Abstract
Aspergillus floridensis and A. trinidadensis spp. nov. are described as novel uniseriate species of Aspergillus section Nigri isolated from air samples. To describe the species we used phenotypes from 7-d Czapek yeast extract agar culture (CYA), creatine agar culture (CREA) and malt extract agar culture (MEA), with support by molecular analysis of the β-tubulin, calmodulin, RNA polymerase II (RPB2), and translation elongation factor-alpha (TEF) gene amplified and sequenced from 56 air isolates and one isolate from almonds belonging to Aspergillus sectionNigri. Aspergillus floridensis is closely related to A. aculeatus, and A. trinidadensis is closely related to A. aculeatinus. Aspergillus brunneoviolaceus (syn. A. fijiensis) and A. uvarum are reported for the first time from the USA and from the indoor air environment. The newly described species do not produce ochratoxin A.
Keywords: Aspergillus brunneoviolaceus, Aspergillus floridensis, Aspergillus trinidadensis, Aspergillus uvarum, Aspergillus violaceofuscus, black aspergilli, environment, phylogeny, ochratoxin A
INTRODUCTION
Aspergillus section Nigri (Gams et al. 1985), commonly known as the black aspergilli, contains many common species in the environment (Klich 2009), and some have been implicated in human and animal diseases (de Hoog et al. 2000, Abarca et al. 2004, Klich 2009). They have a worldwide distribution and occur on a large variety of substrates including soil, grains, dairy and forage products, various fruit, vegetables, beans and nuts, cotton textiles and fabrics, and meat products (Raper & Fennell 1965, Pitt & Hocking 2007, 2009). Black aspergilli are used in the fermentation industry to produce various enzymes and organic acids (Raper & Fennell 1965, Varga et al. 2000). Some black aspergilli produce ochratoxin A (Abarca et al. 1994, 2003, 2004, Wicklow et al. 1996, Varga et al. 2000, Cabanes et al. 2002, Sage et al. 2004, Samson et al. 2004).
Although black aspergilli occur in clinical samples, they are much less frequent than A. fumigatus, A. terreus, or A. flavus. Aspergillus species are widely documented as causative pathogens in invasive and non-invasive infections as well as in allergic reactions especially Types III and IV (Richardson 2005). Indeed, the allergic forms of the disease appear to be “almost exclusively caused by Aspergillus species” (Moss 2002, Knutsen 2011). Some strains of black aspergilli are often misidentified as A. niger due to the difficulties of identifying the species in this group (Samson et al. 2007). Perrone et al. (2012a, b) recognized two new species of black aspergilli that may be involved in human disease from Sri Lanka: A. brunneoviolaceus (= A. fijiensis) in pulmonary aspergillosis, and A. aculeatinus in human dacryocystitis.
We collected 56 isolates of black uniseriate Aspergillus species from air (52 homes and four outside samples) from 17 states of the USA, Bermuda, Martinique, Trinidad & Tobago, and one from almonds in the Czech Republic. Using molecular data and macro- and micro-morphological observations, we discovered and describe here two new species related to A. aculeatus and A. aculeatinus.
MATERIALS AND METHODS
Fungal isolates
The provenance of fungal isolates examined is detailed in Table 1.
Table 1. Provenance of fungal isolates characterized in this study.
| *ITEM number | Provenance |
|---|---|
| Aspergillus aculeatus | |
| 14807 | USA: Georgia, iso. ex indoor air sample, 2010, Ž. Jurjević. |
| Aspergillus brunneoviolaceus (syn. A. fijiensis) | |
| 14784 | USA: Florida, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14785 | USA: Florida, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14786 | USA: Florida, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14790 | Bermuda: isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14791 | USA: Florida, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14794 | USA: Texas, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14795 | USA: Florida, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14796 | USA: Florida, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14799 | USA: Florida, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14802 | USA: Arizona, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14804 | Trinidad and Tobago: Tunapuna, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14806 | USA: Alabama, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14809 | USA: Florida, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14812 | USA: Alabama, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14820 | USA: Florida, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14825 | USA: Missouri, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14831 | USA: Alabama, isol. ex outside air sample, 2011, Ž. Jurjević. |
| 14832 | USA: Alabama, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| Aspergillus floridensis sp. nov. | |
| 14783⊤ | USA: Florida, isol. ex indoor air sample, 2010, Ž. Jurjević, ex-type. |
| ** (=NRRL 62478⊤) | |
| ***CCF 4046 | Czech Republic: Brno, isol. ex almonds in shells imported from USA, 2010, V. Ostrý. |
| CCF 4236 | Martinique: Fort de France, isol. ex outside air sample, 2004, N. Desbois. |
| ****CRI 323-04 | Thailand: Phi Phi Islands, isol. ex Xestospongia testudinaria, 2006, T.S. Bay. |
| *****IFM 55703 | Japan: isol. ex soil from grapery, 2007, K. Yokoyama. |
| Aspergillus violaceofuscus (syn. A. japonicus) | |
| 14787 | USA: Florida, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14788 | USA: Kentucky, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14789 | USA: New Jersey, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14792 | USA: Florida, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14793 | USA: Louisiana, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14797 | USA: New York, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14798 | USA: Florida, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14800 | USA: Alabama, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14801 | USA: Alabama, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14803 | USA: Georgia, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14805 | USA: Louisiana, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14808 | USA: South Carolina, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14810 | USA: Tennessee, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14811 | USA: Hawaii, isol. ex indoor air sample, 2010, Ž. Jurjević. |
| 14813 | USA: Alabama, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14814 | USA: Florida, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14815 | USA: Delaware, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14816 | USA: Maryland, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14817 | USA: Alabama, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14818 | USA: Alabama, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14822 | USA: Missouri, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14823 | USA: Missouri, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14824 | USA: Missouri, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14827 | USA: Missouri, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14828 | USA: Georgia, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14830 | USA: Texas, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14834 | USA: New Jersey, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14835 | USA: North Carolina, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14836 | USA: New York, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14837 | USA: Texas, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| Aspergillus trinidadensis sp. nov. | |
| 14821⊤ (=NRRL 62479⊤) | Trinidad and Tobago: Tunapuna, isol. ex indoor air sample, 2011, Ž. Jurjević, ex-type. |
| 14829 (=NRRL 62480) | USA: California, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| Aspergillus uvarum | |
| 14819 | USA: Florida, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14826 | USA: Missouri, isol. ex indoor air sample, 2011, Ž. Jurjević. |
| 14833 | USA: New Jersey, isol. ex outside air sample, 2011, Ž. Jurjević. |
*ITEM, Agri-Food Toxigenic Fungi Culture Collection, Institute of Sciences of Food Production, Bari, Italy; **NRRL (Northern Regional Research Laboratory), the National Center For Agricultural Utilization Research, USA; ***CCF (Culture Collection of Fungi), Department of Botany, Faculty of Science, Charles University, Prague, Czech Republic; ****CRI (Chulabhorn Research Institute), Vibhavadi-Rangsit Road, Laksi, Bangkok, Thailand; *****IFM (Medical Mycology Research Center) Chiba University, Chiba, Japan.
⊤= ex-type strain
Culture methods
Observations were made on Czapek yeast extract agar (CYA), CYA with 20 % sucrose (CY20S), malt extract agar (MEA), oatmeal agar (OA), and creatine agar (CREA), (Pitt 1980, Samson et al. 2004) cultures incubated at 25 °C for 7 d in darkness, and CYA cultures incubated at 5 °C, 35 °C and 37 °C for 7 d. The cultures were grown on one plate as a three-point inoculation and on another plate as a single center-point inoculation on each medium in 9 cm diam Petri dishes. Colony diameters and appearance were recorded and photographs were made from 7 d culture plates incubated at 25 °C.
Microscopy
Microscopic examination was performed by gently pressing a ca. 20 × 5 mm piece of transparent tape onto a colony, rinsing the tape with one or two drops of 70 % ethanol and mounting the tape in lactic acid with fuchsin dye. Additional microscopic samples were made by teasing apart a small amount of mycelium in a drop of water containing 0.5 % Tween 20. A Leica DM 2500 microscope with bright field, phase contrast and DIC optics was used to view the slides. A Spot camera with Spot imaging software was mounted on the microscope and used for photomicrography. A Nikon digital SLR camera with a D70 lens was used for colony photography. Photographs were resized and fitted into plates using Microsoft PowerPoint 2010.
