not HEY1, knockout mice have defects in neurulation and have premature differentiation of neuronal precursors, suggesting that HES1 has a more prominent role in cells that have a neuronal lineage. It is notable that Rv1 and LNCaP cells have neuroendocrine characteristics. An interaction of HES1 with the AR has not been reported, and the different levels of these two transcriptional co-regulators may be in part responsible for the differences in AR-dependent transcription in the two cell lines.
Mitotic genes identified as DHT targets in LNCaPabl (Wang et al. 2009) were not DHT regulated in R1 or Rv1 cells. However, five of the ten transcripts (CEBPD, KCNN2, NAT1, TSC22D1, and ZBTB16) commonly regulated by DHT in R1 and Rv1 cells were also DHT regulated in C4-2B cells (Jia et al. 2008). A comparison of R1 and Rv1 DHT-regulated genes with DHT-regulated transcripts in PC3 cells showed that CEBPD, TSC22D1, and ACTG2 were commonly regulated (Lin et al. 2009). This indicates that CEBPD and TSC22D1 are commonly regulated in four different CRCaP cell lines. A further analysis found that NAT1, TSC22, and ZBTB16 (also know as PLZF) are DHT regulated in LNCaP cells or in rat prostatic tissue (Nelson et al. 2002, Jiang & Wang 2004, Butcher et al. 2007). Previous studies found that the CEBPD was androgen repressed in rat prostatic tissue, but androgen activated in CWR22 cells (Yang et al. 2001). It is notable that CEBPD is considered a tumor suppressor, since CEBPD silencing has been detected in cervical and hepatocellular carcinomas and its overexpression is associated with a growth arrest (Ko et al. 2008). The expression of CEBPD in CRCaP tumors has not been studied.
So far, the genome-wide studies of AR chromosomal binding have used the androgen-dependant LNCaP cell line, CRCaP LNCaP-derived cell lines (Takayama et al. 2007, Wang et al. 2007, Jia et al. 2008), and PC3 cells transiently expressing the AR (Lin et al. 2009). Studies of AR binding in PC3 cells transiently transfected with AR used ChIP followed by sequencing (ChIP-seq) to identify AR-binding sites associated with DHT-dependent gene regulation. The AR-binding sites had varying distances from the TSS but were preferentially located near the TSS of genes that were androgen regulated; 22.4% of the AR sites mapped were within 2 kb of the TSS and ~40% were within 12 kb of the TSS. The current study used the human promoter array with coverage of ~10 kb upstream/downstream of TSS, thus scanned regions proximal to the TSS of known genes throughout the genome. Therefore, although our analysis could not identify all AR-binding sites, it focused on known transcripts throughout the genome. The majority of AR-binding sites were located more than 2 kb upstream of the TSS in both R1 and Rv1 cells, and more AR binding was detected in Rv1 cells than in R1 cells. This correlates with our results that Rv1 cells have a greater number of DHT-regulated transcripts than R1 cells. Most of the AR sites in R1 cells were identical or similar to the sites in Rv1 cells. Consistent with previous findings, the majority of the AR-binding sites did not contain the canonical AREs. However, a significant number of the sites contained an AR half-site motif, and in many cases had more than one half-motif. Therefore, all of the studies so far indicate that the AR half-site is associated with AR binding, where the canonical ARE is rare. As reported earlier, a number of AR binding sites have either a canonical or half-site ARE. The AR may bind directly previously unidentified sequences as has been proposed by Lin et al. (2009), the AR may be binding to sites that deviate from the consensus ARE sequence, but binding is stabilized by adjacent co-regulator(s) or, alternatively, the AR may be binding indirectly by interacting with another DNA-binding protein.
An analysis of motifs co-present with the AR identified several transcription factors including GRE, GATA, Sp-1, and FoxJ2 in both R1 and Rv1. The GATA motif has been identified by all previous AR-binding studies (Wang et al. 2007, Jia et al. 2008, Lin et al. 2009). Sp1 is a very common transcription factor-binding site found in many promoter sequences. Moreover, previous studies have shown that Sp1 and the AR interact to promote transcription (Lu et al. 2000); therefore, the presence of Sp1 may serve to enhance AR-dependent gene expression. Studies by Jia et al. (2008) also found that GRE sites were co-present with AR-binding sites. The FoxJ2, a member of the forkhead family of transcription factors, has a core sequence that is common to other family members, including FoxA1. Therefore, all of the AR-binding studies indicate that GATA and forkhead transcription factor-binding sites are co-present with AR-binding sites. Previous studies have suggested that the forkhead and GATA proteins may act as ‘pioneers’ factors that are capable of initiating chromatin opening (Cirillo et al. 2002). GATA proteins have been proposed to play major roles in endocrine function and disease (Viger et al. 2008). Forkhead transcription factors may bind to chromatin before the recruitment of subsequent transcription factors such as AR. The binding of forkhead factors appears to be dependent on histone H3K4 methylation (Lupien et al. 2008). The importance of the forkhead factor in AR-dependent gene expression is further substantiated by a recent report that a single nucleotide polymorphism that is associated with an increased prostate cancer risk resides in a FoxA1 site, and this polymorphism facilitates stronger androgen responsiveness (Jia et al. 2009). The major role of these proteins may be to open the chromatin and allow AR binding, rather than to specifically promote AR binding. Subsequent events, such as stabilizing AR/DNA interaction and recruiting appropriate co-factors to regulate gene transcription, may rely on additional factors.
