Abstract
Endogenous retroviruses (ERVs) are long terminal repeat-containing virus-like elements that have colonized approximately 10 per cent of the present day mammalian genomes. The intracisternal A particles (IAPs) are a class of ERVs that is currently highly active in the rodents. IAP elements can influence the transcription profile of nearby genes by providing functional promoter elements and modulating local epigenetic landscape through changes in DNA methylation and histone (H3K9) modifications. Despite the potential role for IAPs in gene regulation, the precise genomic locations where these elements are integrated are not well understood. To address this issue, we have identified more than 400 novel IAP insertion sites within/near annotated genes by searching the murine genome, which suggests that the impact of IAP elements on local and/or global gene regulation could be more profound than was previously expected. On the basis of our independent analyses and already published reports, here we argue that IAPs and ERV elements in general could have an evolutionary role for modulating phenotypic plasticity upon environmental inputs, and that this could be mediated through specific stages of embryonic development such as placentation during which the epigenetic constraints on IAP elements are partially relaxed.
Keywords: retrotransposons, intracisternal A particles, DNA methylation, environmental cues, adaptive response
1. Introduction
Transposable elements (TEs) are discrete DNA sequences that can jump around the genome and multiply by making their own copies. TEs are present in almost all species that have been analysed upto today, which suggests that such mobile elements could be an integral part of most, if not all, life forms. Indeed, approximately half of our own genome is made up of repetitive DNA that originates from various TEs, an observation that is in stark contrast with the meagre amount of genomic regions that code for exons (approx. 1.5%) [1].
The significance of TEs first became clear from the seminal studies performed by Barbara McClintock in maize, where she showed that the variegated colour pattern in maize kernels arose due to the activity of specific classes of TEs termed as Ds (Dissociation) and Ac (Activator) [2]. Because the insertion of a TE (Ds) could modulate neighbouring gene transcription and thus could control its expression pattern, McClintock coined the word ‘controlling elements’ to describe mobile DNA. Since then, considerable interest has been focused into the study of TEs to understand their distribution in the genome, the mechanisms that regulate these elements and their biological significance. TEs are now generally classified into two main groups, namely, class I and class II, according to their structure and mobilization patterns [3].
Class II TEs comprise the DNA transposons, which move within the host genome through a ‘cut and paste’ mechanism. However, it is widely accepted that at present DNA transposons are all but extinct in humans [1]. For this reason, we have excluded class II TEs from the present study. The long terminal repeat (LTR) and non-LTR retrotransposons, which make up the class I compartment, expand in numbers through a ‘copy and paste’ mechanism. In other words, these elements transcribe their own RNAs (copy) and followed by reverse transcription (RNA to cDNA) integrate (paste) into new sites in the host genome. Non-LTR retrotransposons such as long interspersed nuclear element (LINE) L1 sequences have been highly successful in invading human and mouse genomes. It is estimated that there could be as many as 500 000 L1 elements in either of these species [4]. LTR-containing retrotransposons such as the endogenous retroviruses (ERVs) have also undergone considerable expansion in mammals (for a detailed classification of murine ERVs, see [5]). It has been predicted that there are roughly 100 000 HERVs that comprise approximately 8 per cent of the human genome [1,6]. However, in rodents, the number of ERVs seems to be relatively restricted. For example, in case of intracisternal A particles (IAPs), which is the most active ERV class in mouse, 1000–2500 copies could be present in a haploid genome [7]. Interestingly, although the number of the IAP elements is much smaller that the L1 retrotransposons, these retrotransposons could be more active in the mouse.
A recent study has estimated that there could be at least 32 previously identified germline mutations caused by IAPs in mice, which account for roughly 5 per cent of all such incidents reported so far [8]. These include two well analysed loci, namely, the agouti viable yellow (AVY) and the Axin-fused (AxinFU) alleles, which arise from IAP insertions in the promoter and intronic regions of the agouti and the Axin loci, respectively [9–11]. Importantly, it has been noted that in both these mice, the expression patterns of the IAP elements are controlled by a repressive epigenetic mechanism (i.e. DNA methylation). Furthermore, it has been shown that the hypomethylated phenotypes observed in the offspring of agouti viable yellow and Axin-fused mice could be rescued by feeding methyl-donor supplemented diet to pregnant dams, suggesting the possibility that IAPs might function as potential sensors for environmental (in this case, nutritional) cues during gestation [11–13].
The above observations suggest that IAPs could be an interesting model for studying the biological roles of retrotransposons in mammals, because: firstly, they have a higher propensity to cause mutations in the genome; secondly, aberrant expression of IAP elements can lead to gross phenotypic aberrations in mice; thirdly, IAPs are linked with well-defined epigenetic regulatory mechanisms; and, finally, IAPs could function as key players to mediate environmental/developmental signals during development. In addition, although many excellent reviews have summarized the roles of non-LTR retrotransposons [14] and ERVs [8] in mammals, IAPs have remained mostly unexplored except for a limited number of previous studies [15–17].
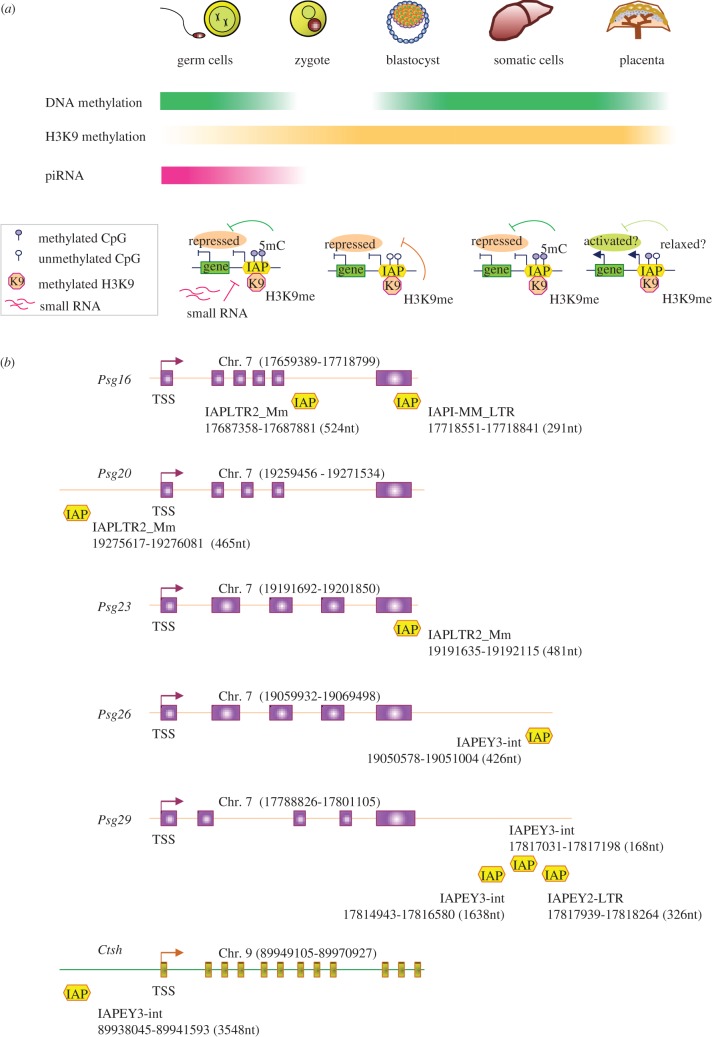
To understand how IAPs could influence the expression of nearby genes, we have performed bioinformatics analysis using the murine genome (mouse genome assembly GRCm38) as a reference and identified more than 400 gene loci that show insertion of IAPs in the intron, exon, upstream (within 5 kb of transcription start site) and downstream (within 5 kb of transcription termination site) regions (see the electronic supplementary material, table S1). In addition, we have summarized the epigenetic regulation pattern of IAP retrotransposons in various tissues including embryonic stem (ES) cells and somatic cells, and based on these observations argued that the environment in the placenta could be more permissible for IAP expression because the level of DNA methylation, which is the major repressor of IAP elements in somatic cells, could be relatively low in the extraembryonic tissues (figure 2a). Further, to support our hypothesis, we have identified IAP insertions within/near multiple placenta-specific genes such as several members of the Psg (pregnancy-specific glycoprotein) cluster (Psg16, Psg20, Psg23, Psg26 and Psg29) and cathepsin H (Ctsh) (figure 2b) [18]. Although all eutherian mammals possess placentas, the structure and the functions of this organ could be dramatically different between different mammalian classes. We have hypothesized that ERVs, because of their species-specific evolutionary origins (for example, HERVs have evolved specifically in humans, while murine ERVs are found only in rodents but not in humans), could have a role to create such divergence of placental functions among different mammalian species. Lastly, we have argued that ERVs could be better suited for creating phenotypic plasticity [19] (i.e. variance in response to a particular environmental cue) because of the species restricted evolution patterns that we have mentioned above.
Figure 2.
