Abstract
Genomic imprinting is widespread in eutherian mammals. Marsupial mammals also have genomic imprinting, but in fewer loci. It has long been thought that genomic imprinting is somehow related to placentation and/or viviparity in mammals, although neither is restricted to mammals. Most imprinted genes are expressed in the placenta. There is no evidence for genomic imprinting in the egg-laying monotreme mammals, despite their short-lived placenta that transfers nutrients from mother to embryo. Post natal genomic imprinting also occurs, especially in the brain. However, little attention has been paid to the primary source of nutrition in the neonate in all mammals, the mammary gland. Differentially methylated regions (DMRs) play an important role as imprinting control centres in each imprinted region which usually comprises both paternally and maternally expressed genes (PEGs and MEGs). The DMR is established in the male or female germline (the gDMR). Comprehensive comparative genome studies demonstrated that two imprinted regions, PEG10 and IGF2-H19, are conserved in both marsupials and eutherians and that PEG10 and H19 DMRs emerged in the therian ancestor at least 160 Ma, indicating the ancestral origin of genomic imprinting during therian mammal evolution. Importantly, these regions are known to be deeply involved in placental and embryonic growth. It appears that most maternal gDMRs are always associated with imprinting in eutherian mammals, but emerged at differing times during mammalian evolution. Thus, genomic imprinting could evolve from a defence mechanism against transposable elements that depended on DNA methylation established in germ cells.
Keywords: marsupials, monotremes, eutherians, retrotransposons, placentation, lactation
1. Introduction
Parent-of-origin gene expression (genomic imprinting) is widespread amongst eutherian mammals and also occurs in marsupials. Most imprinted genes are expressed in the placenta, but the brain is also a favoured site. Although imprinting evolved in therian mammals before the marsupial–eutherian split, the mechanisms have continued to evolve in each lineage to produce the differences in the number and regulation of imprinted genes that now exist between the two groups. There are around 100 genes that are subject to genomic imprinting in eutherian mammals, but there appears to be many fewer in marsupial mammals. Marsupial and eutherian mammals diverged from each other about 160 Ma [1] (figure 1). One hypothesis is that the evolution and diversification of mammals has been driven by a series of chance events, retrotransposition integration and exaptation, to produce novel but essential placental genes [2]. This review addresses these questions on the origin and evolution of genomic imprinting in mammals, and its relevance to viviparity, placentation, lactation and post natal care.
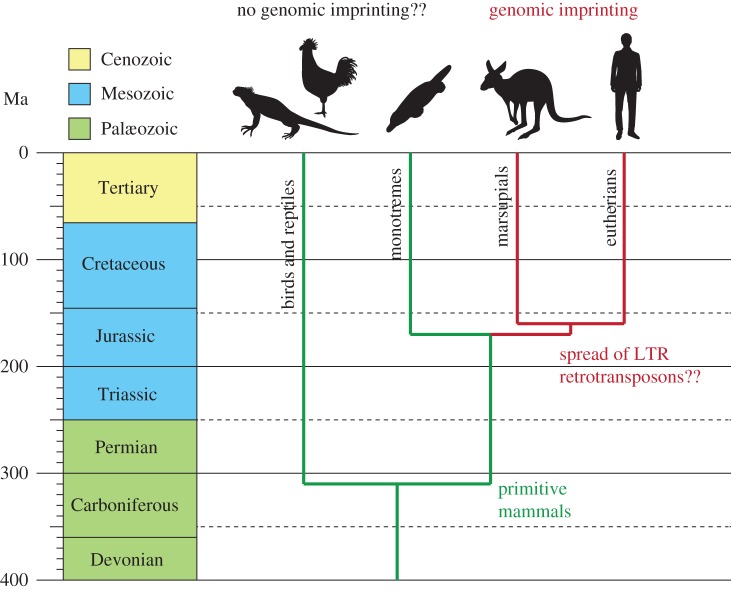
Figure 1.
The timing of genomic imprinting acquisition and of the divergence of birds and reptiles, monotremes, marsupials and eutherians. The vertical axes represent the time line from 400 Ma to the present. The coloured boxes represent each geological period. The green and red lines represent the evolution of the groups with and without genomic imprinting. The silhouettes represent one example species from each group. LTR, long terminal repeat.
2. Evolution of viviparity and the placenta
The placenta is defined as an organ of physiological exchange between the mother and fetus [3]. The placenta is probably the most varied structure in the animal kingdom, and is found in a wide variety of taxa, even among invertebrates such as scorpions and lower vertebrates such as selachian sharks [4,5]. Placentation allows the production of live young, but it is often not recognized that this is not a uniquely mammalian characteristic. In amniotes, there are four fetal membranes: the amnion that surrounds the fetus and protects it from mechanical and physiological shock; the yolk sac that in birds and some reptiles surrounds the egg yolk; the allantois, which is an extension of the embryonic bladder and stores excretory products; and the chorion, which is formed by extraembryonic mesoderm and an outer trophoblast layer and fuses with either the yolk sac to form the chorio-vitelline placenta or with the allantois to form the chorio-allantoic placenta. Most mammals rely on both types of placenta at least for some periods of pregnancy, and even in humans the yolk sac is crucial for the survival of the early embryo [6]. It is the site of fetal blood and blood vessel formation, nutrient and gas exchange before the allantoic placenta is established, and transfer and biosynthesis of cholesterol and proteins [7–10]. Dysfunction or abnormal growth of the yolk sac can negatively influence success of the pregnancy or post natal health.