A closer analysis of well-studied androgen-regulated genes identified AR binding to sequences near the PSA and TMPRSS2 genes in DHT-treated Rv1 cells, but not in DHT-treated R1 cells. However, DHT treatment of Rv1 cells did not transactivate transcription of either gene. AR binding to the 3′-UTR of the NKX3.1 gene was detected in DHT-treated R1 and Rv1 cells. A recent report showed that androgen-responsive element of this gene resides in the 3′-UTR (Thomas et al. 2010). AR binding to this site was more extensive in Rv1 than in R1 cells, yet NKX3.1 transcription was transactivated only in R1 cells. This indicates that although AR binding is required, it is not sufficient for AR androgen-dependent gene expression and that increased binding does not ensure increased gene expression.
By coupling gene expression profile with ChIP-on-chip analysis, we found that 42% of the differentially expressed transcripts identified in R1 contained AR-binding sites, indicating that they are most likely direct AR targets. Some of the AR-binding sites were actually in adjacent genes that were not AR regulated. In contrast, only 6% of the transcripts identified in Rv1 cells had AR-binding sites. Previous studies have shown that AR-binding sites can be far away from transcription start sites (Takayama et al. 2007, Wang et al. 2007). The coverage of the promoter array used for this study is limited within ~10 kb of transcription start sites. Therefore, the actual direct AR targets in R1 and Rv1 cells are most likely higher than what we found. Although the number of DHT-regulated genes was much higher in Rv1 cells, the number of genes that are DHT regulated and are associated with an AR-binding site is more comparable in R1 and Rv1 cells. This suggests that AR binding or AR/DNA complex stability in Rv1 cells is greater or that a large number of the DHT-regulated transcripts in Rv1 cells are indirect AR targets. Several mechanisms may account for this discrepancy. The presence of a 39aa insertion mutation in the Rv1 AR that results in the duplication of the DNA domain may facilitate DNA binding, or the interactions with other DNA-binding protein. However, previous studies have found that the 39aa insertion does not increase the sensitivity of the receptor to ligand (Tepper et al. 2002). The different complement of AR co-regulators in Rv1 and R1 cells is likely to govern AR binding and AR-dependent gene regulation. Since there are extensive differences in the expression of AR co-regulators in the two cell lines, further analysis will be required to identify the role of the specific co-regulators in regulating transcription of specific genes.
In summary, our study of androgen-responsive CRCaP cells lines that were derived from a common progenitor exhibits similar AR-binding profiles. The GATA, GRE, Foxj2, and Sp1-binding motifs are co-present with AR-binding sites in both cell lines. However, the DHT-dependent gene expression profile of the two cell lines is completely different. The AR is regulating a different program in the two cell lines. The combined ChIP-on-chip with microarray analysis also revealed that only a subset of genes adjacent to AR-binding sites showed differential expression in response to DHT arguing that 1) binding of AR to the vicinity of these genes is insufficient for transcriptional regulation in certain cell context or under the specific experimental conditions applied; or 2) the binding sites are indeed nonfunctional. Similarly, other groups have reported that only a subset of AR-binding sites in LNCaP cells (Wang et al. 2007, Jia et al. 2008, Lin et al. 2009) or ER in MCF-7 cells (Carroll et al. 2006) are functional, as there are many more binding sites identified than differentially regulated genes. It is apparent that the presence of a half-ARE or AR binding is not sufficient for androgen-dependent gene regulation, and AR co-regulators are important in controlling AR-mediated transcription. As more studies of AR binding coupled with expression microarray analysis are conducted in different cellular contexts, the rules that govern AR-dependent gene expression in specific cellular context will become more apparent.