Potential roles for IAPs in the placenta. (a) Tissue-specific regulation of the IAP elements is shown in a schematic figure (top panel). The DNA methylation level is higher in the germ and somatic cells (deep green, top bar) but is erased in the early embryo (colourless). The global level of DNA methylation in the placenta (light green) is lower than the somatic and germ cells. H3K9 methylation levels are demonstrated in orange (middle bar). The small RNA pathway represses IAPs in the germ cells (dark pink, bottom bar) but presumably not in the early embryo or somatic cells (colourless). The bottom panel delineates a putative model which shows how activation (or repression) of an IAP element can influence the transcriptional level of a nearby gene. IAP elements are repressed by silencing mechanisms such as DNA methylation (5mC), histone H3K9 methylation (K9) and small RNAs, which also blocks the activation of the nearby gene. Releasing the constraint on the IAP element (i.e. partial demethylation) causes derepression of both the IAP and the nearby gene. (b) Schematic illustrating the approximate positions of the IAP sequences within or near placenta-specific genes. The pregnancy-specific glycoprotein (Psg) genes (Psg16, Psg20, Psg23, Psg26, Psg29) and cathepsin H (Ctsh) are shown. Transcription start site (TSS) for each gene is shown with an arrow. The chromosome number and the start/end of the gene loci are given at the top of each figure. IAP insertions are denoted with yellow boxes and the type, start/end, length of each individual IAP sequence are exhibited beside a box.
In the following sections of this work, we will discuss and elaborate on these themes.
2. Detection of IAP elements in the genic regions of the murine genome
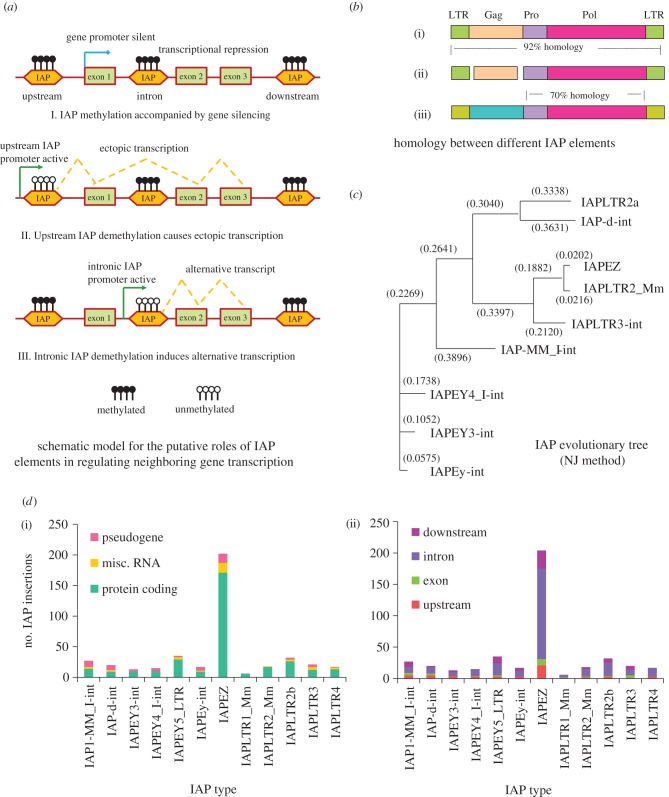
ERVs possess their own promoters, coding regions and splicing sites. Because of these characteristic features, insertion of an ERV element within a genomic locus could affect the transcription pattern of nearby genes by providing de novo promoter elements and alternative splicing sites (figure 1a). IAPs are a highly active class of rodent-specific retroelements and it has previously been estimated that there could be 1000–2500 sites in the murine genome in which these elements have integrated during the course of evolution [7]. Previous studies have shown that the insertion of an IAP element in the upstream region could be sufficient to disrupt the normal transcription profile of a gene and place it under the control of the IAP-derived LTR promoter (see figure 1a, II) [9]. It has also been demonstrated that intronic IAP sequences can cause alternative splicing of a gene (figure 1a, III) [10]. In addition, as IAP retrotransposons are rigorously targeted by repressive epigenetic mechanisms, their insertions within genic loci could cause induction of local heterochromatinization that might in turn suppress nearby genes (shown in figure 1a, I) [20].
Figure 1.
Characterization of IAPs present in the mouse genic regions. (a) Schematic model to show potential IAP-mediated pathways for regulating transcription of nearby genes: (I) demonstrates the general effect of IAP insertion into a genic region. IAP elements heavily attract repressive epigenetic machineries such as DNA methylation (methylated DNA is shown with filled lollipops), which could in turn silence the transcription of nearby genes. In contrast, demethylation of an upstream IAP element could cause ectopic transcription of a gene that originates from a putative promoter element within the IAP sequence ((II); unmethylated CpG sites are shown with open lollipops); (III) exhibits the scenario where demethylation of an intronic IAP sequence could lead to ectopic splicing events. (b) Homology search performed between three IAP elements, namely, IAPLTR2_Mm (i), IAPEZ (ii) and IAPLTR3-int (iii). The IAPLTR2_Mm and IAPEZ sequences are highly similar (92% homology), while the IAPLTR3-int element exhibits approximately 70 per cent homology in the putative Pro and Pol coding regions but not in the Gag locus or the flanking LTR sites. (c) Evolutionary tree constructed by the neighbour joining method for different IAP elements used in this study. The IAPs are clustered into three putative groups, the first comprising IAPLTR2a and IAP-d-int, the second consisting of IAPLTR2_Mm, IAPEZ and IAPLTR3-int, and the last group made up of the remaining four IAP sequences (IAPEy-int, IAPEY3-int, IAPEY4_I-int and IAP-MM_I-int). (d) Bar plots showing the type of genes associated with respective IAP elements: (i) The vertical axis represents the number of insertion sites and the horizontal axis demonstrates the IAP families. The majority of the inserts were linked with protein coding genes, the rest comprising RNA-associated loci and pseudogenes. IAPEZ class of elements were the most abundant, exhibiting more than 200 inserts (approx. half of the sites identified in this study). (ii) Shows the detailed positions of the IAP insertions. Intronic insertions were predominant for all the IAP sequences.
Because of the potential insertional effects of the IAP retrotransposons on neighbouring genes, it is essential to know the genomic positions where these elements are integrated. However, although a limited number of previous studies have examined the distribution pattern of IAPs in the genome [15,17], detailed mapping of these elements have not been performed. To address this question, we retrieved 12 IAP-associated sequences from the Ensembl mouse database (http://www.ensembl.org/Mus_musculus/Info/Index) (also see electronic supplementary material, table S2 for detailed list of the sequences). We aligned these sequences against the mouse GRCm38 reference genome using the Ensembl BLAST/BLAT software (http://www.ensembl.org/Mus_musculus/blastview) and extracted the hits that were located within (exon, intron) or near (up to 5 kb from the transcription start or termination sites) annotated genes. We could identify more than 400 sites that fulfilled the above criteria (see figure 1d and electronic supplementary material, table S1). Our analysis revealed that the IAPEZ class of IAPs was the most abundant elements found within/near the murine genes exhibiting approximately 200 insertions. In contrast, insertions for the other 11 IAP sequences were less frequent, the number of hits ranging from six (IAPLTR1_Mm) to 35 (IAPEY5_LTR). The larger IAP elements (5–8 kb in length), such as IAPEZ, IAPLTR2_Mm and IAPLTR3-int, contained double LTRs in both 5′ and 3′ flanking regions (data not shown). However, we noticed that more than one third (approx. 70 inserts) of the IAPEZ sequences did not possess any LTRs. It is not clear whether the LTR flanking regions were lost during the original integration event or were deleted through negative selection during evolution. For other IAP classes such as IAP1-MM_I-int, IAP-d-int and IAPEy-int, most inserts showed only a single LTR and the average length of these elements were also smaller (2–5 kb, data not shown). In contrast, the IAPLTR4, IAPLTR3, IAPLTR2b, IAPLTR1_Mm and IAPEY5_LTR sequences were present as solitary LTRs exhibiting an average length around 300 nucleotides. As it is generally believed that the promoter activity of the IAP elements are mainly regulated by the flanking LTR sequences, it should be interesting to examine whether there are some functional differences between the double LTR, single LTR and LTR-less IAP sequences.
For most IAP families, more than half of inserts were associated with introns. For example, in the case of IAPEZ sequences about 70 per cent (144 out of 202) of the hits were located within introns. Approximately 20 per cent of the IAPs exhibited integration in the downstream (up to 5 kb from the transcription termination site) and approximately 15 per cent were enriched in the upstream (up to 5 kb from the transcription start site) regions. In contrast, only a small percentage (approx. 6%) of the IAPs overlapped with exons (see figure 1d (ii)). Insertion of retrotransposon-associated sequences within exons might disrupt protein coding genes that are essential for cellular functions, and to avoid such events exonic IAP insertions might have undergone purifying selection (i.e. selective removal of the alleles that are deleterious for the host). To gain insight into the type of genes that the IAP elements are associated with, we categorized the hits into protein coding genes, miscellaneous RNA genes and pseudogenes. We observed that most of the IAPs were linked with protein coding genes. For example, 85 per cent of the IAPEZ inserts (171 out of 202) were found within/near such genes. Approximately 10 per cent of the IAPs were located in RNA (long non-coding RNAs: LincRNAs, antisense RNAs, etc.) coding loci and the rest were associated with pseudogenes. The percentage of pseudogene-related IAPs were relatively higher (approx. 35%) for four smaller IAP classes, namely, IAP1-MM_I-int, IAPEY3-int, IAPEY4_I-int and IAPEy-int (figure 1d (i)).