Although the majority of marsupials depend on a chorio-vitelline placenta, which is often considered somehow a less important placenta, it is fully functional and not only is responsible for the nutrients and gaseous exchange but also induces a maternal recognition of pregnancy [11,12] and elaborates many hormones [13,14]. The term placental mammal (preferably known as eutherian mammal) gives the incorrect impression that the placenta is found only in that mammalian infraclass. The invasiveness of the placenta varies from superficial to highly invasive but in all cases is critical for the survival of the fetus [13,15]. In general, allantoic placentae are more invasive of the uterine endometrium and non-invasive or less invasive placentas were originally considered to be more primitive than highly invasive placentas [16]. This paradigm is now generally not accepted, since a common feature of the placenta of viviparous elasmobranchs, amphibians and reptiles is invasion and haemotrophic nutrition [4,15]. Indeed, in certain marsupials a greater reliance on histotrophic nutrition of a non-invasive placenta can be a derived (or more recently evolved) character [17]. A few marsupials, such as the bandicoot, dasyurids and wombats, have a chorio-allantoic placenta, which supplements the placental functions of the yolk sac or chorio-vitelline placenta [17–20]. In these species, the placenta is invasive to a variable degree, and in the bandicoot a maternal homokaryon is formed which later in development fuses with the trophectoderm to form a feto-maternal syncytium at the site of the chorio-allantois [21].
Every mammalian species has a yolk sac that develops differently and has slightly different functions as a placenta. However, the nutritive, biosynthetic and hematopoietic functions of the human yolk sac are not new inventions specific to humans, but ancestral features shared with other viviparous mammals. The rodent visceral yolk sac (inverted yolk sac) is an important organ of maternal–embryonic exchange of amino acids, transferrin, vitamin B12, calcium and other ions [22,23]. The rodent visceral yolk sac is also the route by which passive immunity (immunoglobulins) is passed to the embryo [24,25]. Agents that cause visceral yolk sac dysfunction during organogenesis can result in embryotoxicity and its damage leads to embryonic malformation [26]. In virtually all eutherian species studied, there is evidence of synthetic activity by the yolk sac and it serves as hematopoietic organ in all mammalian species studied (reviewed by King & Enders [22]). These synthetic activities may occur throughout gestation, influencing implantation as well as embryonic and fetal growth (reviewed by King & Enders [22]). In marsupials, the yolk sac forms the definitive placenta, being responsible for physiological exchange, biosynthesis and endocrinology [17,20,27,28].
There are about 106 origins of viviparity within the squamates (lizards and snakes) [29,30]. Many of the viviparous reptiles have a highly invasive chorio-allantoic placenta, and the developing young depend on receiving nutrients via this placental attachment. Four independent origins of complex placentae have been identified in only one modern reptilian lineage, the lizard family Scincidae [31–34]. Some species have small eggs, complex, eutherian-like chorio-allantoic placentae and are highly placentotrophic [35]. There is significant uptake of nutrients across the placenta of species with complex chorio-allantoic placentae [36]. Indeed, in the viviparous skink of central Africa, placentation is endotheliochorial and is strikingly convergent on features of the placenta of viviparous mammals [37]. As yet, there is no evidence of genomic imprinting in reptiles, but the new data on reptile genes and genomics is awaited with interest [34,38].
3. Transfer of nutrients: a common theme in imprinting
(a). Placentation and pre natal imprinting
Most imprinted genes exhibit placental expression [39]. Several genetic experiments indicated that there are a number of imprinted genes that are essential for placental development and growth. Partial uniparental duplication of certain chromosome regions causes several developmental and growth abnormalities, probably owing to placental defects. Importantly, in mice, seven out of 21 imprinted regions are related to such phenotypes, such as early embryonic, mid-fetal or late fetal lethalities and/or prenatal growth retardation/stimulation [40]. Subsequent knockout mice studies of many candidate imprinted genes confirmed that there are several essential placental genes in such imprinted regions. Among seven such imprinted regions, three imprinted regions are linked to embryonic lethality: proximal chromosome 6, distal chromosome 7 and distal chromosome 12. Embryonic lethality is observed in mice with maternal duplication of proximal chromosome 6 (MatDp(prox6)). Peg10 (paternally expressed 10) is a major gene responsible for this early embryonic lethal phenotype. Peg10 knockout (KO) mice show early embryonic lethality owing to severe placental abnormality with almost complete lack of labyrinth and spongiotrophoblast layers, the essential part of the placenta where nutrient and gas exchange occur between fetal and maternal blood cells [41]. Peg10 encodes a protein with homology to the Gag and Pol proteins of certain long terminal repeat (LTR) retrotransposons [42]. Although its biochemical function is yet unknown, Peg10 is an essential placenta-specific gene expressed paternally in mice. Paternal duplication of distal chromosome 7 (PatDp(dis7)) also causes early embryonic lethality. Mash2/Ascl2 (mammalian achaete scute homologue 2/achaete scute complex like 2), one of the maternally expressed genes in this imprinted gene cluster, is responsible for the phenotype. There is severe placental abnormality in Mash2 KO mice, such as a lack of labyrinth and spongiotrophoblast layers like Peg10 KO mice in addition to an overgrowth of trophoblast giant cells, and they died on day 10 of gestation [43]. Mash2/Ascl2 encodes a basic helix–loop–helix transcription factor specifically expressed in the placenta. Peg10 KO and Mash2 KO mice have a similar placental phenotype, although Mash2 is a maternally expressed gene while Peg10 is a paternally expressed gene. It should be noted that Peg10 and Mash2/Ascl2 are the genes responsible for parthenogenetic and androgenetic death of mouse embryos, respectively [44–46]. Interestingly, mice with (MatDp(dis7)) have a disrupted phenotype and die at mid-fetal stages rather than the early embryonic lethality seen in (PatDp(dis7)). It is highly probable