Supplementary Material
Acknowledgements
We thank Dr Elizabeth Wilson for the CWR-R1 cells and greatly appreciate the expert technical assistance and consultation of Ryan R Davis.
Funding
This study was supported by DOD grants PC051049 (M Mudryj), VA Merit (M Mudryj), and DOD predoctoral award PC073557 (H Chen).
Footnotes
Supplementary data
This is linked to the online version of the paper at http://dx.doi.org/10.1677/ERC-10-0081.
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
References
- Beheshti B, Vukovic B, Marrano P, Squire JA, Park PC. Resolution of genotypic heterogeneity in prostate tumors using polymerase chain reaction and comparative genomic hybridization on microdissected carcinoma and prostatic intraepithelial neoplasia foci. Cancer Genetics and Cytogenetics. 2002;137:15–22. doi: 10.1016/s0165-4608(02)00540-x. [DOI] [PubMed] [Google Scholar]
- Belandia B, Powell SM, Garcia-Pedrero JM, Walker MM, Bevan CL, Parker MG. Hey1, a mediator of Notch signaling, is an androgen receptor corepressor. Molecular and Cellular Biology. 2005;25:1425–1436. doi: 10.1128/MCB.25.4.1425-1436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- van Bokhoven A, Varella-Garcia M, Korch C, Johannes WU, Smith EE, Miller HL, Nordeen SK, Miller GJ, Lucia MS. Molecular characterization of human prostate carcinoma cell lines. Prostate. 2003;57:205–225. doi: 10.1002/pros.10290. [DOI] [PubMed] [Google Scholar]
- Borgono CA, Diamandis EP. The emerging roles of human tissue kallikreins in cancer. Nature Reviews. cancer. 2004;4:876–890. doi: 10.1038/nrc1474. [DOI] [PubMed] [Google Scholar]
- Butcher NJ, Tetlow NL, Cheung C, Broadhurst GM, Minchin RF. Induction of human arylamine N-acetyltransferase type I by androgens in human prostate cancer cells. Cancer Research. 2007;67:85–92. doi: 10.1158/0008-5472.CAN-06-2635. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nature Genetics. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Chen H, Libertini SJ, Wang Y, Kung HJ, Ghosh P, Mudryj M. ERK regulates calpain 2-induced androgen receptor proteolysis in CWR22 relapsed prostate tumor cell lines. Journal of Biological Chemistry. 2010;285:2368–2374. doi: 10.1074/jbc.M109.049379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Molecular Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Desai SJ, Ma AH, Tepper CG, Chen HW, Kung HJ. Inappropriate activation of the androgen receptor by nonsteroids: involvement of the Src kinase pathway and its therapeutic implications. Cancer Research. 2006;66:10449–10459. doi: 10.1158/0008-5472.CAN-06-2582. [DOI] [PubMed] [Google Scholar]
- Devlin HL, Mudryj M. Progression of prostate cancer: multiple pathways to androgen independence. Cancer Letters. 2009;274:177–186. doi: 10.1016/j.canlet.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Fischer A, Gessler M. Delta-Notch – and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Research. 2007;35:4583–4596. doi: 10.1093/nar/gkm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes RF. Carcinoma of the prostate. New England Journal of Medicine. 1991;324:236–245. doi: 10.1056/NEJM199101243240406. [DOI] [PubMed] [Google Scholar]
- Gnanapragasam VJ, Robson CN, Neal DE, Leung HY. Regulation of FGF8 expression by the androgen receptor in human prostate cancer. Oncogene. 2002;21:5069–5080. doi: 10.1038/sj.onc.1205663. [DOI] [PubMed] [Google Scholar]
- Gregory CW, Hamil KG, Kim D, Hall SH, Pretlow TG, Mohler JL, French FS. Androgen receptor expression in androgen-independent prostate cancer is associated with increased expression of androgen-regulated genes. Cancer Research. 1998;58:5718–5724. [PubMed] [Google Scholar]
- Gregory CW, He B, Wilson EM. The putative androgen receptor-A form results from in vitro proteolysis. Journal of Molecular Endocrinology. 2001;27:309–319. doi: 10.1677/jme.0.0270309. [DOI] [PubMed] [Google Scholar]
- He WW, Sciavolino PJ, Wing J, Augustus M, Hudson P, Meissner PS, Curtis RT, Shell BK, Bostwick DG, Tindall DJ, et al. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics. 1997;43:69–77. doi: 10.1006/geno.1997.4715. [DOI] [PubMed] [Google Scholar]