To understand how these different IAP sequences diverged during the evolution of the mouse genome, we undertook an evolutionary tree analysis using the neighbour joining (NJ) method proposed by Saitou & Nei [21]. This analysis revealed that the IAP elements could roughly be categorized into three groups, the first containing IAPLTR2a and IAP-d-int sequences, the second comprising IAPEZ, IAPLTR2_Mm and IAPLTR3-int families and the last being enriched for IAP1-MM_I-int, IAPEY3-int, IAPEY4_I-int and IAPEy-int elements (figure 1c). The evolutionary distance for each element in the tree is given within parentheses. Interestingly, we noted that the IAP sequences clustered in the last group were the ones that exhibited abundance for pseudogene associations (i.e. IAP1-MM_I-int, IAPEY3-int, IAPEY4_I-int and IAPEy-int, and see figure 1c,d). Clustering different IAP families according to their evolutionary history might be useful to gain insight into their functional roles in the murine genome.
We tested the degree of sequence similarity among similar families of IAPs by aligning IAPEZ, IAPLTR2_Mm and IAPLTR3-int, which were clustered near each other in the evolutionary tree (figure 1b). To this end, we used the Lalign web browser (http://www.ch.embnet.org/software/LALIGN_form.html) and found that IAPEZ and IAPLTR2 were the most homologous (approx. 92%), which was in harmony with the close proximity of these sequences revealed by the NJ method. However, the other sequence, namely IAPLTR3-int, showed only partial similarity to IAPEZ and IAPLTR2, the putative Pro and Pol regions being the most homologous (approx. 70% match, see figure 1b). This suggested that there could be a certain degree of divergence between the various IAP families present in the murine genome today.
Lastly, we undertook gene ontology (GO) analyses to examine whether the IAP-associated genes were enriched for specific functions or pathways. For this purpose, we used the g:Profiler (http://biit.cs.ut.ee/gprofiler/) web resource and performed tests to obtain GO terms that showed significant enrichments for all IAPs or a specific class of IAPs. Surprisingly, genes linked with all IAPs did not show any significant association with any GO terms, suggesting that the enrichment of IAPs into genic loci could be either highly random (not showing preference for a certain group of genes) or there might be some complex pattern in the interactions between IAPs and the host genes that could not be clearly discerned by using conventional techniques such as GO. However, when we broke down the list of genes according to specific IAP families, we found statistically significant GO enrichment such as ‘amino acid binding (GO:0016597)’ (p-value 7.22 × 10−3) and `glutamate binding (GO:0016595)’ (p-value 9.11 × 10−3) for IAPLTR4 linked genes. IAPEY5_LTR, IAPEy-int and IAPLTR2_Mm families also showed enrichment for GO-associated terms (see the electronic supplementary material, table S3).
Collectively, in this study, we have shown by using genomewide mapping analysis that the possible incidences of IAP integration into a genic locus could be abundant (more than 400 sites). It is tempting to hypothesize that at least some of these insertions might have functional effects to regulate local gene transcription and epigenetic landscape. Detailed investigation of the IAP elements in the murine genome might provide novel insights into the complex relationship between the retrotransposons and their hosts, and how these interactions might have shaped the present day mammalian genome.
3. Epigenetic regulation patterns of IAPs differ among tissues
As other retrotransposons, IAPs are generally maintained in the silenced state by repressive epigenetic modifications (reviewed in [8]). A major contributor for IAP silencing is DNA (5-cytosine) methylation. In vitro methylation of the IAP LTR inhibits its promoter activity [22]. Treating murine cells with the (DNA) hypomethylating agent 5-azacytidine releases the repression of endogenous IAP elements [23]. Furthermore, deletion of the maintenance DNA methyltransferase enzyme Dnmt1 causes a dramatic (50–100-fold) upregulation of IAP elements in early mouse embryos [24]. In addition, IAPs are demethylated and upregulated in Dnmt3L (a germ cell-specific DNA methyltransferase-like protein) deficient murine male germ cells [25]. However, in this case, the level of activation for the IAP elements appears to be around 4–6-fold, which is much lower than the drastic upregulation observed in Dnmt1−/− murine embryos [25]. This could be owing to the fact that although the 5′ region of the IAP LTR is extensively demethylated in these mice, the 3′ area mostly retains DNA methylation. IAP methylation is not grossly affected in Dnmt3a or Dnmt3b (methyltransferases associated with de novo methylation activity) knocked out male germ cells [25]. In our previous study, we have reported that deletion of Np95/Uhrf1, which is the recruiter of Dnmt1 protein into hemimethylated DNA, causes extensive demethylation of the IAP elements in mice [26]. However, we observed only about a sixfold increase of IAP transcription in Np95−/− embryos, which is much lower than that of Dnmt1−/− foetuses. The reason for this discrepancy is not yet understood. Deletion of the Mbd1 (methyl-CpG-binding domain protein 1) gene does not elicit strong derepression of IAPs. In the hippocampus of the Mbd1−/− mice only weak derepression (approx. twofold) is observed [27]. Taken together, although DNA methylation plays a major role in repression of the IAP elements, the detailed mechanism by which this is mediated is not well understood.
Several germ cell-specific factors have a role for silencing IAP retrotransposons in the germline. The male germ cell-specific protein Gtsf1 (gametocyte specific factor 1), a molecule that is specifically expressed in the cytoplasm of male and female germ cells during development, is required to maintain IAP methylation and transcriptional silencing [28]. In addition, male germ cells lacking the Mili and Miwi proteins exhibit demethylation and activation of the IAP elements [29]. The mitochondrial protein MITOPLD has a role for maintaining the DNA methylation of specific IAP elements, but not the bulk of other IAPs. Probably for this reason, the deletion of this protein does not greatly affect the overall expression of IAPs in germ cells [30]. The Mili, Miwi and MITOPLD proteins are associated with the piRNA biogenesis pathway in mouse, which indicates that IAPs are a target of the germline-specific defence mechanism-mediated small RNAs. Indeed, a recent study has reported that many piRNAs generated in the germline bear high sequence homology with known IAP elements [31], and, therefore, could function to silence IAPs in germ cells. A previous paper reported that IAP sequences are more likely to be activated in the germ cells than somatic tissues [32]. Transgenic mice harbouring LacZ alleles that were placed under the control of a full length IAP element, an IAP promoter or a solo IAP-LTR, exhibited LacZ expression patterns specifically in male germ cells. Therefore, the antagonistic nature of germline-specific factors and piRNAs against IAPs could be a mechanism by which the host genome holds the IAP elements in check during germ cell development and reprogramming.
We have previously shown that deletion of Setdb1/ESET, an H3K9me3 enzyme that mainly targets the euchromatinic regions in mice, causes drastic derepression of IAPs and other endogenous retrotransposons in murine ES cells [33]. In addition, we previously noted that knockout ES cells for the H3K9me2 methyltransferase G9a exhibits gross hypomethylation in the IAP loci but this does not lead to the activation of these elements [34]. Kap1/trim28, a KRAB-zinc finger repressor protein that binds methylated H3K9 residues, has a critical role to repress IAP elements in the early embryo [35]. Knocking out this gene in mice causes 1000-fold derepression of IAPs in the early embryo and nearly 100-fold activation in ES cells. Although H3K9 trimethylation plays a vital role for IAP repression in the early embryo, it seems that this pathway is dispensable in somatic cells because the loss of Setdb1 in mouse embryonic fibroblast (MEF) cells does not result in derepression of IAP retrotransposons [33].
The above reports demonstrate that IAPs are silenced by not one but multiple mechanisms which could act in tissue-specific manners (see figure 2a). For example, in ES cells repression of IAPs are mediated by both DNA methylation and H3K9me3-dependent pathways, while in somatic cells the latter appears to be dispensable. In addition, piRNA-directed silencing is required along with DNA methylation in the germline. DNA methylation undergoes drastic changes during specific stages of development. The global DNA methylation patterns in the paternal and maternal genomes are erased after fertilization and are re-established in the inner cell mass (ICM) of the blastocyst by the activity of the de novo methyltransferase enzymes (reviewed in [36]). The H3K9 trimethylation-mediated silencing pathway might function to maintain the IAP elements in silenced state during this early developmental window when global DNA methylation is reprogrammed (reviewed in [37]). It is interesting to note that although the genomic DNA methylation levels remain constantly high in the embryo proper throughout development (once it is rewritten in the blastocyst), the extraembryonic tissues exhibit lower amount of DNA methylation, a phenomenon that appears to have a critical role in the embryonic and extraembryonic lineage commitment process (reviewed in [38]). Recent studies have further shed light on this issue by showing that the observed undermethylation in the extraembryonic lineage could be noted mostly in the intergenic and repeat regions, rather than displaying a pervasive pattern all over the genome [39]. The extraembryonic cells give rise to the trophoblast and the placenta, which are the organs essential for nutrient and gas transfer from the maternal to the foetal interface in the eutherian mammals. It should be, therefore, important to understand whether the relatively lower DNA methylation status in the placenta is more permissible for expression of retrotransposons such as the IAPs.