that both lack of paternally expressed Igf2 (insulin-like growth factor 2) expression and overexpression of two maternally expressed genes, Cdkn1c (cyclin-dependent kinase inhibitor 1C, also known as p57Kip2) and H19 are attributable to this phenotype. Igf2 plays a role as one of major growth factors in fetal development. Igf2 KO mice have severe growth retardation but the KO does not cause lethality alone [47]. Cdkn1c functions as a growth inhibitor. Both functional loss of CDKN1C and the overexpression of IGF2 are attributable to Beckwith–Wiedemann syndrome, characterized by fetal overgrowth [48,49]. Cdkn1c also affects the growth of placental trophoblast giant cells [50]. H19 is a well-conserved non-coding RNA (ncRNA) in both marsupials and eutherians [51], suggesting that it has some unknown important role in mammalian development although KO mice have no significant abnormal phenotypes [52]. H19 may have tumour-suppressing activity when combined with overexpression of Igf2 [53]. H19 is highly expressed in both embryos and placentas, so its overexpression disturbs the gene expression profiles of many other genes that may affect on embryonic growth [54]. Phenotypes of both maternal and paternal duplication of distal chromosome 12 (MatDp(dis12)) are also related to the placental function. Mice with MatDp(dis12) have late embryonic lethality or neonatal lethality associated with growth retardation. Peg11/Rtl1 (paternally expressed 11/retrotransposon-like 1) is a major gene responsible for these phenotypes. Half of Peg11/Rtl1 KO mice have late fetal lethality and another half have neonatal lethality associated with late fetal growth retardation [55]. This is because of the fetal capillary abnormality in the labyrinth layer. Endothelial cells in the fetal capillaries are phagocytosed by surrounding trophoblast cells and clogged at many sites, indicating that Peg11/Rtl1 is essential for the maintenance of the feto-maternal interface of the placenta during gestation. Dlk1/Peg12 may also contribute to these phenotypes because in KO mice there is partial neonatal lethality associated with growth retardation [56,57]. Mice with PatDp(dis12) have late embryonic lethality associated with abnormal fetal morphology and placental enlargement [58,59]. In this case, the major cause is overexpression of Peg11/Rtl1 due to loss of maternally expressed antiPeg11/antiRtl1. AntiPeg11/antiRtl1 is ncRNA but involves at least six micro RNAs targeting to Peg11/Rtl1 by RNA interference. Thus, loss of antiPeg11/antiRtl1 leads to 2 to 4 fold accumulation of Peg11/Rtl1 mRNA. AntiPeg11/anti/Rtl1 KO mice exhibited neonatal lethality and placental overgrowth [55]. In the labyrinth layer of antiPeg11/anti/Rtl1 KO mice, expansion of the fetal capillary size associated with severely damaged surrounding trophoblast cell layers was observed. In the case of paternal duplication (or paternal disomy), double dosage of Peg11/Rtl1 without maternally expressed antiPeg11/antiRtl1 leads to 4–6 fold increment of Peg11/Rtl1 mRNA, causing more severe abnormal phenotypes. Thus, both the lack and overexpression of Peg11/Rtl11 are also responsible for the various phenotypes observed in MatDp(dis12) and PatDp(dis12), respectively. The same is true for human patients with MatDp and PatDp of chromosome 14, orthologous to mouse chromosome 12 [57].
Several imprinted genes so far described are important in the transfer of nutrients across the placenta. Genes that increase growth are usually paternally expressed, such as Igf2, Peg1/Mest, Peg10, Peg3/Pw1, Kcnqlot1/Lit1, Rasgrf1, Zac1, Peg11/Rtl1, Dlk1, while genes that tend to restrict growth are maternally expressed, such as Phlda2, Igf2r, Meg1/Grb10. This was the basis of the original parental conflict theory, which proposed that the paternally inherited genome would be modified to increase the growth of his offspring to increase his genetic fitness [60]. Conversely, the maternal genome would be modified to restrict resources to any one young or litter, allowing her to carry many successive pregnancies, thus increasing her genetic contribution to the next generation.
Another theory, the co-adaptation hypothesis, explains how paternally expressed imprinted genes have been maintained at some loci due to co-adaptation of the maternal hypothalamus and the placenta [61–63]. This is supported by the observations that Peg3 (paternally expressed gene 3), which is involved in maternal care, placental nurturing and regulating milk let-down in the female, also functions in the hypothalamus of the neonate to regulate attachment to the nipple and sucking behaviour [61–66]. The hypothalamus may be a more important site for genomic imprinting than previously recognized, and involved in long-term programming of hypothalamic functions [67]. Further studies are awaited with interest.
Recently, the paternal transmission of X-linked RLIM/Rnf12 gene encoding a nuclear ubiquitin ligase is reported to be essential for mammary gland development because the paternal X chromosome is selectively activated in the mammary gland [68]. Usually, in eutherian mammals random X chromosome inactivation occurs in female tissues and organs while paternal X chromosome is selectively inactivated in the placenta. In contrast, in marsupials, X-inactivation is paternal [69]. Interestingly, certain regions in female brain have a bias to silence the paternal X chromosome [70,71]. The biological meaning of this phenomenon is unclear at the moment and will be addressed when such an organ-specific paternal X chromosome activation/inactivation does occur in other reproductive organs and tissues, such as uterus, ovary and specific sites of the brain. As PEG3 is an autosomal imprinted gene showing constant paternal expression in all the tissue and organs, the situation may be different in this case.
In marsupials, about 18 genes that are imprinted in eutherians have so far been tested (table 1), not including X-inactivation. Of these 18, only six have genomic imprinting: H19 and PEG10 (see below for more details) have differentially methylated regions (DMRs) and IGF2, IGF2R, PEG1 and INS do not [51,73,74,76,78,80,100]. All of these are expressed in the marsupial placenta except for IGF2R which as yet has not been examined in the placenta. Insulin (INS) and insulin-like growth factor regulate growth of the placenta and the young. Tammar wallaby INS, like INS in humans and Ins2, is expressed in the yolk sac placenta [78]. This was thought to be the exclusive site in which this gene is imprinted [77]. However, we have demonstrated imprinting in other tissues (see the next section).
Table 1.