- Heemers HV, Regan KM, Schmidt LJ, Anderson SK, Ballman KV, Tindall DJ. Androgen modulation of coregulator expression in prostate cancer cells. Molecular Endocrinology. 2009;23:572–583. doi: 10.1210/me.2008-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biology. 2003;4:P4. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huggins C, Hodges CV. The effects of castration, of estrogen and of androgen injection on the serum phosphatases in metastatic carcinoma of the prostate. Cancer Research. 1941;1:293–297. [Google Scholar]
- Ji H, Wong WH. TileMap: create chromosomal map of tiling array hybridizations. Bioinformatics. 2005;21:3629–3636. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nature Biotechnology. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Berman BP, Jariwala U, Yan X, Cogan JP, Walters A, Chen T, Buchanan G, Frenkel B, Coetzee GA. Genomic androgen receptor-occupied regions with different functions, defined by histone acetylation, coregulators and transcriptional capacity. PLoS ONE. 2008;3:e3645. doi: 10.1371/journal.pone.0003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Landan G, Pomerantz M, Jaschek R, Herman P, Reich D, Yan C, Khalid O, Kantoff P, Oh W, et al. Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000597. e1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Wang Z. Identification and characterization of PLZF as a prostatic androgen-responsive gene. Prostate. 2004;59:426–435. doi: 10.1002/pros.20000. [DOI] [PubMed] [Google Scholar]
- Ko CY, Hsu HC, Shen MR, Chang WC, Wang JM. Epigenetic silencing of CCAAT/enhancer-binding protein delta activity by YY1/polycomb group/DNA methyltransferase complex. Journal of Biological Chemistry. 2008;283:30919–30932. doi: 10.1074/jbc.M804029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers KA, Samaratunga H, Teng L, Buck M, Burger MJ, Scells B, Lavin MF, Gardiner RA. Identification of claudin-4 as a marker highly overexpressed in both primary and metastatic prostate cancer. British Journal of Cancer. 2008;99:491–501. doi: 10.1038/sj.bjc.6604486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Yu X, Ge K, Melamed J, Roeder RG, Wang Z. Heterogeneous expression and functions of androgen receptor co-factors in primary prostate cancer. American Journal of Pathology. 2002;161:1467–1474. doi: 10.1016/S0002-9440(10)64422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, Hood L, Nelson PS. Prostate-localized and androgen-regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Research. 1999;59:4180–4184. [PubMed] [Google Scholar]
- Lin B, Wang J, Hong X, Yan X, Hwang D, Cho JH, Yi D, Utleg AG, Fang X, Schones DE, et al. Integrated expression profiling and ChIP-seq analyses of the growth inhibition response program of the androgen receptor. PLoS ONE. 2009;4:e6589. doi: 10.1371/journal.pone.0006589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu AY, Roudier MP, True LD. Heterogeneity in primary and metastatic prostate cancer as defined by cell surface CD profile. American Journal of Pathology. 2004;165:1543–1556. doi: 10.1016/S0002-9440(10)63412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie MC, Yang HQ, Ma AH, Xu W, Zou JX, Kung HJ, Chen HW. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor-p160 coactivator complex. PNAS. 2003;100:2226–2230. doi: 10.1073/pnas.0437824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Jenster G, Epner DE. Androgen induction of cyclin-dependent kinase inhibitor p21 gene: role of androgen receptor and transcription factor Sp1 complex. Molecular Endocrinology. 2000;14:753–760. doi: 10.1210/mend.14.5.0461. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagabhushan M, Miller CM, Pretlow TP, Giaconia JM, Edgehouse NL, Schwartz S, Kung HJ, de Vere White RW, Gumerlock PH, Resnick MI, et al. CWR22: the first human prostate cancer xenograft with strongly androgen-dependent and relapsed strains both in vivo and in soft agar. Cancer Research. 1996;56:3042–3046. [PubMed] [Google Scholar]
- Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J, Hood L, Lin B. The program of androgen-responsive genes in neoplastic prostate epithelium. PNAS. 2002;99:11890–11895. doi: 10.1073/pnas.182376299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nwosu V, Carpten J, Trent JM, Sheridan R. Heterogeneity of genetic alterations in prostate cancer: evidence of the complex nature of the disease. Human Molecular Genetics. 2001;10:2313–2318. doi: 10.1093/hmg/10.20.2313. [DOI] [PubMed] [Google Scholar]