However, as we have mentioned above, DNA methylation is not the only mechanism that silences IAPs, because it has been shown that the H3K9 trimethylation enzyme Setdb1 has a role to repress IAPs at least in ES cells [33]. Therefore, although the decrease of DNA methylation should be a major cue for tissue-specific upregulation of these elements, similar effects could also be observed if there is loss of H3K9 methylation. This topic should be examined in future to elucidate how the IAP elements are silenced/expressed by the establishment/loss of H3K9 methylation during development.
4. Potential roles for IAPs in shaping placenta-specific transcriptional landscape
Investigation in 10 different rodent tissues revealed that the IAP retrotransposons are expressed only in the placenta but not in other tissues, which suggested that the epigenetic environment of the placenta could allow IAP expression [40]. Indeed, IAP insertion affecting gene expression in the placenta has been reported in multiple studies. Integration of an IAP LTR in the 5′ regions of the MIPP gene (now known as Ipp: IAP promoted placental gene) has been linked with abundant transcription of this transcript in the placenta but not in other tissues [20]. A similar case has been reported for the pregnancy-specific glycoprotein 23 (Psg23) gene [41]. In addition to Psg23, we have newly identified IAP-associated sequences near/inside four other Psg genes, namely, Psg16, Psg20, Psg26 and Psg29 (figure 2b). Detailed analysis revealed that there were two IAP solo-LTR insertions within the Psg16 gene, one in the exon (IAPI-MM_LTR, 291nt) and the other in the intron (IAPLTR2_Mm, 524nt). An IAP LTR (IAPLTR2, 465nt) was found approximately 4 kb upstream of the Psg20 transcription start site, while three IAP-related sequences (IAPEY3-int, 1638nt; IAPEY3-int, 168nt; IAPEY2-LTR, 326nt) were identified about approximately 10 kb downstream of the Psg29 gene. Finally, a 426nt long IAPEY3-int fragment was noted in the approximately 10 kb downstream region of the Psg26 gene. We also found a partially mutated IAPY3 related sequence in approximately 7.5 kb upstream region of the cathepsin H (Ctsh) gene, a molecule that has placenta-specific roles (reviewed in [18]). These insights suggest a model in which insertion of an IAP element could attract DNA methylation (and other repressive modifications such as H3K9 methylation) to silence a neighbouring gene in somatic tissues, but allows the activation of that gene specifically in the placenta where the epigenetic constraints might be partially relaxed.
Interestingly, previous studies that have addressed the roles of retrotransposons in human placenta support our above hypothesis by showing that these elements could indeed contribute to activating the transcriptional level of neighbouring genes. It has been reported that the placenta-specific expression pattern of the human insulin-family gene INSL4 (insulin-like 4) is regulated by an ERV (HERV-K)-derived promoter element [42,43]. The LTR region of the HERV-K element is heavily (approx. 90%) methylated in the tissues where INSL4 is not transcribed (for example, in the adult and foetal tissues, whole blood and the umbilical cord), but is hypomethylated (approx. 20%) in the placental tissues where this gene is expressed [42]. A similar connection between HERV methylation state and the transcriptional level of a nearby gene has been indicated for several other loci such as EDNRB (endothelin receptor type B), which is expressed from a hypomethylated (approx. 20%) HERV-E derived promoter in the placenta but exhibits transcriptional suppression in other tissues in which the LTR-associated promoter is hypermethylated (approx. 90%) [42,44]. The PTN (pleiotrophin) gene is expressed exclusively in the human trophoblast and has functions for cell growth, migration and differentiation. The promoter region of PTN harbours an HERV-E element which shows low DNA methylation levels (approx. 20%) in the placental tissues but heavy methylation (approx. 75%) in other tissues [42,45]. The MID1 (midline 1) gene is linked with the Opitz syndrome—a genetic disorder that causes various types of midline defects such as clefts in the lips and larynx and heart defects in humans. Similar to EDNRB and PTN, the MID1 promoter contains an HERV-E retrotransposon and the LTR region of this retrotransposon is hypomethylated in the placenta which correlates to the elevated expression pattern of this gene exclusively in the placental tissues [42,46]. In addition, other studies that have examined the tissue-specific expression patterns of HERVs found that many of these elements (HERV-E, HERV-R, HERV-W, HERV-K and HERV-L) are specifically expressed in the placenta but not in other embryonic or adult tissues [47]. Intriguingly, previous studies have suggested that the placenta-specific expression pattern of a gene homologous to several mammalian species could be regulated in a similar fashion, namely, by acquisition of a retrotransposon in the promoter region. The retrotransposon derived loci in the gene promoter attract transcriptional silencing in non-placental tissues but allow gene expression specifically in the trophoblast [48]. The prolactin (PRL) gene, which has a role for regulating lactation in mammals, is expressed in humans specifically in the endometrial tissues of the placenta owing to the activity of an endogenous retrotransposon element known as MER39 (a member of the ERV1 class), located in the promoter region. Notably, in mice the same gene is also expressed in the placenta but is instead driven by the LTR of the rodent-specific ERV MER77. Taken together, these insights lend support to our hypothesis that the environment in the placenta could be more permissible for the expression and function of retrotransposons which could affect the transcriptional landscape of the nearby genes.
It is worthy of mention that ERV-associated genes such as the syncytin factors have been previously linked with the acquisition of placental functions in mammals and other species. Syncytins are conserved in primates (syncytin-1 and -2), rodentia (syncytin-A and -B), lagomorpha (syncytin-Ori1) and have also been identified recently in the carnivore (syncytin-Car1) [49]. These discoveries also stress the possibility that the endogenous retroviral sequences might have an evolutionarily conserved role for functions in the placenta.
5. Do IAP insertions sensitize host genes to environmental cues during gestation?
Above, we have discussed that the global epigenetic levels in the placenta could be permissive for IAP expression. We have also showed that IAP insertions could be found within or near several genes that have been previously associated with placenta-specific functions. On the basis of these notions, we will proceed to contemplate the biological significance of IAP elements in regulation of the placenta. To this end, we will first consider the following two cases where IAP insertion within/near a gene has been linked with well-known phenotypes in mice. The first is the agouti viable yellow (AVY) and the second is the axin-fused (AxinFu) mice [9–11].
The agouti allele is linked with determining the coat colour in mice. In the agouti viable yellow mice, an IAP insertion is observed in the upstream region of the agouti transcription start site [9]. When this region is methylated, transcriptional activity from the IAP sequence is repressed and the agouti gene is transcribed from its original promoter which leads to the normal brown coat colour. However, upon loss of DNA methylation, a cryptic promoter located within the IAP retrotransposon is activated and this results in ectopic initiation of agouti transcripts from the upstream locus, simultaneously giving rise to yellow coat colour in these mice. Furthermore, although agouti is normally expressed only in the hair follicles, demethylation and activation of the IAP promoter causes this gene to be transcribed throughout all cells. This leads to pleiotropic phenotypes including the readily observable yellow coat colour in these mice and other additional effects such as diabetes, obesity and tumorigenesis (reviewed in [50]). Interestingly, the occurrence of the hypomethylated IAP phenotype in the offspring can be exacerbated by administering nutritional inputs. One study showed that feeding a methyl-donor enriched diet to the pregnant dams can reduce the occurrence of yellow fur and obesity in the offspring [12]. Apart from nutritional cues, the IAP allele appears to be sensitive to environmental stimuli such as various chemical products. Bisphenol A (BPA) is a chemical reagent used in the production of plastic and a potential environmental pollutant. It was shown that administering BPA to pregnant mice could cause reduction of DNA methylation of IAP elements present in epigenetically metastable loci such as agouti viable yellow (AVY) and the CDK5 activator-binding protein (CabpIAP) [51].
In addition to the agouti viable yellow (AVY) mice, the axin-fused (AxinFu) mice are examples of IAP insertion in a specific allele that leads to a visible phenotypic aberration [10]. Integration of the IAP element into intron 6 of the axin gene causes the appearance of a kinked tail in mouse. Interestingly, similar to the AVY allele, administration of a methyl donor supplemented diet helped us to maintain DNA methylation of the IAP element in the AxinFu locus and reduced the appearance of kinked tail by half in these mice [13]. These studies demonstrate that, like the AVY allele, the IAP element in the AxinFu locus is sensitive to environmental/nutritional inputs.