List of genes that are imprinted in eutherian mammals tested for imprinting in marsupial mammals.
| gene | eutherian reference | marsupial reference |
|---|---|---|
| imprinted in marsupials | ||
| with a DMR | ||
| H19 | Zhang & Tycko [72] | Smits et al. [51] |
| PEG10 | Ono et al. [42] | Suzuki et al. [73] |
| without a DMR | ||
| IGF2 | DeChiara et al. [47] | O'Neill et al. [74] |
| IGF2R | Barlow et al. [75] | Killian et al. [76] |
| INS | Moore et al. [77] | Ager et al. [78] |
| PEG1/MEST | Kaneko-Ishino et al. [79] | Suzuki et al. [80] |
| not imprinted in marsupials | ||
| PHLDA2 (IPL) | Qian et al. [81] | Suzuki et al. [82] |
| PPP1R9A (Ppp1r9a) | Nakabayashi et al. [83] | Suzuki et al. [73] |
| SGCE | Müller et al. [84] | Suzuki et al. [73] |
| SNRPN | Leff et al. [85] | Rapkins et al. [86] |
| UBE3A | Herzing et al. [87] | Rapkins et al. [86] |
| DLK | Wylie et al. [88], da Rocha et al. [89] | Weidman et al. [90], Edwards et al. [91] |
| DIO3 | Tsai et al. [92], da Rocha et al. [89] | Edwards et al. [91] |
| not expressed in marsupials | ||
| RTL1 (Rtl1;PEG11) | Seitz et al. [93] | Edwards et al. [91] |
| no orthologue found in marsupials | ||
| Air/Airn | Lyle et al. [94] | Weidman et al. [95] |
| MEG3 | Miyoshi et al. [96] | Weidman et al. [90] |
| NNAT | Kagitani et al. [97] | Evans et al. [98] |
| PEG3 | Kaneko-Ishino et al. [79] | Suzuki et al. [99] |
A number of other genes imprinted in eutherians are not imprinted in the marsupial placenta (table 1). PHILDA2, for example, is another gene, like Rtl1, which is essential for placental formation and function, yet neither gene is imprinted in marsupials and Rtl1 is not expressed [55,82,91]. The DLK1–DIO3 imprinting cluster appears to have arisen with the introduction of a new gene only in the eutherian cluster [91]. The marsupial locus is twice the size of the eutherian one, because of the accumulation of LINE repeats. SNRPN in the tammar and UBE3A in the tammar and in the platypus are not imprinted [86], and the rearrangement that brought UBE3A and SNRPN together must have occurred in the eutherian ancestor after the two groups of therian mammals diverged from each other.
No orthologue has been found for the ncRNA Air/Airn, or for the genes NNAT or MEG3 [90,95,98]. PEG3 cannot be identified in the tammar genome possibly because of poor genomic coverage of this region [99,101]. These differences in the extent of imprinting may reflect the different reproductive strategies of these two infraclasses of therian mammals, since marsupials have a uniformly short gestation. However, they have an extremely sophisticated and lengthy lactation, during which milk composition changes dramatically [20], and the maternal control of post natal growth is absolute [102,103]. Perhaps there are as yet undiscovered imprinted genes in marsupials that are not imprinted in eutherian mammals.
(b). Post natal imprinting and behaviour
Viviparity results in the birth of live young. In mammals, the maternal support of the young extends beyond birth into a period in which there is extensive post natal care of the young, supported by the secretion of milk. Nutrient transfer via a modification of the maternal physiology may be the key mammalian characteristic, using both placenta and mammary gland to do so. However, in the pigeon there is a crop milk produced from the oesophagus that is rich in lipids and protein and which sustains the young squab. Similarly, flamingos and especially penguins also produce crop milk. However, these secretions are produced by both sexes, and so are not in any way analogous to mammalian milk. Thus, in mammals it is the convergence of viviparity, placentation and lactation in the female that has led to their successful mode of reproduction.
The mammary gland is thought to have evolved initially as a small integumental gland that synthesized antimicrobial secretions, and that the nutritional role evolved subsequent to its protective function [104]. It now seems probable that it evolved from the innate immune system [105]. The mammary gland regulates post natal nutritional transfer by a positive feedback loop with the mother's brain in response to the sucking stimulus, in a similar way to that observed between the placenta, fetus and the maternal hypothalamus. There are monoallelically expressed imprinted genes in the mammary gland that are mis-regulated in breast cancer [106,107]. We have recently shown that two genes intimately involved in growth and metabolism, INS and IGF2, are both tissue-specifically imprinted in the marsupial mammary gland and liver [108]. GRB10 binds to the INS and IGF1 receptors and inhibits the growth-promoting activities of INS and IGF1 and 2 [109]. Disruption of GRB10 in mice causes fetal and placental overgrowth [110–112]. Thus, GRB10 may also regulate mammary gland growth and ultimately milk production through its inhibition of INS and IGF2. Genomic imprinting in the mammary gland therefore may be as critical for regulating post natal growth as it is for regulating pre natal growth in the placenta. These data also support the co-adaptation hypothesis. It is tempting to speculate that the acquisition of imprinting in the mammary gland might have contributed to the development and elaboration of lactation as a key reproductive strategy amongst mammals.