Ochratoxin A (OTA) assay
Aspergillus strains were grown in duplicate in 100-ml stationary liquid cultures (5 cm diam) of Czapek w/20 % Sucrose Broth (200 g l−1 Sucrose, 1 g l−1 K2HPO4, 3 g l−1 NaNO3, 0.5 g l−1 KCl, 0.05 g MgSO4·7•H2O, 0.01 g l−1 FeSO4 7•H2O) (Health Link®, Jacksonville, FL) and Yeast Extract Broth {2 g l−1 Yeast extract, 15 g l−1 Sucrose, 0.5 g l−1 MgSO4·7•H2O, 1 ml l−1 trace metal solution (1 g l−1 ZnSO4·7•H2O, 0.5 g l−1 CuSO4 5•H2O)} (Health Link®, Jacksonville, FL) in 250 ml flasks for 7 d at 25 °C± 0.2 °C in the dark. Each sample was inoculated with 106 spores counted by hemocytometer, previously grown on MEA.
Sample preparation (extraction and cleanup)
100 ml of liquid culture was homogenized (Waring®, USA) for 2 min. Two millilitre (ml) aliquots were diluted with 2 ml of acetonitrile/water (50/50, v/v) containing 0.5 % acetic acid, vortex mixed for 30 sec and then filtered through Acrodisc syringe filters with 0.45 μm PTFE membrane (Pall Corporation, http://www.pall.com/main/home.page) before LC/MS analysis.
Standard
Ochratoxin A standard was purchased from Sigma (http://www.sigmaaldrich.com/united-states.html) and stored at −20 °C.
LC/MS equipment and parameters
Analyses were performed on an Agilent 6330 series ion trap LC/MS system (http://www.home.agilent.com/), equipped with an ESI interface and an 1100 series LC system comprising a quaternary pump and an auto-sampler, from Agilent Technologies.
The analytical column was an Allure Bi-Phenyl column 30 mm × 2.1 mm with 5 μm particle sizes (http://www.restek.com/). The column oven was set at 40 °C. The flow rate of the mobile phase was 250 μl min l−1 and the injection volume was 10 μ l−1 . The column effluent was directly transferred into the ESI interface, without splitting.
Eluent A was 95 % water: 5 % acetonitrile, and eluent B was 95 % acetonitrile: 5 % water, both containing 0.5 % acetic acid. Gradient elution was performed starting with 100 % eluent A, the proportion of eluent B was linearly increased to 100 % over a period of 5 min and then kept constant for 5 min. The column was re-equilibrated with 100 % eluent A for 5 min. For LC/MS analyses, the ESI interface was used in positive ion mode, with parameters set at: DRY TEMP 350 °C; NEBULIZER 40 psi, nitrogen, DRY GAS 10 l min−1, Capillary voltage −3500 V. The mass spectrometer operated in MRM (multiple reaction monitoring) mode, by monitoring three transitions (1 quantifier, 2 qualifiers) for each compound, with a dwell time of 200 ms. Quantification of ochratoxin A was performed by measuring peak areas in the MRM chromatogram, and comparing them with the relevant calibration curve.
Tuning experiments were performed by direct infusion at a flow rate of 0.6 ml h −1 of 1μg l−1 standard solutions in acetonitrile/water (50/50, v/v) containing 0.5 % acetic acid. The infusion was performed by using a model KDS100CE infusion pump (KDS Scientific Holliston, MA).
Interface parameters were: DRY TEMP 350 °C; NEBULIZER 10 psi nitrogen, DRY GAS 5L/min, Capillary voltage −3500 V, spacer was removed for flow infusion.
Fungal cultures, DNA extraction and DNA sequencing
Monoconidial isolates of each fungal strain were deposited at the ITEM Collection (CNR-ISPA, Bari, Italy) and received an ITEM accession number (Table 1). Supplemental information about the isolates can be recovered from the ITEM electronic catalogue (http: www.ispa.cnr.it/Collection).
For mycelium production, a suspension of spores from each fungal strain was grown in Wickerham’s medium (glucose 40 g, peptone 5 g, yeast extract 3 g, malt extract 3 g and distilled water to 1 l). Mycelia were filtered and lyophilized for total DNA isolation. The fungal DNA was extracted with mechanical grinding using 5 mm iron beads in a Mixer Mill MM 400 (http://www.retsch.com/), and a “Wizard® Magnetic DNA Purification System for Food” kit (Promega, http://www.promega.com/), starting from 10 mg of lyophilized mycelium. The quality of genomic DNA was determined by electrophoresis and it was quantified using a ND-1000 (Nano Drop) spectrophotometer.
Beta-tubulin (BenA, ca. 400 nt) was amplified using BT2a and BT2b primers and PCR conditions described by Glass & Donaldson (1995), calmodulin (CaM, ca. 650 nt) was amplified using CL1 and CL2A primers (O’Donnell et al. 2000), translation elongation factor-1 alpha (TEF-1α, ca. 700 nt) was amplified using A-EF_F/A-EF_R primers (Perrone et al. 2011) and RNA polymerase II (RPB2, ca. 950 nt) was amplified using primers 5F and 7CR (Liu et al. 1999). After amplification, the products were purified with the enzymatic mixture EXO/SAP (Exonuclease I, Escherichia coli / Shrimp Alkaline Phosphatase; Fermentas International, http://www.fermentas.com/en/home).
Bidirectional sequencing was performed for all loci and isolates. Sequence reactions were performed with the Big Dye Terminator Cycle Sequencing Ready Reaction Kit for both strands, purified by gel filtration through Sephadex G-50 (Amersham Pharmacia Biotech) and analyzed on the “ABI PRISM 3730 Genetic Analyzer” (Applied Biosystems, http://www.appliedbiosystems.com/absite/us/en/home.html).
The preliminary alignments of sequences from each of the four loci was performed using the software package BioNumerics 5.1 from Applied Maths (http://www.applied-maths.com/bionumerics/bionumerics.htm) with manual adjustments where judged necessary.
Sequence Data Analysis
DNA sequences were aligned using the Clustal W algorithm (Thompson et al. 1994) in MEGA version 5 (Tamura et al. 2011). Sequences were deposited in GenBank (Tables 2 & 3). Each locus was aligned separately and then concatenated in a super-gene alignment used to generate the phylogenetic tree. Phylogenetic analysis was performed in MEGA5 using both Neighbor-Joining (NJ) (Saitou & Nei 1987) and Maximum Likelihood (ML) methods and the Tamura-Nei model (Tamura & Nei 1993). Evolutionary distances for NJ were computed using the Tamura-Nei method of the package and are in units of number of base substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (Complete deletion option). Bootstrap values (Felsenstein 1985, 1995) were calculated from 1000 replications of the bootstrap procedure using programs within MEGA5.
Table 2. GenBank accession numbers of reference and ex-type strains. Sequence IDs in red are the sequences deposited for this manuscript.