In humans, foetuses that experienced undernutrition (or similar adverse environmental cues) during gestation are more prone to develop cardiovascular disease, impaired insulin resistance and neurological disorders in adult life (reviewed in [52,53]). The examples from agouti viable yellow and Axin-fused mice suggest that IAPs might be linked with the sensing process of such gestational inputs. However, it is not understood whether IAPs have a role to regulate genes that are associated with metabolism, energy consumption and neural functions. To investigate this issue, we analysed the list of IAP inserted genes in mice (see the electronic supplementary material, table S2) and asked whether these genes were associated with any of the above functions based on the previously published reports (in humans or mice). Interestingly, we found many genes that have been connected with adult life diseases such as diabetes, insulin resistance and neurological disorders. For example, Zfp69, Agtr1a, Dlg2, Pla2g4e and Lrp1b52 have been linked with diet-induced obesity in previous studies [54–58]. Further, several factors such as Tll2, Vegfc, Sesn1 and Sirt5 have been implicated in phenotypes such as recovery from nutritional restriction and food deprivation [59–62]. Another group of genes including Rab3gap1, Cntn6, Dld, Dlg2, Grm5, Hmox2, Accn1, Cntnap2, Hpse2, Il1rapl2, Immp2l, Kalrn and Park2 are associated with neurological diseases such as autism, speech disorder and craniofacial abnormalities [63–75]. Lastly, several genes such as Ninj2, Fhit and Dlg2 have been connected with heart rate, blood pressure and ischaemic stroke [56,76]. However, although these analyses might suggest a role for IAP elements for regulating genes with neurological functions, caution is needed to extrapolate a general biological significance from such results. It is necessary to remember that the GO analysis for IAP elements did not show any statistically significant enrichment for neural genes. Further, it has also been suggested that the genes associated with neuronal functions could be larger than the non-neuronal genes [77], which could be one reason why we saw many IAP insertions within such loci.
Taken together, these findings show that a group of IAP inserted genes that we have identified in this study are associated with metabolic and neural functions. We have also mentioned that previous studies suggest that nutritional and environmental challenges during the gestational period in humans could have long lasting impacts in terms of metabolic and psychological health [52,53]. It should be interesting to see whether these IAP elements are functionally linked for modulating the transcriptional status of these genes. Such functions for endogenous retrotransposons for regulating the expression pattern of neighbouring genes could be advantageous for the host during the course of evolution. In the previous section, we have argued that ERVs could be more active in the placenta because of the partial relaxation of epigenetic constraints that limit their transcription in other tissues. Here, we extend this idea by suggesting that placenta could play a vital role to link long-term environmental cues with the reproductive success of a species by mediating adaptive phenotypic responses. During gestation, the transfer of nutrients and gas from the mother to the foetus is mediated through the foeto-maternal interface (i.e. the placenta). In other words, in the intrauterine environment the survival and growth of the foetus is heavily dependent on the supply of nutrition and oxygen from the mother. Because of this reproductive strategy, the placental structures exhibit great variation among different classes of mammals—an issue that we will discuss in the ensuing section of the work, owing to their separate evolutionary origins. The insertions of retrotransposons could alter the transcriptional landscape by providing novel enhancers, promoters, alternative splicing sites and poly-A sequences in the genome. That is to say, they could be highly effective tools to create instant variations among a population by contributing to the long-term reproductive success of a species. Of course, among many retrotransposon insertional events taking place in the genome of a given species during evolution, most will probably be deleterious or neutral for the host. To test which insertion is beneficial for the host in terms of future reproductive fitness, the intrauterine conditions could be an ideal environment for evaluation because this is where the foetus is exposed to a myriad array of developmental cues, which could be beneficial, harmful or neutral. To achieve an increased level of fitness for the individual in adult life (and for the survival of the species in the long run), it is of utmost importance to correctly interpret the gestational signals and programme the foetus accordingly for the future life. As most (if not all) of these environmental cues should be passed on through the placenta during gestation, the activity of the endogenous retrotransposons permitted specifically in the extraembryonic tissues could be an evolutionary strategy for the species to select which retrotransposon insertion could be beneficial and which could be deleterious for the host.
6. Is there a role for ERVs to link environmental stimuli with divergence of placental functions?
In the previous sections of this work, we have suggested that IAPs could have specific roles in the placenta. One line of reasoning might suggest that the activation of IAPs in the placenta could occur purely by chance, because the global DNA methylation is lower in the extraembryonic tissues. However, other features of the IAP elements, namely their rodent-specific nature, imply that IAPs could also have important roles for a placenta-specific function. Eutherian mammals are commonly characterized by the placenta, and the structure and function of this organ exhibits great variation between different classes of mammals [18,78]. Placentas could be classified by the number and types of layers between the maternal and the foetal blood. According to this categorization, humans and mice possess hemochorial placentas: that is, in these species, the foetal chorion (a layer that separates the mother and the foetus) is directly bathed in the maternal blood. In contrast, cows and horses exhibit epitheliochorial placentas that contain three extra layers of membranes between the foetal chorion and the maternal blood preventing a direct contact. Furthermore, the gross phenotype of the placenta could differ drastically from one class of mammals to another. In both humans and mice, the placenta is shaped like a flattened disc (discoid), while in horses and pigs the structure of the same organ is quite different (diffuse placenta). In addition, even among the mammals that share similar classes of placentas, there could be additional levels of diversity. For example, although both humans and mice possess hemochorial and discoid placentas, the pattern of nutrient and gas transfer between the mother and the foetus is different among these species [79]. The placenta in mouse forms a labyrinthine structure between the foetal blood vessels and maternal blood spaces, which is more invasive than the villous placentas observed in humans. The invasive nature of the murine placenta could account for an increased necessity of nutrient and gas exchange between the mother and the foetus, because mice have larger litter size but much shorter gestation time compared with humans.
Because of these differences in the placental structures and functions among the mammals, it is conceivable that each group of mammals might employ different strategies (i.e. a different set of genes expressed) for the growth and differentiation of the placenta. In our previous work, we had observed a link between the evolution of placenta and genes that have CpG islands (CGIs) in their promoters [80]. Comparison of three mammalian species (human, mouse, rat) with seven other non-mammals revealed that the genes unique to the mammals, especially that are associated with placental development, were enriched in CGIs. This could suggest that a specific set of genes (i.e. CGI possessing genes) might function as ancestral elements to promote the part of placental development that is common to many mammalian species (human, mouse, rat). The necessity for additional genes that are unique to a species will probably emerge at a later step of placental development, during which the species-specific nature of the placenta will take centre stage. The endogenous retrotransposons such as the IAPs are unique to certain species and for this reason could be ideal for such purposes. Interestingly, previous investigations support this line of reasoning. A recent study suggested that the types of genes expressed during the earlier and later stages of placental development could differ dramatically among species [81]. The genes that are expressed during the early development of the placenta have a common ancestral origin and are linked with growth and metabolism. After implantation, the extraembryonic tissue has to undergo rapid development to ensure the transfer of nutrients to the foetus, which should be a common theme for all mammals during gestation. Therefore, it is not surprising to see that such growth and metabolism-associated genes are enriched in the ancestral gene category. However, during the later stages of placental development the species-specific metabolic need could take priority and during this time-frame genes that are unique to a certain species might be employed. Furthermore, closer inspection reveals that these genes include a disproportionate ratio of duplicated genes, a fact that is reminiscent of the evolutionary expansion strategy of endogenous retrotransposons by duplication. Recent studies suggest that such sequential activation of CGI containing (common) genes, followed by species-specific (unique) placental factors could be a strategy employed by the mammals for placental development. In humans, the CGI-containing transcription factors HOXA11 (homeobox A11) and FOXO1A (forkhead box O1), which have important roles in the placenta, appear to function in tandem to regulate the transcription of PRL (prolactin) gene [82], which is a retrotransposon-driven trophoblast specific gene found in several but not all mammalian species [48].
In summary, we argue that there could be a role for endogenous retrotransposons in the placenta because this is an organ which is associated with supplying the foetus with nutritional and environmental cues that might be useful for the postnatal life. This role should be consistent with the previously identified function of such endogenous retrotransposons as sensors for nutritional inputs. In addition, the relaxed epigenetic constraint in the extraembryonic tissue could allow the retrotransposons (and their neighbourhood genes) to be expressed in a developmental stage-dependent manner. We have discussed that there is great variety among the structures and functions of the placentas in different groups of mammals. It is conceivable that to achieve such diversity each class of mammals might chose to use tool-kit genes that are unique to that species. Previous studies suggest that recently duplicated genes could be important for species-specific functions in the placenta. As the endogenous retrotransposon compartment in each species has been enhanced by multiple duplication events, these elements fit the category for recently duplicated genes. Furthermore, there could be hundreds, if not thousands, of endogenous retrotransposons in mouse that belong to a same class. Insertions of such similar DNA sequences near placenta-associated genes could provide a cue for the onset of a specific transcriptional programme, in the placenta that could contribute to the diversity of placental functions among species.