4. Evolution of mammalian imprinting
(a). Germline DMR, a master key for imprinting control
Imprinted genes are most often seen in clusters termed imprinted regions or domains. Imprinted expression of multiple genes in an imprinted domain is coordinately regulated by a single genomic element called the gDMR or ICR (imprinting control region), even in the case that an imprinted region consists of both paternally and maternally expressed genes (PEGs and MEGs). gDMRs are CpG-rich and differential DNA methylation is observed between two parental alleles. The difference in DNA methylation on gDMRs is established during gametogenesis and is maintained throughout development. There are two types of gDMRs, one is paternally methylated and the other is maternally methylated. Eighteen loci are associated with maternal methylation and all these maternal gDMRs are located at promoters while only three loci have paternally methylated gDMRs and are located in intergenic regions (The Gpr1–Zdbf2 locus, which was previously thought to be the fourth paternally imprinted region, has recently been reported as a maternally imprinted region) [113,114]. In the Igf2–H19 domain, the paternally methylated gDMR represses H19 expression from the paternal allele, but on the other hand it induces upstream Igf2 expression from the paternal allele inhibiting the binding of an insulator protein, CTCF, which blocks the action of downstream enhancer (figure 2). Thus, DNA methylation of a single gDMR can induce the Igf2 (Peg) expression and the H19 (Meg) repression at the same time in this regulatory mechanism known as the insulator model (figure 2). Large ncRNA-mediated epigenetic silencing is another important regulatory mechanism to control domain-wide imprinted expression by a single gDMR. In the Kcnq1 and Igf2r domains, the maternally methylated gDMRs induce paternal expression of the large ncRNAs named Lit1/Kcnq1ot1 and Airn/Air, respectively, and these ncRNAs mediate domain-wide silencing only on the paternal allele by recruiting repressive epigenetic modifiers such as G9a [115,116]. Because differential methylation of gDMRs plays the most critical role in the imprinting regulatory mechanism, the acquisition of gDMRs in the genome must be a pivotal event for the evolution of imprinting in mammals. The emergence of imprinted regions must have been related to the emergence of gDMR in the mammalian genome. It is therefore quite important to elucidate how the gDMRs emerged in the imprinted region and what the origin of gDMR sequences is.
Figure 2.
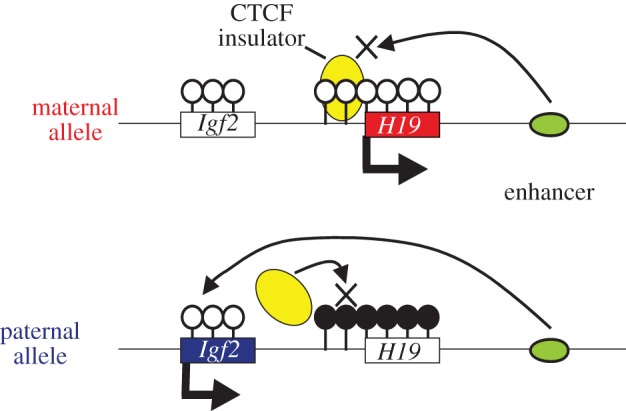
Regulation of the paternal and maternal expression in the Igf2–H19 imprinted domain by a single gDMR. The open lollipops represent unmethylated CpG islands and the black lollipops represent methylated CpG islands. The boxes represent Igf2 and H19 genes and the arrows from the boxes indicate expression of the genes. The yellow and green circles represent the CTCF insulator protein and a downstream enhancer, respectively.
(b). Biological importance of the regulatory mechanisms
Both Pegs and Megs are often contained in a single imprinted domain as described above. Regardless of the mechanisms directly regulating imprinted expression of each gene, it is true that only Pegs or Megs are expressed from the chromosome with a methylated or unmethylated gDMR. Therefore, the only way that the complete set of Pegs and Megs can be expressed in a single diploid cell is that each parental chromosome has a reciprocal methylation pattern of gDMRs. The imprinted regions are unique regions where epigenetic complementation by two parental chromosomes is necessary. As many imprinted genes are essential for mammalian development, this is the reason why mammals cannot lose genomic imprinting once the complex gene expression pattern has evolved at imprinted regions [117].
(c). Retrotransposons, novel CpG islands and gDMRs
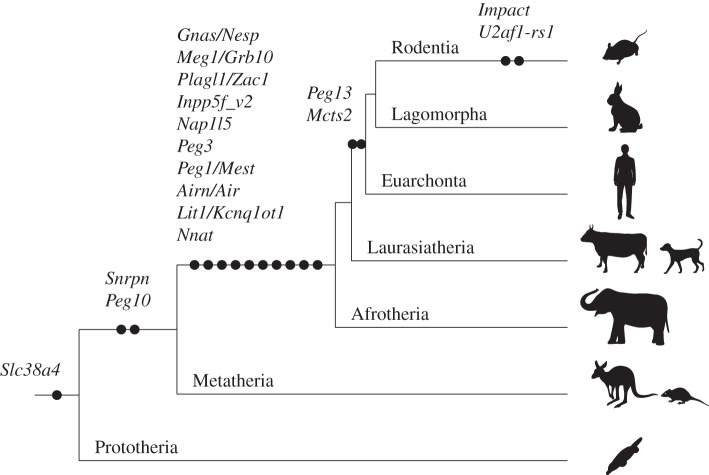
In therian genomes, almost half of the genomic sequences contain traces of transposable elements such as transposons and retrotransposons [118]. It has long been suggested that the molecular mechanisms underlying genomic imprinting evolved from host defence epigenetic mechanisms including small RNAs, large ncRNAs, DNA methylation and chromatin modification against these transposable elements [119–122]. Genomic imprinting is found in some plants (eg. Arabidopsis sp., Zea mays), some insects and in some mammals. However, it is most widespread in mammals, and almost all of the imprinted genes described are expressed in the placenta. Interestingly, a failure of the genome defence mechanism to regulate the expression of retroelements can lead to placental defects, as recently demonstrated in hybrids between Mus musculus and Mus caroli, in which the DNA methylation is lost [123]. In both plants and animals there appears to be a correlation of imprinting with the ratio of transposable elements. Once imprinting is established, imprinted domains can develop in several ways [124]. Imprinting can spread to adjacent regions in a stepwise manner, or can develop by genome reorganization. Divergent evolution of the domain can also influence the acquisition of imprinting (reviewed in [125]). In monotremes, the arrangements of the gene clusters that are imprinted in eutherians are conserved in other vertebrates [126,127], but the distribution of repeats including LTR and DNA elements has significantly expanded in only therian mammals [126]. There are several reports suggesting that retrotransposition is involved in