- Riegman PH, Vlietstra RJ, van der Korput JA, Brinkmann AO, Trapman J. The promoter of the prostate-specific antigen gene contains a functional androgen responsive element. Molecular Endocrinology. 1991;5:1921–1930. doi: 10.1210/mend-5-12-1921. [DOI] [PubMed] [Google Scholar]
- Ruizeveld de Winter JA, Janssen PJ, Sleddens HM, Verleun-Mooijman MC, Trapman J, Brinkmann AO, Santerse AB, Schroder FH, van der Kwast TH. Androgen receptor status in localized and locally progressive hormone refractory human prostate cancer. American Journal of Pathology. 1994;144:735–746. [PMC free article] [PubMed] [Google Scholar]
- Shah RB, Mehra R, Chinnaiyan AM, Shen R, Ghosh D, Zhou M, Macvicar GR, Varambally S, Harwood J, Bismar TA, et al. Androgen-independent prostate cancer is a heterogeneous group of diseases: lessons from a rapid autopsy program. Cancer Research. 2004;64:9209–9216. doi: 10.1158/0008-5472.CAN-04-2442. [DOI] [PubMed] [Google Scholar]
- Takayama K, Kaneshiro K, Tsutsumi S, Horie-Inoue K, Ikeda K, Urano T, Ijichi N, Ouchi Y, Shirahige K, Aburatani H, et al. Identification of novel androgen response genes in prostate cancer cells by coupling chromatin immunoprecipitation and genomic microarray analysis. Oncogene. 2007;26:4453–4463. doi: 10.1038/sj.onc.1210229. [DOI] [PubMed] [Google Scholar]
- Takayama K, Tsutsumi S, Suzuki T, Horie-Inoue K, Ikeda K, Kaneshiro K, Fujimura T, Kumagai J, Urano T, Sakaki Y, et al. Amyloid precursor protein is a primary androgen target gene that promotes prostate cancer growth. Cancer Research. 2009;69:137–142. doi: 10.1158/0008-5472.CAN-08-3633. [DOI] [PubMed] [Google Scholar]
- Tan J, Sharief Y, Hamil KG, Gregory CW, Zang DY, Sar M, Gumerlock PH, deVere White RW, Pretlow TG, Harris SE, et al. Dehydroepiandrosterone activates mutant androgen receptors expressed in the androgen-dependent human prostate cancer xenograft CWR22 and LNCaP cells. Molecular Endocrinology. 1997;11:450–459. doi: 10.1210/mend.11.4.9906. [DOI] [PubMed] [Google Scholar]
- Tepper CG, Boucher DL, Ryan PE, Ma AH, Xia L, Lee LF, Pretlow TG, Kung HJ. Characterization of a novel androgen receptor mutation in a relapsed CWR22 prostate cancer xenograft and cell line. Cancer Research. 2002;62:6606–6614. [PubMed] [Google Scholar]
- Thomas MA, Preece DM, Bentel JM. Androgen regulation of the prostatic tumour suppressor NKX3.1 is mediated by its 3′ untranslated region. Biochemical Journal. 2010;425:575–583. doi: 10.1042/BJ20091109. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Viger RS, Guittot SM, Anttonen M, Wilson DB, Heikinheimo M. Role of the GATA family of transcription factors in endocrine development, function, and disease. Molecular Endocrinology. 2008;22:781–798. doi: 10.1210/me.2007-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainstein MA, He F, Robinson D, Kung HJ, Schwartz S, Giaconia JM, Edgehouse NL, Pretlow TP, Bodner DR, Kursh ED, et al. CWR22: androgen-dependent xenograft model derived from a primary human prostatic carcinoma. Cancer Research. 1994;54:6049–6052. [PubMed] [Google Scholar]
- Wang Q, Li W, Liu XS, Carroll JS, Janne OA, Keeton EK, Chinnaiyan AM, Pienta KJ, Brown M. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Molecular Cell. 2007;27:380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitbread AK, Veveris-Lowe TL, Lawrence MG, Nicol DL, Clements JA. The role of kallikrein-related peptidases in prostate cancer: potential involvement in an epithelial to mesenchymal transition. Biological Chemistry. 2006;387:707–714. doi: 10.1515/BC.2006.089. [DOI] [PubMed] [Google Scholar]
- Xu LL, Shi Y, Petrovics G, Sun C, Makarem M, Zhang W, Sesterhenn IA, McLeod DG, Sun L, Moul JW, et al. PMEPA1, an androgen-regulated NEDD4-binding protein, exhibits cell growth inhibitory function and decreased expression during prostate cancer progression. Cancer Research. 2003;63:4299–4304. [PubMed] [Google Scholar]
- Yang G, Gregory CW, Shang Q, O’Brien DA, Zhang YL. Differential expression of CCAAT/enhancer-binding protein-delta (c/EBPdelta) in rat androgen-dependent tissues and human prostate cancer. Journal of Andrology. 2001;22:471–480. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.