| Species | Source* | RPB2 | TEF | CaM | BenA |
|---|---|---|---|---|---|
| Aspergillus acidus | ITEM 4507 = CBS 564.65 | EF661052 | FN665410 | AM419749 | AY585533 |
| Aspergillus aculeatus | ITEM 7046⊤ = CBS 172.66⊤ | EF661046 | HE984381 | AJ964877 | AY585540 |
| Aspergillus ‘aculeatus’ | ITEM 4760 = CBS 620.78=NRRL 2053 | EF661044 | HE984382 | EF661145 | EU982087 |
| Aspergillus ‘aculeatus’ | ITEM 15927 = NRRL 359 | EF661043 | HE984383 | EF661146 | EF661106 |
| Aspergillus aculeatinus | CBS 121060⊤ = IBT 29077⊤ | HF559233 | HF559230 | EU159241 | EU159220 |
| Aspergillus aculeatinus | ITEM 13553 | HE984359 | HE984385 | HE984422 | HE984407 |
| Aspergillus awamori | ITEM 4509⊤ = CBS 557.65⊤ | HE984360 | FN665395 | AJ964874 | AY820001 |
| Aspergillus brasiliensis | ITEM 7048 = CBS 101740⊤ | EF661063 | FN665411 | AM295175 | AY820006 |
| Aspergillus brunneoviolaceus | ITEM 7047 ⊤ = CBS 621.78 ⊤ | EF661045 | HE984384 | EF661147 | EF661105 |
| Aspergillus carbonarius | ITEM 4503⊤ = CBS 556.65⊤ | EF661068 | FN665412 | AJ964873 | AY585532 |
| Aspergillus costaricaensis | ITEM 7555⊤ = CBS 115574⊤ | HE984361 | FN665409 | EU163268 | AY820014 |
| Aspergillus ellipticus | ITEM 4505⊤ = CBS 482.65⊤ | EF661051 | HE984386 | AM117809 | FJ629279 |
| Aspergillus fijiensis | ITEM 7037⊤ = CBS 119.49⊤ | HE984362 | HE984387 | HE818081 | HE818086 |
| Aspergillus helicothrix | ITEM 4499 = CBS 677.79 | HE984363 | HE984389 | AM117810 | FJ629279 |
| Aspergillus heteromorphus | ITEM 7045 = CBS 117.55 | - | HE984388 | AM421461 | FJ629284 |
| Aspergillus homomorphus | ITEM 7556⊤ = CBS 101889⊤ | HE984365 | HE984390 | AM887865 | AY820016 |
| Aspergillus ibericus | ITEM 4776⊤ = IMI 391429⊤ | EF661065 | HE984391 | AJ971805 | AM419748 |
| Aspergillus indologenus | ITEM 7038 = CBS 114.80 | HE984366 | HE984392 | AM419750 | AY585539 |
| Aspergillus japonicus | ITEM 7034⊤ = CBS 114.51⊤ | EF661047 | HE984393 | AJ964875 | HE577804 |
| Aspergillus japonicus | ITEM 15926 = NRRL 35494 | EU021639 | HE984394 | EU021690 | EU021665 |
| Aspergillus lacticoffeatus | ITEM 7559⊤ = CBS 101883⊤ | HE984367 | FN665406 | EU163270 | AY819998 |
| Aspergillus niger | ITEM 4501⊤ = CBS 554.65⊤ | XM_001395124 | FN665404 | AY585536 | AJ964872 |
| Aspergillus pulverulentus | ITEM 4510⊤ = CBS 558.65⊤ | HE984368 | HE984395 | HE984423 | HE984408 |
| Aspergillus saccharolyticus | ITEM 16159⊤ = CBS 127449 | HF559235 | HF559232 | HM853554 | HM853553 |
| Aspergillus sclerotioniger | ITEM 7560⊤ = CBS 115572⊤ | HE984369 | HE984396 | EU163271 | AY819996 |
| Aspergillus tubingensis | ITEM 7040⊤ = CBS 134.48⊤ | EF661055 | FN665407 | AJ964876 | AY820007 |
| Aspergillus uvarum | ITEM 4834⊤ = IMI 388523⊤ | HE984370 | HE984397 | AM745755 | AM457751 |
| Aspergillus vadensis | ITEM 7561⊤ = CBS 113.365⊤ | HE984371 | FN665408 | EU163269 | AY585531 |
| Aspergillus violaceofuscus | ITEM 16177⊤ = CBS 102.23⊤ | HF559234 | HF559231 | FJ491698 | HE577805 |
Table 3. GenBank accession numbers of Aspergillus strains isolated from air.
| Species | Source | RPB2 | TEF | CaM | BenA |
|---|---|---|---|---|---|
| Aspergillus aculeatus | ITEM 14807 | HE984372 | HE984398 | HE984424 | HE984409 |
| Aspergillus brunneoviolaceus | ITEM 14784 | HE984374 | HE984400 | HE984426 | HE984411 |
| Aspergillus brunneoviolaceus | ITEM 14785 | HE984375 | HE984427 | ||
| Aspergillus brunneoviolaceus | ITEM 14795 | HE984428 | |||
| Aspergillus brunneoviolaceus | ITEM 14802 | HE984401 | |||
| Aspergillus brunneoviolaceus | ITEM 14804 | HE984402 | |||
| Aspergillus floridensis sp. nov. | ITEM 14783⊤ =NRRL 62478⊤ | HE984376 | HE984403 | HE984429 | HE984412 |
| Aspergillus violaceofuscus | ITEM 14787 | HE984377 | HE984404 | HE984430 | HE984413 |
| Aspergillus violaceofuscus | ITEM 14788 | HE984431 | |||
| Aspergillus violaceofuscus | ITEM 14789 | HE984432 | HE9844174 | ||
| Aspergillus violaceofuscus | ITEM 14793 | HE984415 | |||
| Aspergillus violaceofuscus | ITEM 14801 | HE984433 | |||
| Aspergillus violaceofuscus | ITEM 14805 | HE984378 | HE984416 | ||
| Aspergillus violaceofuscus | ITEM 14814 | HE984417 | |||
| Aspergillus violaceofuscus | ITEM 14822 | HE984418 | |||
| Aspergillus violaceofuscus | ITEM 14834 | HE984405 | |||
| Aspergillus violaceofuscus | ITEM 14835 | HE984419 | |||
| Aspergillus trinidadensis sp.nov. | ITEM 14821⊤ =NRRL 62479⊤ | HE984379 | HE984406 | HE984434 | HE984420 |
| Aspergillus trinidadensis | ITEM 14829 =NRRL 62480 | HE984373 | HE984399 | HE984425 | HE984410 |
| Aspergillus uvarum | ITEM 14819 | HE984380 | HE984435 | HE984421 | |
| Aspergillus uvarum | ITEM 14826 | HE984364 | HE984437 | ||
| Aspergillus uvarum | ITEM 14833 | HE984436 |
* The sequences were deposited only for the strains that differ in their sequences from the sequence of the type strain for a specific locus/gene.
The evolutionary history was inferred by using the Maximum Likelihood method based on the Tamura-Nei model implemented in MEGA5. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically as follows. When the number of common sites was < 100 or less than one fourth of the total number of sites, the maximum parsimony method was used; otherwise the BIONJ method with MCL distance matrix was used. A discrete Gamma distribution was used to model evolutionary rate differences among sites (five categories; +G, parameter = 0.2036). All positions containing gaps and missing data were eliminated. There were 2329 positions in the final dataset. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
A Markov Chain Monte Carlo (MCMC) algorithm was used to generate phylogenetic trees with Bayesian probabilities using MrBayes v3. 2 (Huelsenbeck & Ronquist 2001, Ronquist & Huelsenbeck 2003) for the combined sequences datasets. The analysis was run in duplicate with four MCMC chains and setting random trees for 107 generations sampled every 100 generations. A total of 15 738 trees were read in the two runs, 7869 for each, and the first 1967 trees (25 %) were discarded in each run as the burn-in phase of the analysis and posterior probabilities were determined from the remaining trees (5902 in each run).
Maximum Parsimony analysis (MP) was performed for all data sets using the heuristic search option and Close-Neighbor-Interchange algorithm (with search level 1 in which the initial trees were obtained with the random addition of sequences). To assess the robustness of the topology, 1000 bootstrap replicates were run. The tree is drawn to scale, with branch lengths calculated using the average pathway method and are in units of the number of changes over the whole sequence (Nei & Kumar 2000). The analysis involved data from 86 isolates and all positions containing gaps and missing data were eliminated. There were a total of 2329 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 (Tamura et al. 2011).
RESULTS
Phylogenic analysis of sequence data
The multilocus analysis was performed on 56 isolates collected from air (52 homes and 4 outside samples) from 17 states of the United States, Bermuda, Martinique, Trinidad and Tobago, and one isolated from almonds in the Czech Republic (Table 1), along with 28 reference and ex-type strains from Aspergillus section Nigri (Table 4). The ex-type strain of Aspergillus flavus (ITEM 7526) was used as outgroup. The percentage of variable sites and parsimony informative sites for each locus differ, the benA sequences have the highest percentage of variable and parsimony informative sites, the CaM sequences have the highest nucleotide diversity, TEF sequences have the lowest variability and RPB2 has lower sequence diversity than CaM and benA but the highest number of informative sites (Table 5). After a preliminary analysis using MEGA5 Neighbour-Joining, the best substitution model among the evolutionary models in MEGA5 was calculated. The best model was Tamura-Nei with Gamma distribution (TN93 + G). Evolutionary history was inferred using the Neighbor-Joining method. The tree with the highest log likelihood is shown (Fig 1). Bootstrap proportions are shown next to the branches. The tree is drawn to scale, with branch lengths reflecting evolutionary distance computed using the Maximum Composite Likelihood method as number of base substitutions per site (MEGA5). The rate variation among sites was modeled with a gamma distribution (shape parameter = 0.3). Phylogenetic analysis was conducted first on the four single locus alignments and subsequently the combined alignment of the four loci. The single locus and four locus combined data trees contained the same topology fulfilling the requirements of genealogical concordance phylogenetic species recognition (GCPSR, Taylor et al. 2000).