the acquisition of a gDMR. We previously reported that the insertion of Peg10, a retrotransposon-derived imprinted gene essential for placental development in the mouse, must have occurred in therian ancestors after the divergence of marsupials and eutherian mammals from monotremes [73]. The CpG island (CGI) forming the PEG10 DMR has also newly emerged in the therian ancestor [73]. The IGF2–H19 imprinted region is also conserved in both marsupials and eutherians and H19 genes as well as H19 DMRs emerged in the therian ancestor [51]. These genomic data demonstrated that PEG10 and IGF2–H19 imprinted regions are the oldest among all the imprinted regions during therian evolution and both the maternal gDMR (PEG10 DMR) and paternal gDMR (H19 DMR) were established when the genomic imprinting started at least 160 Ma. Importantly, they are essential for embryonic and placental growth and development, as mentioned above. Also some of the small imprinted genes that reside in an intron of other genes, such as Mcts2, Nap1l5, Inpp5f_v2, U2af1-rs1 and Nnat, are thought to be inserted into their present positions by retrotransposition [98,128] (figure 3). Interestingly, in every case, the CGIs forming the gDMR probably emerged as novel CGIs at the same time as the retrotransposition of each gene occurred. Therefore, we recently examined the generality of the hypothesis that the CGIs forming gDMRs were newly acquired during mammalian evolution by reviewing the time of novel CGI emergence for all the maternal gDMR loci using new and published data [99]. The comparative sequence analyses suggested that emergence of novel CGIs occurred universally in the maternal gDMR loci at different time points during mammalian evolution (figures 3 and 4). In most loci, the novel CGIs emerged in introns but in Slc38a4, Snrpn and Gnas loci CGIs were unlikely to have emerged in introns in the ancestral mammal and differential methylation was acquired only in the eutherian lineage. In the eutherian lineage, interestingly, the location of the CGIs became internalized within the transcription unit by the acquisition of the IC transcript and Nesp to Snrpn and Gnas loci, respectively. This is consistent with the recent studies showed that transcription is required to establish maternal imprinting at these loci [129,130]. Thus, the emergence of a novel CGI in an intron of an existing gene or the acquisition of upstream transcript over an existing CGI might be part of the evolutionary pathway for emergence of gDMRs. Not all loci have evidence of involvement of retrotransposition events for the acquisition of gDMRs, but considering the number of gDMRs associated with retrogenes, retrotransposition appears to be a key mechanism by which novel CGIs have been acquired. However, the mechanistic link between retrotransposition, novel CGI acquisition and differential methylation is largely unknown. The mechanism of novel CGI emergence may be more complex than the suggestion that CpG sequences originated solely by insertion of GC-rich retrotransposons [99]. Further investigations will be required to clarify how these phenomena can be connected.
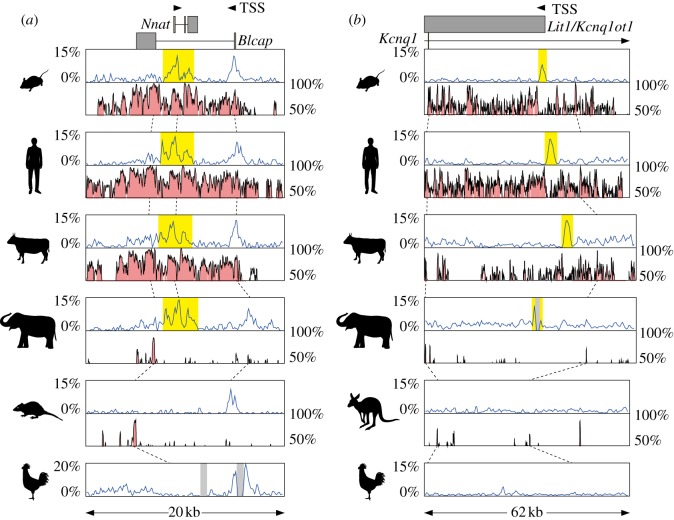
Figure 3.
(a) Comparison of CpG contents and conservation among the orthologous genomic regions around the Nnat gDMR. The upper blue graphs show the CpG content in the genomic sequences. The lower pink graphs show conserved regions in the genomic sequences between one species and the other species located just below (eg. the pink graph seen in the mouse row is the comparison of mouse (base) and human and the graph in the human row is the comparison of human (base) and cow). The broken lines indicate where some conservation peaks in the upper row correspond in the next lower row. The arrowhead indicates the transcription start site (TSS) with the direction and the grey box shows the exon. Gaps in the sequences are represented by the light grey shadows in graph regions. The CGI forming gDMR in the mouse and the corresponding CGI in other species are highlighted in yellow. (b) Comparison of CpG contents and conservation among the orthologous genomic regions around the Lit1/Kcnq1ot1 gDMR. Explanations for the each component are the same as (a).
Figure 4.
The timing of the novel CpG island emergence in each maternal gDMR locus during mammalian evolution. The black circles represent the acquisition of novel CGI to the locus. The genes associated with novel CGI emergence are shown above the black circles. The silhouettes represent several example species from each group.
5. Conclusions
Genomic imprinting in mammals is essentially a conserved process, but has greatly been expanded in the Eutheria. It is clear that imprinting arose at many different time points in mammalian evolution due to different selective pressures at different loci and that it is continuing to evolve. It is also clear that there is a great diversity of roles that imprinted genes play. Acquisition of novel CGIs appears to be a key genomic change for the evolution of genomic imprinting that generally occurred in the maternal gDMR loci. Novel CGIs may also have emerged in genomic loci other than in imprinted domains, but only became differentially methylated under certain conditions, contributing to the diversification of mammals. This might explain why imprinted genes are often associated with fetal–maternal nutrient transfer, placental development, viviparity, lactation and the mammalian-specific maternal behaviour associated with suckling young that must have evolved for mammals to survive.