7. Concluding remarks
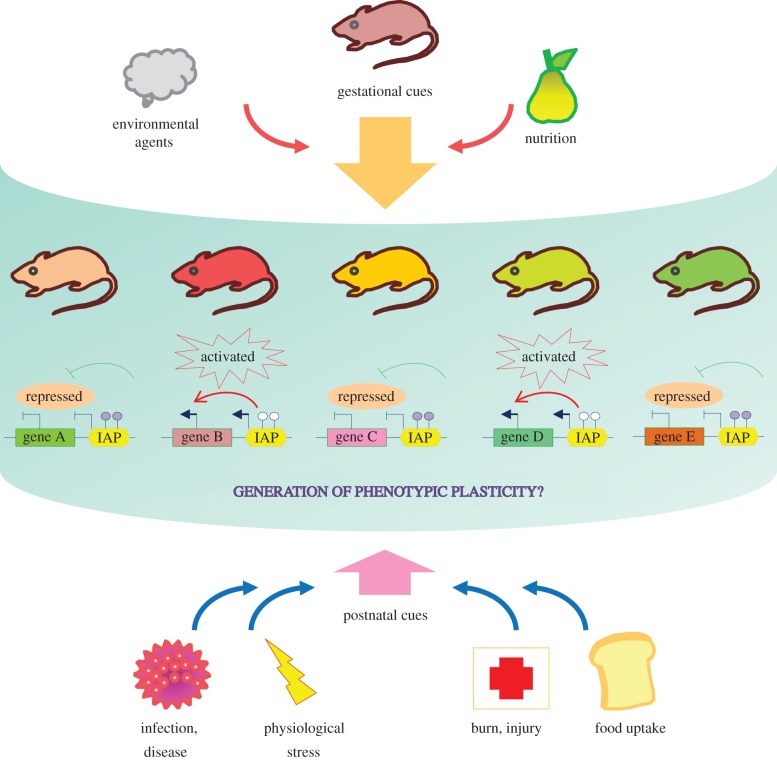
The potential of the IAP elements as sensors and mediators for environmental cues during critical stages of development could have the impact of creating far-reaching effects including phenotypic plasticity among a population (see figure 3). The ability of a genotype to exhibit variable phenotypes under different environmental circumstances is interpreted as phenotypic plasticity [19]. Phenotypic plasticity is mediated through various physiological and cellular mechanisms such as gene transcription, translation and hormonal activities to mount systematic responses to specific environmental cues that might positively contribute to the fitness and reproductive success of an individual (or a group of individuals). Previous studies suggest that IAPs could be mediators of such phenotypic plasticity because the epigenetic status of the IAP elements can be modified by exogenous inputs such as nutrition [11–13]. Furthermore, the stochastic epigenetic state of such IAP elements can influence the gene expression pattern and in extreme cases can translate to gross phenotypic changes in an animal [10]. Importantly, variable and plastic phenotypic responses created by epigenetic changes do not require the alteration of the genetic sequence, and, therefore, should pose a lesser amount of risk to a population. In addition, the epigenetic information could potentially be rewritten and modified through each new generation by genome reprogramming (such as erasure and re-establishment of the epigenome during the pre-implantation development stage) to suit the need of the changing environment. The link between retrotransposons, the prenatal and postnatal environment and plastic response of the phenotype should be a rewarding area of research for the future.
Figure 3.
Schematic model showing a role for ERVs in generating phenotypic plasticity. During gestation, nutritional and environmental cues establish unique epigenetic signatures for each (or a group) of ERVs (shown as IAPs) in an individual (lower panel, schematic showing hypomethylated ones as ‘activated’ and hypermethylated ones as ‘repressed’). Although each individual has the same genotype, the prenatal programming generates a broad range of variety by modulating the epigenome (individual animals having a unique epigenetic pattern, shown in different colours, upper panel). Upon reception of exogenous cues such as viral infection, burn or other injuries, deregulation in calorie uptake and other physiological stresses, robust and plastic response is generated among a group of individuals which is based on their variable epigenetic phenotypes established during gestation. Such plasticity and stochastic response against exogenous stimuli could improve the health and reproductive phenotype of a group of individuals and their overall chance of survival. Endogenous retroviruses, because of their species restricted nature and ability to modulate local (and perhaps global) gene expression, are attractive candidates for such a role.
Acknowledgements
The authors thank Dr Meiji Kit-Wan Ma for discussion and critical reading of the manuscript. J.S. is supported by the RIKEN FPR fellowship and the RIKEN Incentive Research Grant 2012.
References
- 1.Lander ES, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409, 860–921 10.1038/35057062 (doi:10.1038/35057062) [DOI] [PubMed] [Google Scholar]
- 2.McClintock B. 1950. The origin and behavior of mutable loci in maize. Proc. Natl Acad. Sci. USA 36, 344–355 10.1073/pnas.36.6.344 (doi:10.1073/pnas.36.6.344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wicker T, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982 10.1038/nrg2165 (doi:10.1038/nrg2165) [DOI] [PubMed] [Google Scholar]
- 4.Martin SL. 2006. The ORF1 protein encoded by LINE-1: structure and function during L1 retrotransposition. J. Biomed. Biotechnol. 2006, 45621. 10.1155/JBB/2006/45621 (doi:10.1155/JBB/2006/45621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stocking C, Kozak CA. 2008. Murine endogenous retroviruses. Cell Mol. Life Sci. 65, 3383–3398 10.1007/s00018-008-8497-0 (doi:10.1007/s00018-008-8497-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ryan PF. 2004. Human endogenous retroviruses in health and disease: a symbiotic perspective. J. R. Soc. Med. 97, 560–565 10.1258/jrsm.98.3.134-a (doi:10.1258/jrsm.98.3.134-a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y, Maksakova IA, Gagnier L, van de Lagemaat LN, Mager DL. 2008. Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet. 4, e1000007. 10.1371/journal.pgen (doi:10.1371/journal.pgen) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maksakova IA, Mager DL, Reiss D. 2008. Keeping active endogenous retroviral-like elements in check: the epigenetic perspective. Cell Mol. Life Sci. 65, 3329–3347 10.1007/s00018-008-8494-3 (doi:10.1007/s00018-008-8494-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. 1999. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 23, 314–318 10.1038/15490 (doi:10.1038/15490) [DOI] [PubMed] [Google Scholar]
- 10.Rakyan VK, Chong S, Champ ME, Cuthbert PC, Morgan HD, Luu KV, Whitelaw E. 2003. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc. Natl Acad. Sci. USA 100, 2538–2543 10.1073/pnas.0436776100 (doi:10.1073/pnas.0436776100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolff GL, Kodell RL, Moore SR, Cooney CA. 1998. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 12, 949–957 [PubMed] [Google Scholar]
- 12.Cropley JE, Suter CM, Beckman KB, Martin DI. 2006. Germ-line epigenetic modification of the murine A vy allele by nutritional supplementation. Proc. Natl Acad. Sci. USA 103, 17 308–17 312 10.1073/pnas.0607090103 (doi:10.1073/pnas.0607090103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waterland RA, Dolinoy DC, Lin JR, Smith CA, Shi X, Tahiliani KG. 2006. Maternal methyl supplements increase offspring DNA methylation at axin fused. Genesis 44, 401–406 10.1002/dvg.20230 (doi:10.1002/dvg.20230) [DOI] [PubMed] [Google Scholar]
- 14.Ostertag EM, Kazazian HH., Jr 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35, 501–538 10.1146/annurev.genet.35.102401.091032 (doi:10.1146/annurev.genet.35.102401.091032) [DOI] [PubMed] [Google Scholar]
- 15.Kuff EL, Lueders KK. 1988. The intracisternal A-particle gene family: structure and functional aspects. Adv. Cancer Res. 51, 183–276 (doi:10.1016/S0065–230X(08)60223-7) [DOI] [PubMed] [Google Scholar]
- 16.Dewannieux M, Dupressoir A, Harper F, Pierron G, Heidmann T. 2004. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat. Genet. 36, 534–539 10.1038/ng1353 (doi:10.1038/ng1353) [DOI] [PubMed] [Google Scholar]
- 17.Qin C, et al. 2010. Intracisternal A particle genes: distribution in the mouse genome, active subtypes, and potential roles as species-specific mediators of susceptibility to cancer. Mol. Carcinog. 49, 54–67 10.1002/mc.20576 (doi:10.1002/mc.20576) [DOI] [PubMed] [Google Scholar]
- 18.Rawn SM, Cross JC. 2008. The evolution, regulation, and function of placenta-specific genes. Annu. Rev. Cell Dev. Biol. 24, 159–181 10.1146/annurev.cellbio.24.110707.175418 (doi:10.1146/annurev.cellbio.24.110707.175418) [DOI] [PubMed] [Google Scholar]
- 19.Johnson LJ, Tricker PJ. 2010. Epigenomic plasticity within populations: its evolutionary significance and potential. Heredity (Edinb.) 105, 113–121 10.1038/hdy.2010.25 (doi:10.1038/hdy.2010.25) [DOI] [PubMed] [Google Scholar]
- 20.Chang-Yeh A, Mold DE, Huang RC. 1991. Identification of a novel murine IAP-promoted placenta-expressed gene. Nucleic Acids Res. 19, 3667–3672 10.1093/nar/19.13.3667 (doi:10.1093/nar/19.13.3667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 10.1093/molbev/msl072 (doi:10.1093/molbev/msl072) [DOI] [PubMed] [Google Scholar]