Table 4. Provenance of Aspergillus section Nigri isolates used as reference strains
| Species | Source* | Provenance |
|---|---|---|
| Aspergillus acidus | ITEM 4507⊤ = IMI 104688⊤ = CBS 564.65⊤ = NRRL 4796⊤ | JAPAN: unknown, R. Nakazawa. |
| Aspergillus aculeatus | ITEM 7046⊤ = IMI 211388⊤ = CBS 172.66⊤ = NRRL 5094⊤ | USA: isol. ex tropical soil, 1962, K. B. Raper. |
| Aspergillus ‘aculeatus’ | ITEM 4760 = CBS 620.78 = NRRL 2053 = IMI 358696 | New Guinea: isol. ex canvas tent, 1946, received from D. L. White |
| Aspergillus ‘aculeatus’ | ITEM 15927 = NRRL 359 | Thom and Raper 1945 received it from Dr. A. F. Blakeslee. |
| Aspergillus brunneoviolaceus | ITEM 7047⊤ = CBS 621.78⊤ = IMI 312981⊤ = NRRL 4912⊤ | BRAZIL: culture contaminant, A. C. Batista and H. Maia. |
| Aspergillus aculeatinus | ITEM 16172⊤ = CBS 121060⊤ = IBT 29077⊤ | THAILAND: Chumporn Prov.: isol. ex arabica coffee, P. Noonim. |
| Aspergillus aculeatinus | ITEM 13553 | SRI LANKA: isol. ex human dacryocystitis. |
| Aspergillus awamori | ITEM 4509⊤ = CBS 557.65⊤ = NRRL 4948⊤ = IMI 211394⊤ | Unknown - Raper and Fennel 1965 received it from the Instituto Ozwaldo Cruz |
| Aspergillus brasiliensis | ITEM 7048 = IMI 381727⊤ = CBS 101740⊤ = NRRL 26652⊤ | BRAZIL: Pedreira: isol. ex soil, J. H. Croft. |
| Aspergillus carbonarius | ITEM 4503⊤ = IMI 016136⊤ = CBS 556.65⊤ = NRRL 369⊤ | Unknown: paper, A. F. Blakeslee. |
| Aspergillus costaricaensis | ITEM 7555⊤ = CBS 115574⊤ | COSTA RICA: Taboga island: isol. ex soil, 2000, M. Christensen. |
| Aspergillus ellipticus | ITEM 4505⊤ = IMI 172283⊤ = CBS 482.65⊤ = NRRL 5120⊤ | COSTA RICA: isol. ex soil, 1962, K. J. Kwon. |
| Aspergillus fijensis | ITEM 7037⊤ = CBS 119.49⊤ | INDONESIA: Palembang: Lactuca sativa, 1949. |
| Aspergillus heteromorphus | ITEM 7045⊤ = CBS 117.55⊤ = IMI 172288⊤ = NRRL 4747⊤ | BRAZIL: Recife: culture contaminant, A. C. Batista. |
| Aspergillus homomorphus | ITEM 7556⊤ = CBS 101889⊤ | ISRAEL: isol. ex soil 2 km away from Dead Sea. |
| Aspergillus ibericus | ITEM 4776⊤ = CBS 121594⊤ = IMI 391429⊤ = NRRL 35644⊤ | PORTUGAL : Iberian Peninsula: isol. ex grapes, 2001, R. Serra. |
| Aspergillus indologenus | ITEM 7038⊤ = CBS 114.80⊤ = IBT 3679⊤ | INDIA: isol. ex soil. |
| Aspergillus japonicus | ITEM 7034⊤ = CBS 114.51⊤ | Unknown, K. Saito. |
| Aspergillus japonicus | ITEM 15926 = NRRL 35494 | Unknown. |
| Aspergillus lacticoffeatus | ITEM 7559⊤ = CBS 101883⊤ | INDONESIA: South Sumatra: isol. ex coffee bean, J. M. Frank. |
| Aspergillus niger | ITEM 4501⊤ = IMI 050566⊤ = CBS 554.65⊤ = NRRL 326⊤ | USA: Connecticut: tannin-gallic acid fermentation, 1913, A. Hollander. |
| Aspergillus pulverulentus | ITEM 4510⊤ = CBS 558.65⊤ = NRRL 4851⊤ = IMI 211396⊤ | AUSTRALIA: Victoria: isol. ex Phaseolus vulgaris, ~1907, D. McAlpine. |
| Aspergillus sclerotioniger | ITEM 7560⊤ = CBS 115572⊤ = IBT 22905⊤ | INDIA: Karnataka: isol. ex green arabica coffee J. M. Frank. |
| Aspergillus tubingensis | ITEM 7040⊤ = CBS 134.48⊤ = NRRL 4875⊤ | Unknown: 1948, deposited by R. Mosseray. |
| Aspergillus uvarum | ITEM 4834⊤ = IMI 388523⊤ = CBS 127591⊤ = IBT 26606⊤ | ITALY: Brindisi: isol. ex grapes, 2001, P. Battilani. |
| Aspergillus uvarum | ITEM 4685 = IMI 387209 | PORTUGAL: Régua, Douro Region: isol. ex grapes. |
| Aspergillus uvarum | ITEM 4962 = IMI 3888715 | SPAIN: isol. ex grapes. |
| Aspergillus uvarum | ITEM 4997 = IMI 388670 | ISRAEL: Lichron: isol. ex grapes. |
| Aspergillus uvarum | ITEM 5020 = IMI 388660 | ITALY: Brindisi, Apulia: isol. ex grapes. |
| Aspergillus uvarum | ITEM 5321 = IMI 389195 | FRANCE: Narbonne, Languedoc: isol. ex grapes. |
| Aspergillus uvarum | ITEM 5350 = IMI 389166 | ISRAEL: Pdaya: isol. ex grapes. |
| Aspergillus vandensis | ITEM 7561⊤ = IMI 313493⊤ = CBS 113365⊤ | EGYPT: air, A. H. Moubasher. |
| Aspergillus violaceofuscus | ITEM 16159⊤ = CBS 102.23⊤ | FRANCE: Strassbourg: received by D Borrel, 1923 |
| Aspergillus saccharolyticus | ITEM 16177⊤ = CBS 127449⊤ = IBT 28509⊤ | DENMARK: Gentofte: ex under a toilet seat made of treated oak wood, P. J. Teller. |
*CBS, Centraalbureau voor Scimmelcultures, Utrecht, The Netherlands; IMI, CABI Bioscience Genetic Resource Collection, Egham, United Kingdom; ITEM, Agri-Food Toxigenic Fungi Culture Collection, Institute of Sciences of Food Production, Bari, Italy; NRRL (Northern Regional Research Laboratory), the National Center For Agricultural Utilization Research, USA.
⊤= ex-type strain
Table 5. Sequence characteristics and phylogenetic information for RPB2, TEF, CaM, BenA and combined MLS.
| Locus | Region | Sites | Net Sites | % GC | No. of variable sites | No. of informative sites | No. of mutations (Eta) | Nucleotide diversity |
|---|---|---|---|---|---|---|---|---|
| BenA | 1-369 | 369 | 326 | 58 | 180 | 149 | 229 | 0.101 |
| CaM | 370-937 | 568 | 496 | 53 | 269 | 224 | 349 | 0.103 |
| RPB2 | 938-1922 | 985 | 878 | 52 | 370 | 271 | 442 | 0.063 |
| TEF | 1923-2552 | 630 | 629 | 57 | 93 | 55 | 105 | 0.023 |
| MLS | 1-2560 | 2552 | 2329 | 55 | 912 | 699 | 1125 | 0.067 |
Fig. 1.
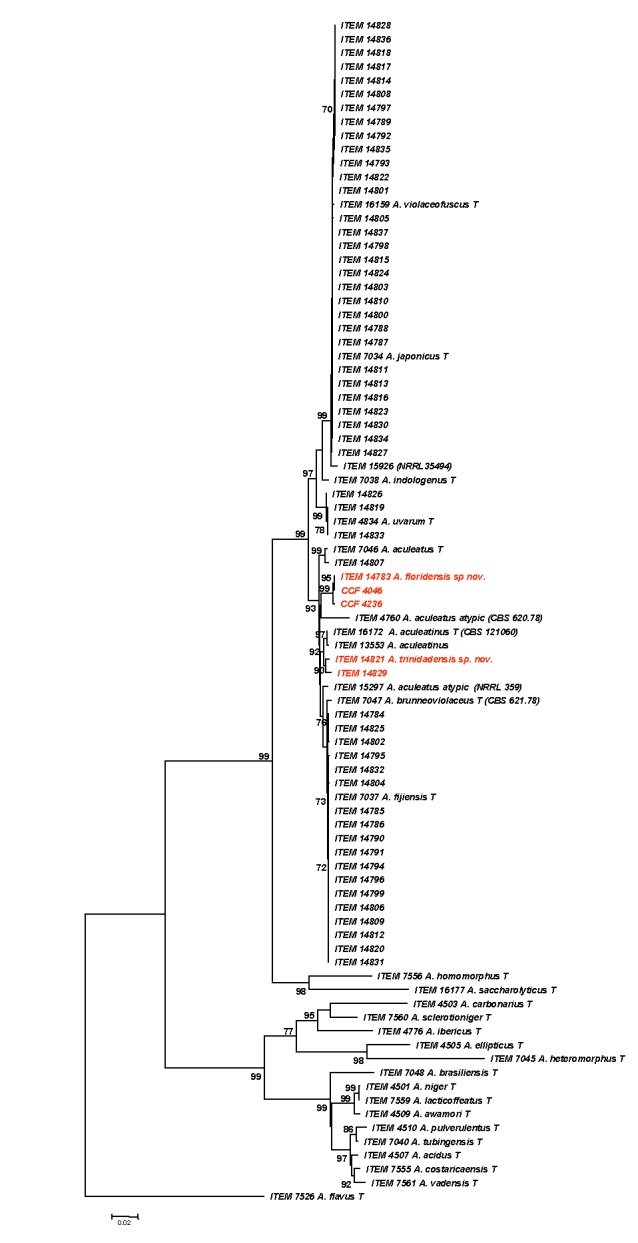
Phylogenetic trees produced from the combined sequence data of four loci (CaM, benA, RPB2 and TEF) of 57 strains of uniseriate black Aspergillus, 28 reference strains of species belonging to Aspergillus section Nigri, and A. flavus (ITEM 7526) as outgroup. Numbers above branches are bootstrap values. Only values above 70 % are indicated. The evolutionary history was inferred using the Neighbour-Joining method computed with the Maximum Likelihood Evolutionary method.