References
- 1.Luo ZX, Yuan CX, Meng QJ, Ji Q. 2011. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature 476, 442–445 10.1038/nature10291 (doi:10.1038/nature10291) [DOI] [PubMed] [Google Scholar]
- 2.Kaneko-Ishino T, Ishino F. 2010. Retrotransposon silencing by DNA methylation contributed to the evolution of placentation and genomic imprinting in mammals. Dev. Growth Differ. 52, 533–543 10.1111/j.1440-169X.2010.01194.x (doi:10.1111/j.1440-169X.2010.01194.x) [DOI] [PubMed] [Google Scholar]
- 3.Mossman HW. 1987. Vertebrate fetal membranes. New Brunswick, NJ: Rutgers University Press [Google Scholar]
- 4.Amoroso EC, Heap RB, Renfree MB. 1979. Hormones and the evolution of viviparity. In Hormones and evolution (ed. Barrington JW.), pp. 925–989 New York, NY: Academic Press [Google Scholar]
- 5.Renfree MB. 1982. Implantation and placentation. In Reproduction in mammals (eds Austin CR, Short RV.), pp. 26–69, 2nd edn Cambridge, UK: Cambridge University Press [Google Scholar]
- 6.Jones CJ, Jauniaux E. 1995. Ultrastructure of the materno-embryonic interface in the first trimester of pregnancy. Micron 26, 145–173 10.1016/0968-4328(95)00002-L (doi:10.1016/0968-4328(95)00002-L) [DOI] [PubMed] [Google Scholar]
- 7.Enders AC, King BF. 1993. Development of the human yolk sac. In The human yolk sac and yolk sac tumors (ed. Nogales FF.), pp. 33–47 Berlin, Germany: Springer [Google Scholar]
- 8.Takashina T. 1993. Histology of the secondary human yolk sac with special reference to hematopoiesis. In The human yolk sac and yolk sac tumors (ed. Nogales FF.), pp. 48–69 Berlin, Germany: Springer [Google Scholar]
- 9.Palis J, Yoder MC. 2001. Yolk-sac hematopoiesis: the first blood cells of mouse and man. Exp. Hematol. 29, 927–936 10.1016/S0301-472X(01)00669-5 (doi:10.1016/S0301-472X(01)00669-5) [DOI] [PubMed] [Google Scholar]
- 10.Migliaccio G, Migliaccio AR. 1993. Development of the human yolk sac. In The human yolk sac and yolk sac tumors (ed. Nogales FF.), pp. 70–83 Berlin, Germany: Springer [Google Scholar]
- 11.Renfree MB. 1972. Influence of the embryo on the marsupial uterus. Nature 240, 475–477 10.1038/240475a0 (doi:10.1038/240475a0) [DOI] [PubMed] [Google Scholar]
- 12.Renfree MB. 2000. Maternal recognition of pregnancy in marsupials. Rev. Reprod. 5, 6–11 10.1530/ror.0.0050006 (doi:10.1530/ror.0.0050006) [DOI] [PubMed] [Google Scholar]
- 13.Renfree MB. 2010. Marsupials: placental mammals with a difference. Placenta 31(Suppl), S21–S26 10.1016/j.placenta.2009.12.023 (doi:10.1016/j.placenta.2009.12.023) [DOI] [PubMed] [Google Scholar]
- 14.Menzies BR, Pask AJ, Renfree MB. 2011. Placental expression of pituitary hormones is an ancestral feature of therian mammals. Evodevo 2, 16. 10.1186/2041-9139-2-16 (doi:10.1186/2041-9139-2-16) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wooding P, Burton G. 2008. Comparative placentation: structures, functions and evolution. Berlin, Germany: Springer [Google Scholar]
- 16.Amoroso EC. 1952. Placentation. In Marshall‘s physiology of reproduction (ed. Parkes AS.), pp. 127–311 New York, NY: Longmans Green [Google Scholar]
- 17.Freyer C, Zeller U, Renfree MB. 2003. The marsupial placenta: a phylogenetic analysis. J. Exp. Zool. A Comp. Exp. Biol. 299, 59–77 10.1002/jez.a.10291 (doi:10.1002/jez.a.10291) [DOI] [PubMed] [Google Scholar]
- 18.Freyer C, Renfree MB. 2009. The mammalian yolk sac placenta. J. Exp. Zool. B Mol. Dev. Evol. 312, 545–554 10.1002/jez.b.21239 (doi:10.1002/jez.b.21239) [DOI] [PubMed] [Google Scholar]
- 19.Freyer C, Zeller U, Renfree MB. 2007. Placental function in two distantly related marsupials. Placenta 28, 249–257 10.1016/j.placenta.2006.03.007 (doi:10.1016/j.placenta.2006.03.007) [DOI] [PubMed] [Google Scholar]
- 20.Tyndale-Biscoe CH, Renfree MB. 1987. Reproductive physiology of marsupials. Cambridge, UK: Cambridge University Press [Google Scholar]
- 21.Padykula HA, Taylor JM. 1982. Marsupial placentation and its evolutionary significance. J. Reprod. Fertil. Suppl. 31, 95–104 [PubMed] [Google Scholar]
- 22.King BF, Enders AC. 1993. Comparative development of the mammalian yolk sac. In The human yolk sac and yolk sac tumors (ed. Nogales FF.), pp. 1–32 Berlin, Germany: Springer [Google Scholar]
- 23.Beckman DA, Brent RL, Lloyd JB. 1996. Sources of amino acids for protein synthesis during early organogenesis in the rat. 4. Mechanisms before envelopment of the embryo by the yolk sac. Placenta 17, 635–641 10.1016/S0143-4004(96)80082-8 (doi:10.1016/S0143-4004(96)80082-8) [DOI] [PubMed] [Google Scholar]
- 24.Brambell FWR. 1970. The transmission of passive immunity from mother to young. Amsterdam, The Netherlands: North-Holland Publishing Company [Google Scholar]
- 25.Merad Z, Wild AE. 1992. The route of maternal IgM transport to the rabbit fetus. Placenta 13, 291–304 10.1016/0143-4004(92)90044-T (doi:10.1016/0143-4004(92)90044-T) [DOI] [PubMed] [Google Scholar]