- 22.Feenstra A, Fewell J, Lueders K, Kuff E. 1986. In vitro methylation inhibits the promotor activity of a cloned intracisternal A-particle LTR. Nucleic Acids Res. 14, 4343–4352 10.1093/nar/14.10.4343 (doi:10.1093/nar/14.10.4343) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lasneret J, Canivet M, Hojman-Montes de Oca F, Tobaly J, Emanoil-Ravicovitch R, Peries J. 1983. Activation of intracisternal A particles by 5-azacytidine in mouse Ki-BALB cell line. Virology 128, 485–489 10.1016/0042-6822(83)90275-1 (doi:10.1016/0042-6822(83)90275-1) [DOI] [PubMed] [Google Scholar]
- 24.Walsh CP, Chaillet JR, Bestor TH. 1998. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 20, 116–117 10.1038/2413 (doi:10.1038/2413) [DOI] [PubMed] [Google Scholar]
- 25.Hata K, Kusumi M, Yokomine T, Li E, Sasaki H. 2006. Meiotic and epigenetic aberrations in Dnmt3L-deficient male germ cells. Mol. Reprod. Dev. 73, 116–122 10.1002/mrd.20387 (doi:10.1002/mrd.20387) [DOI] [PubMed] [Google Scholar]
- 26.Sharif J, et al. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912 10.1038/nature06397 (doi:10.1038/nature06397) [DOI] [PubMed] [Google Scholar]
- 27.Zhao X, et al. 2003. Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc. Natl Acad. Sci. USA 100, 6777–6782 10.1073/pnas.1131928100 (doi:10.1073/pnas.1131928100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshimura T, Toyoda S, Kuramochi-Miyagawa S, Miyazaki T, Miyazaki S, Tashiro F, Yamato E, Nakano T, Miyazaki J. 2009. Gtsf1/Cue110, a gene encoding a protein with two copies of a CHHC Zn-finger motif, is involved in spermatogenesis and retrotransposon suppression in murine testes. Dev. Biol. 335, 216–227 10.1016/j.ydbio.2009.09.003 (doi:10.1016/j.ydbio.2009.09.003) [DOI] [PubMed] [Google Scholar]
- 29.Kuramochi-Miyagawa S, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 22, 908–917 10.1101/gad.1640708 (doi:10.1101/gad.1640708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe T, et al. 2011. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev. Cell 20, 364–375 10.1016/j.devcel.2011.01.005 (doi:10.1016/j.devcel.2011.01.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oey HM, Youngson NA, Whitelaw E. 2011. The characterisation of piRNA-related 19mers in the mouse. BMC Genom. 12, 315. (doi:10.1186/1471–2164-12-315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dupressoir A, Heidmann T. 1996. Germ line-specific expression of intracisternal A-particle retrotransposons in transgenic mice. Mol. Cell Biol. 16, 4495–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464, 927–931 10.1038/nature08858 (doi:10.1038/nature08858) [DOI] [PubMed] [Google Scholar]
- 34.Dong KB, et al. 2008. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 27, 2691–2701 10.1038/emboj.2008.193 (doi:10.1038/emboj.2008.193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowe HM, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463, 237–240 10.1038/nature08674 (doi:10.1038/nature08674) [DOI] [PubMed] [Google Scholar]
- 36.Reik W, Dean W, Walter J. 2001. Epigenetic reprogramming in mammalian development. Science 293, 1089–1093 10.1126/science.1063443 (doi:10.1126/science.1063443) [DOI] [PubMed] [Google Scholar]
- 37.Leung DC, Lorincz MC. 2012. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem. Sci. 37, 127–133 10.1016/j.tibs.2011.11.006 (doi:10.1016/j.tibs.2011.11.006) [DOI] [PubMed] [Google Scholar]
- 38.Hemberger M. 2010. Genetic–epigenetic intersection in trophoblast differentiation: implications for extraembryonic tissue function. Epigenetics 5, 24–29 10.4161/epi.5.1.10589 (doi:10.4161/epi.5.1.10589) [DOI] [PubMed] [Google Scholar]
- 39.Farthing CR, Ficz G, Ng RK, Chan CF, Andrews S, Dean W, Hemberger M, Reik W. 2008. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 4, e1000116. 10.1371/journal.pgen.1000116 (doi:10.1371/journal.pgen.1000116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Djaffar I, Dianoux L, Leibovich S, Kaplan L, Emanoil-Ravier R, Peries J. 1990. Detection of IAP related transcripts in normal and transformed rat cells. Biochem. Biophys. Res. Commun. 169, 222–231 10.1016/0006-291X(90)91457-4 (doi:10.1016/0006-291X(90)91457-4) [DOI] [PubMed] [Google Scholar]
- 41.Ball M, McLellan A, Collins B, Coadwell J, Stewart F, Moore T. 2004. An abundant placental transcript containing an IAP-LTR is allelic to mouse pregnancy-specific glycoprotein 23 (Psg23): cloning and genetic analysis. Gene 325, 103–113 10.1016/j.gene.2003.10.001 (doi:10.1016/j.gene.2003.10.001) [DOI] [PubMed] [Google Scholar]
- 42.Macaulay EC, Weeks RJ, Andrews S, Morison IM. 2011. Hypomethylation of functional retrotransposon-derived genes in the human placenta. Mamm. Genome 22, 722–735 10.1007/s00335-011-9355-1 (doi:10.1007/s00335-011-9355-1) [DOI] [PubMed] [Google Scholar]
- 43.Bièche I, et al. 2003. Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol. Reprod. 68, 1422–1429 10.1095/biolreprod.102.010322 (doi:10.1095/biolreprod.102.010322) [DOI] [PubMed] [Google Scholar]
- 44.Landry JR, Mager DL. 2003. Functional analysis of the endogenous retroviral promoter of the human endothelin B receptor gene. J. Virol. 77, 7459–7466 10.1128/JVI.77.13.7459-7466.2003 (doi:10.1128/JVI.77.13.7459-7466.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulte AM, Lai S, Kurtz A, Czubayko F, Riegel AT, Wellstein A. 1996. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ-line insertion of an endogenous retrovirus. Proc. Natl Acad. Sci. USA 93, 14 759–14 764 10.1073/pnas.93.25.14759 (doi:10.1073/pnas.93.25.14759) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Landry JR, Rouhi A, Medstrand P, Mager DL. 2002. The Opitz syndrome gene Mid1 is transcribed from a human endogenous retroviral promoter. Mol. Biol. Evol. 19, 1934–1942 10.1093/oxfordjournals.molbev.a004017 (doi:10.1093/oxfordjournals.molbev.a004017) [DOI] [PubMed] [Google Scholar]
- 47.Nelson PN, Hooley P, Roden D, Davari Ejtehadi H, Rylance P, Warren P, Martin J, Murray PG. 2004. Human endogenous retroviruses: transposable elements with potential? Clin. Exp. Immunol. 138, 1–9 10.1111/j.1365-2249.2004.02592.x (doi:10.1111/j.1365-2249.2004.02592.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emera D, Casola C, Lynch VJ, Wildman DE, Agnew D, Wagner GP. 2012. Convergent evolution of endometrial prolactin expression in primates, mice, and elephants through the independent recruitment of transposable elements. Mol. Biol. Evol. 29, 239–247 10.1093/molbev/msr189 (doi:10.1093/molbev/msr189) [DOI] [PubMed] [Google Scholar]
- 49.Cornelis G, Heidmann O, Bernard-Stoecklin S, Reynaud K, Véron G, Mulot B, Dupressoir A, Heidmann T. 2012. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc. Natl Acad. Sci. USA 109, E432–E441 10.1073/pnas.1115346109 (doi:10.1073/pnas.1115346109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dolinoy DC. 2008. The agouti mouse model: an epigenetic biosensor for nutritional and environmental alterations on the fetal epigenome. Nutr. Rev. 66(Suppl. 1), S7–S11 10.1111/j.1753-4887.2008.00056.x (doi:10.1111/j.1753-4887.2008.00056.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dolinoy DC, Huang D, Jirtle RL. 2007. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc. Natl Acad. Sci. USA 104, 13 056–13 061 10.1073/pnas.0703739104 (doi:10.1073/pnas.0703739104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gluckman PD, Hanson MA, Pinal C. 2005. The developmental origins of adult disease. Matern. Child Nutr. 1, 130–141 10.1111/j.1740-8709.2005.00020.x (doi:10.1111/j.1740-8709.2005.00020.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wells JC. 2011. The thrifty phenotype: an adaptation in growth or metabolism? Am. J. Hum. Biol. 23, 65–75 10.1002/ajhb.21100 (doi:10.1002/ajhb.21100) [DOI] [PubMed] [Google Scholar]
- 54.Scherneck S, et al. 2009. Positional cloning of zinc finger domain transcription factor Zfp69, a candidate gene for obesity-associated diabetes contributed by mouse locus Nidd/SJL. PLoS Genet. 5, e1000541. 10.1371/journal.pgen.1000541 (doi:10.1371/journal.pgen.1000541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kouyama R, et al. 2005. Attenuation of diet-induced weight gain and adiposity through increased energy expenditure in mice lacking angiotensin II type 1a receptor. Endocrinology 146, 3481–3489 10.1210/en.2005-0003 (doi:10.1210/en.2005-0003) [DOI] [PubMed] [Google Scholar]