Of the 56 strains collected from air, 30 strains were A. violaceofuscus (syn. A. japonicus), 18 A. brunneoviolaceus (syn. A. fijiensis), three A. uvarum, one (ITEM 14807) was A. aculeatus, two (ITEM 14821 and 14829) were grouped (high bootstrap) in a distinct cluster from A. aculeatinus, and three (ITEM 14783, CCF 4046 and CCF 4236) were phylogenetically isolated (with strong statistical support) from A. aculeatus, and are described as two new species here. In addition, two strains previously characterized by CaM analysis CRI 323-04 (sequence accession number FJ525444, Ingavat et al 2009) and IFM 55703 (sequences from Tetsuhiro Matsuzawa, Chiba University, Japan) resulted to have an homology > 99.5% with ITEM 14783. They also showed a different phylogenetic position from the uniseriate species decribed as A. indologenus (CBS 114.80), A. brunneoviolaceus (ITEM 7047) and from two atypical Aspergillus “aculeatus” (ITEM 4760 and ITEM 15927) strains, not yet well-defined and characterized as belonging in any of the known uniseriate species.
Bayesian inference analysis of the multilocus (benA, CaM, TEF-1α, RPB2) data set produced a phylogenetic tree (log likelihood 14835.94) with high PP values for the same monophyletic group obtained with Maximum Likelihood analysis, the atypical A. aculeatus isolate ITEM 4760 clustered together with ITEM 14873, while the two other atypical A. aculeatus isolates were placed in the A. brunneoviolaceus clade. The results obtained by the Maximum Parsimony analysis are represented by one of the 80 equally most parsimonious trees (Fig. 2). The consistency index is (0.491302), the retention index is (0.863662), and the composite index is 0.466789 (0.424319) for all sites and parsimony-informative sites (in parentheses). The MP phylogenetic analysis agreed with the evolutionary results obtained by the ML and Bayesian analysis. A. violaceofuscus, A. brunneoviolaceus and A. uvarum are the principal black aspergilli “uniseriate” species collected in these indoor air samples with the identification of two possible new species (ITEM 14821 and 14783). The topology of MP, Bayesian phylogenetic trees is concordant, and the two trees are represented together in Fig. 2. The phylogenetic tree obtained by ML analysis has also the same topology of the other two phylogenetic analyses with some minor exception regarding the clades of A. ellipticus and A. heteromorphus (Fig. 1). All three phylogenetic analyses performed give evidence with high bootstrap that the two new species belong to different monophyletic groups (Figs 1–2), and that the atypical strain (ITEM 15297) belongs to the A. brunneoviolaceus clade, and that ITEM 4760 needs further characterization as belonging alone in a clade close to the new species A. floridensis but with no high supported bootstrap (Figs 1–2).
Fig. 2.
Maximum parsimony phylogram derived from the combined sequence data of four loci (CaM, benA, RPB2 and TEF) of 57 strains of uniseriate black Aspergillus, 28 reference strains of Aspergillus section Nigri, and A. flavus (ITEM 7526) as outgroup. Numbers at nodes are bootstrap values/Bayesian posterior probabilities. A dash indicates the support for the branch was < 70 % BP or < 0.80 PP.
TAXONOMY
Previously described species
Aspergillus brunneoviolaceus Batista & Maia, Anais Soc. Biol. Pernambuco 13: 91 (1955).
Synonym: Aspergillus fijiensis Varga et al., Stud. Mycol. 69: 9 (2011).
Fig. 3.
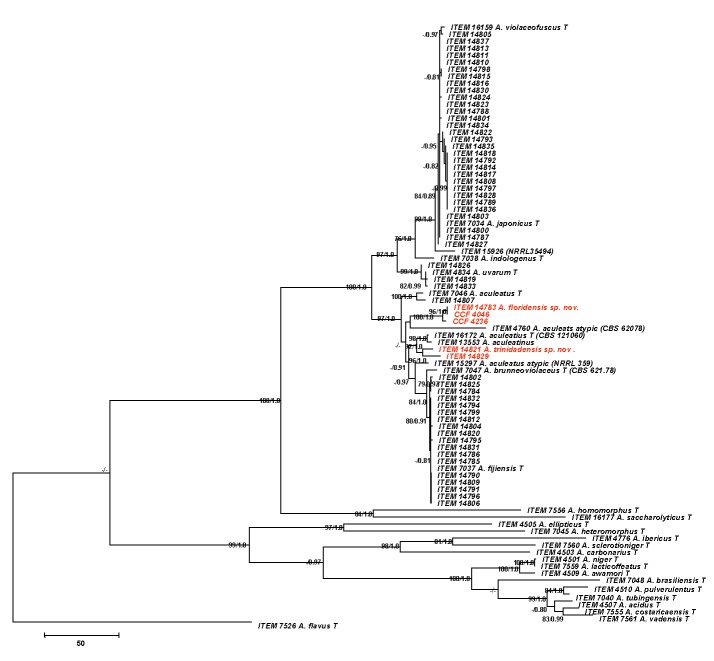
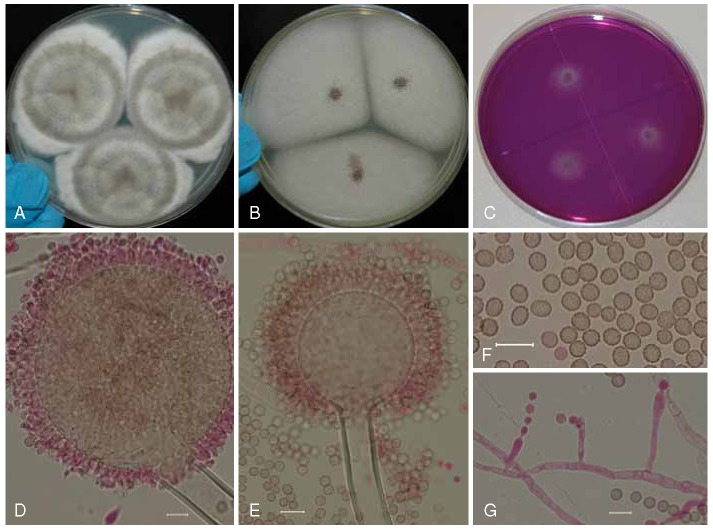
Aspergillus brunneoviolaceus (syn. A. fijiensis; ITEM 7037), culture plates are 9 cm diam, colonies grown at 25 °C for 7 d. A. CYA colonies. B. MEA colonies. C. CREA colonies. D–E. Stipes smooth or with a limited surface granulation just below the vesicle, globose to ellipsoidal vesicle, and conidia. F. Globose to ellipsoidal, conidia, with echinulate surface. Bars = 10 μm.
Type: (CBS 621.78⊤ =NRRL 4912⊤).
Description: Colony diameters after 7 d incubation at 25 °C on CYA (Fig. 3a) > 85 mm (50–75 mm 5 d), MEA (Fig. 3b) 45–75(< 85) mm, CY20S 50–65 mm, OA 55–70 mm, CREA (Fig. 3c) displayed poor sporulation but commonly good to very good acid production, conidial heads on CYA brown to dark brown near black, commonly abundant, velutinous to slightly floccose, white to buff mycelium, commonly moderate radial sulcation, exudate clear to brown, sparse to abundant, soluble pigment not seen, occasionally present and brown at 37 °C, if present sclerotia subglobose to elongate 250–800 μm long, buff to orange-brown, reverse buff to yellow. On MEA conidial heads are brown, sclerotia absent, mycelium white, reverse uncolored to yellowish-gray. Incubation for 7 d on CYA at 5 °C produced no growth or germination of conidia. Incubation for 7 d on CYA at 35 °C and 37 °C produced growth of 35–63 mm, and (12−)17–26 mm diam, respectively.
Stipes (Fig. 3d–e) smooth or with a limited surface granulation just below the vesicle, hyaline or pigmented just below the vesicle, (75−)200–800(−1600) × (8−)10–15(−21) μm, isolate ITEM 7037 has longer stipes (400−)800–2000 (−3400) × (8−)10–15(−18) μm than other A. brunneoviolaceus isolates, vesicles globose to elipsoidal, (30−)35–70(−90) μm diam, conidial heads uniseriate, phialides (6−)7–9(−10) × 3.5–4.5(−5) μm covering entire vesicle, conidia (Fig. 3e) globose to ellipsoidal, 3.5–4.5(−6) × 3.5–4.5(−5) μm, occasionally subglobose to angular 2.5–3.5 μm, brown near black, with coarsely roughened to echinulate surface.
Aspergillus uvarum G. Perrone et al., Int. J. Syst. Evol.Microbiol. 58: 1036 (2008).
MycoBank MB510962.
(Fig. 4a–f).
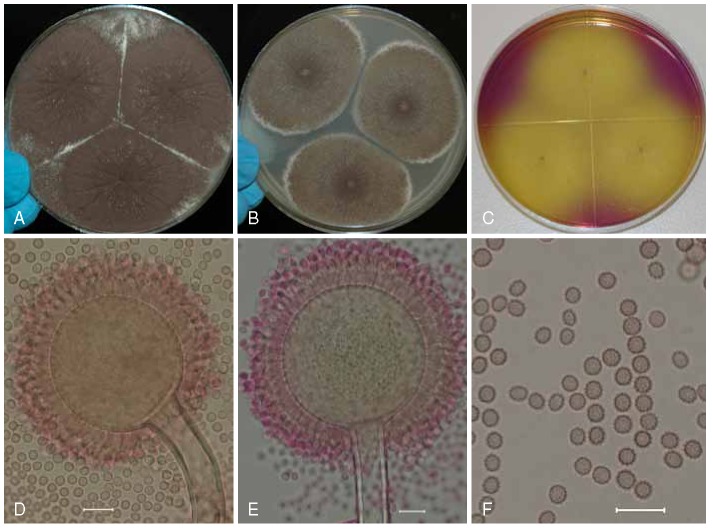
Fig. 4.
Aspergillus uvarum (ITEM 4834⊤), culture plates are 9 cm diam, colonies grown at 25 °C for 7 d. A. CYA colonies. B. MEA colonies. C. OA colony. D–E. Smooth stipes, globose to ellipsoidal vesicle, and conidia. F. Globose to ellipsoidal, conidia, with echinulate surface. Bars = 10 μm.
Type: Italy: Apulia, Brindisi, isol. ex grapes (ITEM 4834⊤; = IMI 388523 ⊤).
Description: Colony diameters after 7 d incubation at 25 °C on CYA (Fig. 4a) > 85 mm (47–88 mm at 5 d), MEA (Fig. 4b) > 85 mm (58–84 mm at 5 d), CY20S 55–65 mm, OA (Fig. 4c) 60-70 mm, CREA produced good growth, acid production ranged from good to very poor to none depending on the isolates, conidial heads on CYA brown to dark brown near black, sporulating abundantly, granular, mycelium white to buff, moderate to deep radial sulcation, exudate clear to brown, soluble pigments when present pale-yellow, brown at 35 °C, at 37 °C rarely present, brown, sclerotia when present abundant in the center of the colony, globose to elongate, buff to brown, reverse wrinkled, white to dull brown occasionally pink-orange. On MEA conidial heads are brown to dark brown, sporulating abundantly, mycelium white and commonly inconspicuous, reverse yellowish-grayish-green. Incubation for 7 d on CYA at 5 °C produced no growth or germination of conidia. Incubation for 7 d on CYA at 35 °C and 37 °C produced growth of 18–27(−46) mm, and (germinate)3–15(−21) mm diam, respectively.
Stipes (Fig. 4d–e) smooth, hyaline, becoming brown with age (250−)600–1800(−3600) × (8−)10–18(−24) μm, vesicles globose to ellipsoidal, (30−)45–100(−121) μm diam, conidial heads uniseriate, phialides 7–10(−12) × (3−)3.5–4.5(−7) μm covering entire vesicle, conidia (Fig. 4f) globose to ellipsoidal, (4−)4.5–7(−9) × 3.5–7 μm, with echinulate surface.
New species
Aspergillus floridensis Ž. Jurjević, G. Perrone & S. W. Peterson, sp. nov.
MycoBank MB802363.
Fig. 5.
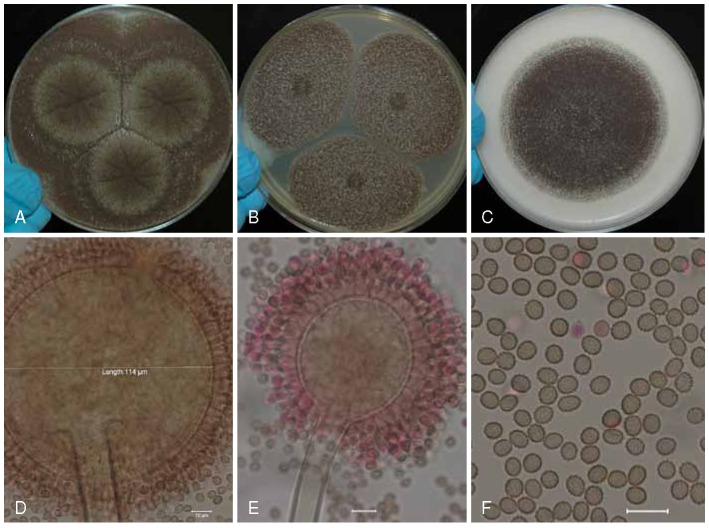
Aspergillus floridensis (ITEM 14783⊤) culture plates are 9 cm diam, colonies grown at 25 °C for 7 d. A. CYA colonies. B. CYA colony, buff-yellowish sclerotia, subglobose to elongate (200−)400–700(−1100) μm long, clear to brown exudates. C. MEA colonies. D. CREA colony. E–F. Smooth stipes, globose to ellipsoidal vesicle, and conidia. G. Globose to ellipsoidal, conidia, with echinulate surface. Bars = 10 μm.
Etymology: Isolated in Florida.
Type: USA: Florida: isol. ex air sample, August 2010, Ž. Jurjević (BPI 883907 – holotype; from dried colonies of ITEM 14783 (=NRRL 62478) grown 7 d at 25 °C on CYA and MEA)
Diagnosis: Stipes uniseriate, mycelium white to yellow on MEA, vesicles globose to subglobose occasionally ellipsoidal (14−)35–65(−105) μm diam, conidia globose to ellipsoidal, 4–5(−6) × 3.5–5.5 μm, with echinulate surface, incubation at 37 °C produced growth of 18–24 mm diam.
Description: Colony diameters after 7 d incubation at 25 °C on CYA (Fig. 5a) 80–85 (> 85) mm, MEA (Fig. 5c) 50–55 (> 85) mm, CY20S 32–53 mm, OA 55–60 mm, CREA (Fig. 5d) displayed poor sporulation but good acid production, conidial heads on CYA dark brown to black, abundantly produced, globose to subglobose at first and later radiate, developing into columns, mycelium white to buff-yellow, velutinous, moderate radial sulcation, exudate clear to brown, sparse to abundant, soluble pigment not seen, occasionally producing buff-yellowish sclerotia (Fig. 5b) subglobose to elongate (200−)400–700(−1100) μm long, reverse brownish-yellow to yellow-brown. On MEA conidial heads are brown, sclerotia absent, mycelium white to yellow, reverse gray to grayish-yellow. Incubation for 7 d on CYA at 5 °C produced no growth or germination of conidia. Incubation for 7 d on CYA at 35 °C and 37 °C produced growth of 40–50 mm and 18–24 mm diam, respectively.
Stipes smooth, hyaline (50−)200–650(−950) × (8−)10–15(−21) μm, vesicles (Fig. 5e–f) globose to subglobose occasionally ellipsoidal (14−)35–65(−105) μm diam, conidial heads uniseriate, phialides (6−)7–9(−11) × 3.5–4.5(−5) μm covering entire vesicle, conidia (Fig. 5g) globose to ellipsoidal, 4–5(−6) × 3.5–5.5 μm, with echinulate surface. No ochratoxin A produced.