- 56.Nikpay M, Seda O, Tremblay J, Petrovich M, Gaudet D, Kotchen TA, Cowley AW, Jr, Hamet P. 2012. Genetic mapping of habitual substance use, obesity-related traits, responses to mental and physical stress, and heart rate and blood pressure measurements reveals shared genes that are overrepresented in the neural synapse. Hypertens. Res. 35, 585–591 10.1038/hr.2011.233 (doi:10.1038/hr.2011.233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Song YB, An YR, Kim SJ, Park HW, Jung JW, Kyung JS, Hwang SY, Kim YS. 2012. Lipid metabolic effect of Korean red ginseng extract in mice fed on a high-fat diet. J. Sci. Food Agric. 92, 388–396 10.1002/jsfa.4589 (doi:10.1002/jsfa.4589) [DOI] [PubMed] [Google Scholar]
- 58.Masson O, Chavey C, Dray C, Meulle A, Daviaud D, Quilliot D, Muller C, Valet P, Liaudet-Coopman E. 2009. LRP1 receptor controls adipogenesis and is up-regulated in human and mouse obese adipose tissue. PLoS ONE 4, e7422. 10.1371/journal.pone.0007422 (doi:10.1371/journal.pone.0007422) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Allen DL, Greybeck BJ, Hanson AM, Cleary AS, Lindsay SF. 2010. Skeletal muscle expression of bone morphogenetic protein-1 and tolloid-like-1 extracellular proteases in different fiber types and in response to unloading, food deprivation and differentiation. J. Physiol. Sci. 60, 343–352 10.1007/s12576-010-0110-2 (doi:10.1007/s12576-010-0110-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mehedint MG, Craciunescu CN, Zeisel SH. 2010. Maternal dietary choline deficiency alters angiogenesis in fetal mouse hippocampus. Proc. Natl Acad. Sci. USA 107, 12 834–12 839 10.1073/pnas.0914328107 (doi:10.1073/pnas.0914328107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen JH, Tarry-Adkins JL, Matharu K, Yeo GS, Ozanne SE. 2010. Maternal protein restriction affects gene expression profiles in the kidney at weaning with implications for the regulation of renal function and lifespan. Clin. Sci. (Lond.) 119, 373–384 10.1042/CS20100230 (doi:10.1042/CS20100230) [DOI] [PubMed] [Google Scholar]
- 62.Chalkiadaki A, Guarente L. 2012. Sirtuins mediate mammalian metabolic responses to nutrient availability. Nat. Rev. Endocrinol. 8, 287–296 10.1038/nrendo.2011.225 (doi:10.1038/nrendo.2011.225) [DOI] [PubMed] [Google Scholar]
- 63.Morris-Rosendahl DJ, et al. 2010. New RAB3GAP1 mutations in patients with Warburg Micro Syndrome from different ethnic backgrounds and a possible founder effect in the Danish. Eur. J. Hum. Genet. 18, 1100–1106 10.1038/ejhg.2010.79 (doi:10.1038/ejhg.2010.79) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Daalen E, et al. 2011. Social responsiveness scale-aided analysis of the clinical impact of copy number variations in autism. Neurogenetics 12, 315–323 10.1007/s10048-011-0297-2 (doi:10.1007/s10048-011-0297-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown AM, Gordon D, Lee H, Caudy M, Hardy J, Haroutunian V, Blass JP. 2004. Association of the dihydrolipoamide dehydrogenase gene with Alzheimer's disease in an Ashkenazi Jewish population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 131B, 60–66 10.1002/ajmg.b.30008 (doi:10.1002/ajmg.b.30008) [DOI] [PubMed] [Google Scholar]
- 66.MacLaren EJ, Charlesworth P, Coba MP, Grant SG. 2011. Knockdown of mental disorder susceptibility genes disrupts neuronal network physiology in vitro. Mol. Cell Neurosci. 47, 93–99 10.1016/j.mcn.2010.12.014 (doi:10.1016/j.mcn.2010.12.014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Elia J, et al. 2011. Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat. Genet. 44, 78–84 10.1038/ng.1013 (doi:10.1038/ng.1013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ayuso P, et al. 2011. A polymorphism located at an ATG transcription start site of the heme oxygenase-2 gene is associated with classical Parkinson's disease. Pharmacogenet. Genom. 21, 565–571 10.1097/FPC.0b013e328348f729 (doi:10.1097/FPC.0b013e328348f729) [DOI] [PubMed] [Google Scholar]
- 69.Gregersen N, et al. 2012. A genome-wide study of panic disorder suggests the amiloride-sensitive cation channel 1 as a candidate gene. Eur. J. Hum. Genet. 20, 84–90 10.1038/ejhg.2011.148 (doi:10.1038/ejhg.2011.148) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Peñagarikano O, Geschwind DH. 2012. What does CNTNAP2 reveal about autism spectrum disorder? Trends Mol. Med. 18, 156–163 10.1016/j.molmed.2012.01.003 (doi:10.1016/j.molmed.2012.01.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Al Badr W, et al. 2011. Exome capture and massively parallel sequencing identifies a novel HPSE2 mutation in a Saudi Arabian child with Ochoa (urofacial) syndrome. J. Pediatr. Urol. 7, 569–573 10.1016/j.jpurol.2011.02.034 (doi:10.1016/j.jpurol.2011.02.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kantojärvi K, Kotala I, Rehnström K, Ylisaukko-Oja T, Vanhala R, von Wendt TN, von Wendt L, Järvelä I. 2011. Fine mapping of Xq11.1-q21.33 and mutation screening of RPS6KA6, ZNF711, ACSL4, DLG3, and IL1RAPL2 for autism spectrum disorders (ASD). Autism Res. 4, 228–233 10.1002/aur.187 (doi:10.1002/aur.187) [DOI] [PubMed] [Google Scholar]
- 73.Swaminathan S, Shen L, Kim S, Inlow M, West JD, Faber KM, Foroud T, Mayeux R, Saykin AJ. 2012. Analysis of copy number variation in Alzheimer's disease: the NIA-LOAD/NCRAD Family Study. Curr. Alzheimer Res. 9, 801–814 10.2174/156720512802455331 (doi:10.2174/156720512802455331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kushima I, et al. 2012. Resequencing and association analysis of the KALRN and EPHB1 genes and their contribution to schizophrenia susceptibility. Schizophr Bull. 38, 552–560 10.1093/schbul/sbq118 (doi:10.1093/schbul/sbq118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yoritaka A, Shimo Y, Shimo Y, Inoue Y, Yoshino H, Hattori N. 2011. Nonmotor symptoms in patients with PARK2 mutations. Parkinsons Dis. 2011, 473640. 10.4061/2011/473640 (doi:10.4061/2011/473640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wan XH, Li SJ, Cheng P, Zhang Q, Yang XC, Zhong GZ, Hu WL, Jin L, Wang XF. 2011. NINJ2 polymorphism is associated with ischemic stroke in Chinese Han population. J. Neurol. Sci. 308, 67–71 10.1016/j.jns.2011.06.011 (doi:10.1016/j.jns.2011.06.011) [DOI] [PubMed] [Google Scholar]
- 77.Raychaudhuri S, Korn JM, McCarroll SA, International Schizophrenia Consortium, Altshuler D, Sklar P, Purcell S, Daly MJ. 2010. Accurately assessing the risk of schizophrenia conferred by rare copy-number variation affecting genes with brain function. PLoS Genet. 6, e1001097. 10.1371/journal.pgen.1001097 (doi:10.1371/journal.pgen.1001097) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wildman DE, Chen C, Erez O, Grossman LI, Goodman M, Romero R. 2006. Evolution of the mammalian placenta revealed by phylogenetic analysis. Proc. Natl Acad. Sci. USA 103, 3203–3208 10.1073/pnas.0511344103 (doi:10.1073/pnas.0511344103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Watson ED, Cross JC. 2005. Development of structures and transport functions in the mouse placenta. Physiology (Bethesda) 20, 180–193 10.1152/physiol.00001.2005 (doi:10.1152/physiol.00001.2005) [DOI] [PubMed] [Google Scholar]
- 80.Sharif J, Endo TA, Toyoda T, Koseki H. 2010. Divergence of CpG island promoters: a consequence or cause of evolution? Dev. Growth Differ. 52, 545–554 10.1111/j.1440-169X.2010.01193.x (doi:10.1111/j.1440-169X.2010.01193.x) [DOI] [PubMed] [Google Scholar]
- 81.Knox K, Baker JC. 2008. Genomic evolution of the placenta using co-option and duplication and divergence. Genome Res. 18, 695–705 10.1101/gr.071407.107 (doi:10.1101/gr.071407.107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lynch VJ, Brayer K, Gellersen B, Wagner GP. 2009. HoxA-11 and FOXO1A cooperate to regulate decidual prolactin expression: towards inferring the core transcriptional regulators of decidual genes. PLoS ONE 4, e6845. 10.1371/journal.pone.0006845 (doi:10.1371/journal.pone.0006845) [DOI] [PMC free article] [PubMed] [Google Scholar]