Aspergillus trinidadensis Ž. Jurjević, G. Perrone & S. W. Peterson, sp. nov.
MycoBank MB802364.
Fig. 6.
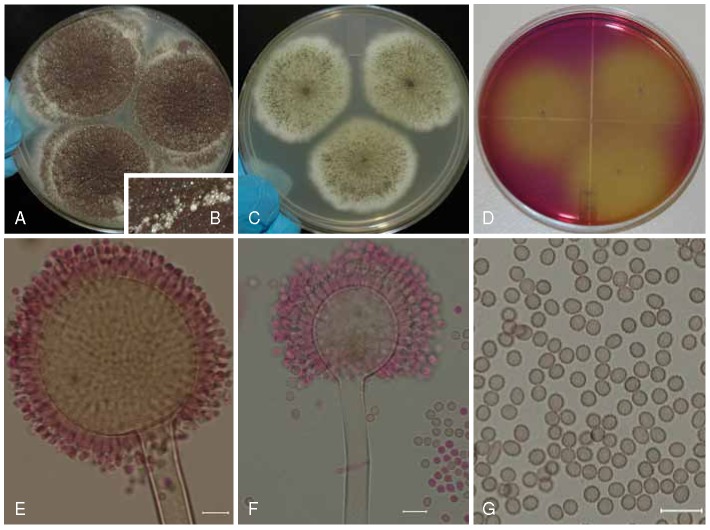
Aspergillus trinidadensis (ITEM 14821⊤) culture plates are 9 cm diam, colonies grown at 25 °C for 7 d. A. CYA colonies. B. MEA colonies. C. CREA colonies. D–E. Smooth stipes, globose to subglobose vesicle, and conidia. F. Globose to ellipsoidal, conidia, with echinulate surface. G. Monophialides and conidia. Bars = 10 μm.
Etymology: Isolated in Trinidad.
Type: Trinidad & Tobago:Tunapuna, isol. ex air sample, July 2011, Ž. Jurjević (BPI 883908 – holotype; from dried colonies of ITEM 14821⊤ (=NRRL 62479⊤) grown 7 d at 25 °C on CYA and MEA).
Diagnosis: Stipes uniseriate, mycelium white to orangish-yellow on CYA, vesicles globose to subglobose occasionally ellipsoidal (10−)30–70(−100) μm diam, conidia large 4–7(−8) × 3.5–7 μm, if borne from monophialides up to 13 × 10 μm, with finely spiny to echinulate surface, and range from no growth to 7 mm diam growth at 37 °C.
Description: Colony diameters after 7 d incubation at 25 °C on CYA (Fig. 6a) 65–78 mm, MEA (Fig. 6b) 57 to > 85 mm, CY20S 50–55 mm, OA 55–60 mm, CREA (Fig. 6c) showed poor sporulation and no acid production, conidial heads on CYA brown to dark brown, globose to subglobose initially, later radiate, then developing into columns, sporulating well, mycelium white to yellowish creamy or orangish-yellow toward the center of the colony, white at margins, floccose, moderate to deep radial sulcation, exudate clear to brownish, soluble pigments and sclerotia not seen, occasionally globose to elongate chlamydospores present, reverse brown to brownish-yellow. On MEA conidial heads brown to dark brown, sporulating well centrally, mycelium white to buff-yellowish-orange, reverse buff. Incubation for 7 d on CYA at 5 °C produced no growth or germination of conidia. Incubation for 7 d on CYA at 35 °C and 37 °C produced growth 4–21 mm and from no growth to 7 mm diam, respectively.
Stipes (Fig. 6d–e) smooth or occasionally with a limited surface granulation just below the vesicle, hyaline or occasionally pigmented just below the vesicle, long if from substrate, short with small vesicles if borne from aerial hyphae (50−)150–800(−1800) × (5−)8–14(18) μm, vesicles globose to subglobose occasionally ellipsoidal (10−)30–70(−100) μm diam, conidial heads uniseriate, phialides (5-)7–9(−12) × (3−)3.5–4.5(−6) μm commonly covering entire vesicle, occasionally producing monophialides (Fig. 6g) 3–42 × 3.5–8 μm, conidia (Fig. 6f) globose to ellipsoidal, rarely pyriform, 4–7(−8) × 3.5–7 μm, if borne from monophialides up to 13 × 10 μm, with finely spiny to echinulate surface. No ochratoxin A produced.
DISCUSSION
In our studies of the indoor environment the dominant species of uniseriate Aspergillus section Nigri were A. violaceofuscus (syn. A. japonicus) (30 of 55 isolates) and A.brunneoviolaceus (syn. A. fijiensis) (18 of 55 isolates). Aspergillus violaceofuscus was isolated from 15 states in the USA, mainly from Southern and Mid-Atlantic states (Table 1). Aspergillus violaceofuscus (syn. A. japonicas) and A. aculeatus have previously only been found in the tropics (Nielsen et al. 2009). Nine isolates (ITEM 14800, 14803, 14805, 14810, 14827, 14828, 14830, 14834, and14837) of the 30 A. violaceofuscus isolates produced sclerotia on CYA or OA, buff to yellowish-orange or orange-brown, subspherical to elongate, 300–1000 μm long. Also, three isolates (ITEM 14794, ITEM 14799, and ITEM 14802) of A. brunneoviolaceus produced abundant buff to orange-brown sclerotia, 250–800 μm long. None of the three sclerotium producing A. brunneoviolaceus isolates produced ochratoxin A. Aspergillus brunneoviolaceus isolates commonly have good to very good acid production. However, two isolates ITEM 14806 and ITEM 14831, did not show acid reactions on CREA agar, nor were they sulcate on CYA. Aspergillus brunneoviolaceus (syn. A. fijiensis) was previously isolated from soil, Fiji (CBS 313.89), Lactuca sativa, Indonesia (CBS 119.49) (Varga et al. 2011), guano, Peru (IHEM 18675), corneal scraping keratitis, India (IHEM 22812), droppings of Coenobita sp., Bahamas (IHEM 4062) (Hendricks at al. 2011), and industrial material, China (CCF 108) (Hubka & Kolarik 2012). This is the first report of A. brunneoviolaceus isolated from the indoor air environment and the first reported isolation in the United States. We found only one isolate of A. aculeatus and three isolates of A. uvarum (Table 1). A. uvarum was previously known only from grapes in the Mediterranean basin (Perrone et al. 2008). This is the first time that A. uvarum was isolated from the indoor air environment and its first isolation in the USA.
A. brunneoviolaceus, A. uvarum, and A. violaceofuscus are the uniseriate black aspergilli occurring in the indoor environment in the USA. The A. brunneoviolaceus clade (Fig. 1) showed the presence of two statistically supported subgroups, one included 15 strains and the ex-type strain of A. fijiensis ITEM 7037, while the other included 3 strain (ITEM 14802, 14825, and 14784). Two strains previously characterized as atypical A. aculeatus (ITEM 7047 – the ex-type strain of A. brunneoviolaceus, and NRRL 359) belong to the same subclade as A. brunneoviolaceus with high bootstrap in all the three phylogenetic analysis conducted (Figs 1–2). These findings confirm the data of Hubka & Kolarik (2012) that suggest treating A. fijiensis as a synonym of A. brunneoviolaceus because they are indistinguishable by multilocus sequence analysis and belong in the same highly supported clade. Then, as A. brunneoviolaceus has been previously described at species level, we suggest treating A. fijiensis as a synonym of it, in agreement with findings of Hubka & Kolarik (2012). The same should be done for A. japonicus and A. violaceofuscus, previously proposed as separate taxa (Varga et al. 2011), as our phylogenetic results do not support this separation and suggest they should be treated as the same taxon; i.e. A. japonicus should be treated as a synonym of A. violaceofuscus which was described earlier.
In the case of the atypical A. aculeatus isolate ITEM 4760, although the molecular difference suggests the possible recognition of further new species, there is no unique topology among the four single locus trees. Two loci indicate it belongs to A. brunneoviolaceus and the other two loci form a clade with the A. floridensis (data not shown). When the combined multilocus alignment was conducted, ML, MP, and PP criteria put it close to A. floridensis (Figs 1–2), but not with a high bootstrap/PP value.
Phenotypically, the atypical A. aculeatus (ITEM 4760) grows slower on CY20S (30 mm diam) and CYA (70-78 mm diam) after 7 d at 25 °C than A. brunneoviolaceus isolates that grow on CYA < 85 mm (50–75 mm diam 5 d), and CY20S 50–65 mm. ITEM 4760 also has slower growth on CYA when compared with Aspergillus floridensis that grows on CYA 80–85(> 85) mm diam after 7 d at 25 °C.
The phylogenetic analysis evidenced both in single locus and in a multilocus analysis showed that the two strains ITEM 14821 and 14829 of A. trinidadensis belong to the A. aculeatinus clade, a black Aspergillus species known only from Thai coffee beans (Noonim et al. 2008). Finally, the newly described A. floridensis was highly supported in both the MP, ML, and Bayesian analysis (Figs 1–2), and in particular the five strains (Table 1) isolated from different world geographic area belonging in the same group by phylogenetic calmodulin analysis (data not shown).
Acknowledgments
We thank Filomena Epifani (ISPA-CNR) for his valuable technical help in growing, DNA extraction, and sequencing of the fungal strains. Frank Robinson (Paul VI High School, Haddonfield, NJ) kindly advised us on Latin usage. J. Varga was partly supported by OTKA grant no. K 84077. Mention of a trade name, proprietary product, or specific equipment does not constitute a guarantee or warranty by the United States Department of Agriculture and does not imply its approval to the exclusion of other products that may be suitable. USDA is an equal opportunity provider and employer.
REFERENCES
- Abarca ML, Accensi F, Bragulat MR, Castella G, Cabanes FJ. (2003) Aspergillus carbonarius as the main source of ochratoxin A contamination in dried wine fruits from the Spanish market. Journal of Food Protection 66: 504–506 [DOI] [PubMed] [Google Scholar]
- Abarca ML, Accensi F, Cano J, Cabanes FJ. (2004) Taxonomy and significance of black aspergilli. Antonie van Leeuwenhoek 86: 33–49 [DOI] [PubMed] [Google Scholar]
- Abarca ML, Bragulat MR, Castella G, Cabanes FJ. (1994) Ochratoxin A production by strains of Aspergillus niger var. niger. Applied and Environmental Microbiology 60: 2650–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabanes FJ, Accensi F, Bragulat MR, Abarca ML, Castella G, Minguez S, Pons A. (2002) What is the source of ochratoxin A in wine? International Journal of Food Microbiology 79: 213–215 [DOI] [PubMed] [Google Scholar]
- de Hoog GS, Guarro J, Figueras MJ, Gené J. (2000) Atlas of Clinical Fungi. Baarn: Centraalbureau voor Schimmelcultures; [Google Scholar]
- Felsenstein J. (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791 [DOI] [PubMed] [Google Scholar]
- Gams W, Christensen M, Onions AHS, Pitt JI, Samson RA. (1985) Infrageneric taxa of Aspergillus. In: Advances in Penicillium and Aspergillus Systematics (Samson RA, Pitt JI, eds): 55–64 New York: Plenum Press; [Google Scholar]
- Glass NL, Donaldson GC. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61: 1323–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx M, Beguin H, Detandt M. (2012) Genetic re-identification and antifungal susceptibility testing of Aspergillus section Nigri strains of the BCCM⁄IHEM collection. Mycoses 55: 148–155 [DOI] [PubMed] [Google Scholar]
- Hubka V, Kolarik M. (2012) β-tubulin paralogue tubC is frequently misidentified as the benA gene in Aspergillus section Nigri taxonomy: primer specificity testing and taxonomic consequences Persoonia 29: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Ingavat N, Dobereiner J, Wiyakrutta S, Mahidol C, Ruchirawat S, Kittakoop P. (2009) Aspergillusol A, an alpha-glucosidase inhibitor from the marine-derived fungus Aspergillus aculeatus. Journal of Natural Products 72: 2049–2052 [DOI] [PubMed] [Google Scholar]
- Klich MA. (2009) Health effects of Aspergillus in food and air. Toxicology and Industrial Health 25: 657–667 [DOI] [PubMed] [Google Scholar]
- Knutsen AP. (2011) Immunopathology and immunogenetics of allergic bronchopulmonary aspergillosis. Journal of Allergy: doi:10.1155/2011/785983 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Liu YL, Whelen S, Hall BD. (1999) Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution 16: 1799–1808 [DOI] [PubMed] [Google Scholar]
- Moss RB. (2002) Allergic bronchopulmonary aspergillosis. Clinical Reviews in Allergy and Immunology 23: 87–104 [DOI] [PubMed] [Google Scholar]
- Nei M, Kumar S. (2000) Molecular Evolution and Phylogenetics. New York: Oxford University Press; [Google Scholar]
- Nielsen KF, Mogensen JM, Johansen Maria, Larsen TO, Frisvad JC. (2009) Review of secondary metabolites and mycotoxins from the Aspergillus niger group. Analytical and Bioanalytical Chemistry 395: 1225–1242 [DOI] [PubMed] [Google Scholar]
- Noonim P, Mahakarnchanakul W, Varga J, Frisvad JC, Samson RA. (2008) Two novel species of Aspergillus section Nigri from Thai coffee beans. International Journal of Systematic and Evolutionary Microbiology 58: 1727–1734 [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Nirenberg HI, Aoki T, Cigelnik E. (2000) A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience 41: 61–78 [Google Scholar]
- Perrone G, Varga J, Susca A, Frisvad JC, Stea G, et al (2008) Aspergillus uvarum sp. nov., an uniseriate black Aspergillus species isolated from grapes in Europe. International Journal of Systematic and Evolutionary Microbiology 58: 1032–1039 [DOI] [PubMed] [Google Scholar]
- Perrone G, Stea G, Epifani F, Varga J, Frisvad JC, Samson RA. (2011) Aspergillus niger contains the cryptic phylogenetic species A. awamori. Fungal Biology 115: 1138–1150 [DOI] [PubMed] [Google Scholar]
- Perrone G, Epifani F, Rambukwelle K, Parahitiyawa N, Wijedasa H, Arseculeratne SN. (2012a) Aspergillus aculeatinus n. sp. in chronic human dacryocystitis; the first report. Journal of Infectious Disease and Antimicrobial Agents 29: 89–97 [Google Scholar]
- Perrone G, Stea G, Kulathunga CN, Wijedasa H, Arseculeratne SN. (2012b) Aspergillus fijiensis isolated from broncial washings in a case of bronchiectasis with invasive aspergillosis; the first report of probable pathogenicity. Journal of Infectious Disease and Antimicrobial Agents (in press). [Google Scholar]
- Pitt JI. (1980) [“1979”] The Genus Penicillium and its teleomorph states Eupenicillium and Talaromyces. London: Academic Press; [Google Scholar]
- Pitt JI, Hocking AD. (2007) Fungi and Food Spoilage. 2rd edn London: Blackie; [Google Scholar]
- Pitt JI, Hocking AD. (2009) Fungi and Food Spoilage. 3rd edn London: Springer; [Google Scholar]
- Raper KB, Fennell DI. (1965) The Genus Aspergillus. Baltimore, MD: Williams & Wilkins; [Google Scholar]
- Richardson MD. (2005) Aspergillosis. In: Topley & Wilson’s Medical Mycology. (Merz WG, Hay RJ, eds): 687–738 10th edn London: Hodder Arnold; [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Sage L, Garon D, Seigle-Murandi F. (2004) Fungal microflora and ochratoxin A risk in French wineyards. Journal of Agricultural and Food Chemistry 52: 5764–5768 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Samson RA, Hoekstra ES, Frisvad JC. (2004) Introduction to Food- and Airborne Fungi. 7th edn Utrecht: Centraalbureau voor Schimmelcultures; [Google Scholar]
- Samson RA, Noonim P, Meijer M, Houbraken J, Frisvad JC, Varga J. (2007) Diagnostic tools to identify black Aspergilli. Studies in Mycology 59: 129–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution 10: 512–526 [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011) MEGA5: Molecular evolutionary genetics analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony methods. Molecular Biology and Evolution 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. (2000) Phylogenetic species recognition and species concepts in fungi. Fungal Genetics and Biology 31: 21–32 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J, Kevei F, Hamari Z, Tóth B, Téren J, Croft JH, Kozakiewicz Z. (2000) Genotypic and phenotypic variability among black aspergilli. In: Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification (Samson RA, Pitt JI, eds): 397–411 Amsterdam: Harwood Academic Publishers; [Google Scholar]
- Varga J, Frisvad JC, Kocsubé S, Brankovics B, Tóth B, Szigeti G, Samson RA. (2011) New and revisited species in Aspergillus section Nigri. Studies in Mycology 69: 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicklow DT, Dowd PF, Alftafta AA, Gloer JB. (1996) Ochratoxin A: an antiinsectan metabolite from the sclerotia of Aspergillus carbonarius NRRL 369. Canadian Journal of Microbiology 42: 1100–1103 [DOI] [PubMed] [Google Scholar]