- 26.Beckman DA, Koszalka TR, Jensen M, Brent RL. 1990. Experimental manipulation of the rodent visceral yolk sac. Teratology 41, 395–404 10.1002/tera.1420410405 (doi:10.1002/tera.1420410405) [DOI] [PubMed] [Google Scholar]
- 27.Renfree MB. 1973. The composition of fetal fluids of the marsupial Macropus eugenii. Dev. Biol. 33, 62–79 10.1016/0012-1606(73)90165-6 (doi:10.1016/0012-1606(73)90165-6) [DOI] [PubMed] [Google Scholar]
- 28.Renfree MB. 1983. Marsupial reproduction the choice between placentation and lactation. Oxford Rev. Reprod. Biol. 5, 1–29 [Google Scholar]
- 29.Blackburn DG. 1982. Evolutionary origins of viviparity in the Reptilia I. Sauria Amphibia-Reptilia 3, 185–205 10.1163/156853882X00419 (doi:10.1163/156853882X00419) [DOI] [Google Scholar]
- 30.Blackburn DG. 2006. Squamate reptiles as model organisms for the evolution of viviparity. Herpetol. Monogr. 20, 131–146 10.1655/0733-1347(2007)20[131:SRAMOF]2.0.CO;2 (doi:10.1655/0733-1347(2007)20[131:SRAMOF]2.0.CO;2) [DOI] [Google Scholar]
- 31.Blackburn DG. 1992. Convergent evolution of viviparity, matrotrophy, and specializations for fetal nutrition in reptiles and other vertebrates. Am. Zool. 32, 313–321 10.1093/icb/32.2.313 (doi:10.1093/icb/32.2.313) [DOI] [Google Scholar]
- 32.Blackburn DG. 1993. Chorioallantoic placentation in squamate reptiles—structure, function, development, and evolution. J. Exp. Zool. 266, 414–430 10.1002/jez.1402660508 (doi:10.1002/jez.1402660508) [DOI] [Google Scholar]
- 33.Stewart JR, Thompson MB. 2000. Evolution of placentation among squamate reptiles: recent research and future directions. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 127, 411–431 10.1016/S1095-6433(00)00273-7 (doi:10.1016/S1095-6433(00)00273-7) [DOI] [PubMed] [Google Scholar]
- 34.Murphy BF, Thompson MB. 2011. A review of the evolution of viviparity in squamate reptiles: the past, present and future role of molecular biology and genomics. J. Comp. Physiol. B 181, 575–594 10.1007/s00360-011-0584-0 (doi:10.1007/s00360-011-0584-0) [DOI] [PubMed] [Google Scholar]
- 35.Blackburn DG, Vitt LJ, Beuchat CA. 1984. Eutherian-like reproductive specializations in a viviparous reptile. Proc. Natl Acad. Sci. USA 81, 4860–4863 10.1073/pnas.81.15.4860 (doi:10.1073/pnas.81.15.4860) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson MB, Stewart JR, Speake BK. 2000. Comparison of nutrient transport across the placenta of lizards differing in placental complexity. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 127, 469–479 10.1016/S1095-6433(00)00277-4 (doi:10.1016/S1095-6433(00)00277-4) [DOI] [PubMed] [Google Scholar]
- 37.Blackburn DG, Flemming AF. 2012. Invasive implantation and intimate placental associations in a placentotrophic african lizard, Trachylepis ivensi (Scincidae). J. Morphol. 273, 137–159 10.1002/jmor.11011 (doi:10.1002/jmor.11011) [DOI] [PubMed] [Google Scholar]
- 38.Brandley MC, Young RL, Warren DL, Thompson MB, Wagner GP. 2012. Uterine gene expression in the live-bearing lizard, Chalcides ocellatus, reveals convergence of squamate and mammalian pregnancy mechanisms. Genome Biol. Evol. 4, 1–18 10.1093/gbe/evr123 (doi:10.1093/gbe/evr123) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaneko-Ishino T, Kohda T, Ishino F. 2003. The regulation and biological significance of genomic imprinting in mammals. J. Biochem. 133, 699–711 10.1093/jb/mvg090 (doi:10.1093/jb/mvg090) [DOI] [PubMed] [Google Scholar]
- 40.MouseBook. 2012. Murine imprinting data and references. See http://www.mousebook.org/
- 41.Ono R, et al. 2006. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 38, 101–106 10.1038/ng1699 (doi:10.1038/ng1699) [DOI] [PubMed] [Google Scholar]
- 42.Ono R, Kobayashi S, Wagatsuma H, Aisaka K, Kohda T, Kaneko-Ishino T, Ishino F. 2001. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 73, 232–237 10.1006/geno.2001.6494 (doi:10.1006/geno.2001.6494) [DOI] [PubMed] [Google Scholar]
- 43.Guillemot F, et al. 1995. Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nat. Genet. 9, 235–242 10.1038/ng0395-235 (doi:10.1038/ng0395-235) [DOI] [PubMed] [Google Scholar]
- 44.Surani MA, Barton SC, Norris ML. 1984. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature 308, 548–550 10.1038/308548a0 (doi:10.1038/308548a0) [DOI] [PubMed] [Google Scholar]
- 45.McGrath J, Solter D. 1984. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell 37, 179–183 10.1016/0092-8674(84)90313-1 (doi:10.1016/0092-8674(84)90313-1) [DOI] [PubMed] [Google Scholar]
- 46.Mann JR, Lovell-Badge RH. 1984. Inviability of parthenogenones is determined by pronuclei, not egg cytoplasm. Nature 310, 66–67 10.1038/310066a0 (doi:10.1038/310066a0) [DOI] [PubMed] [Google Scholar]
- 47.DeChiara TM, Robertson EJ, Efstratiadis A. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64, 849–859 10.1016/0092-8674(91)90513-X (doi:10.1016/0092-8674(91)90513-X) [DOI] [PubMed] [Google Scholar]
- 48.Hatada I, et al. 1996. An imprinted gene p57KIP2 is mutated in Beckwith–Wiedemann syndrome. Nat. Genet. 14, 171–173 10.1038/ng1096-171 (doi:10.1038/ng1096-171) [DOI] [PubMed] [Google Scholar]
- 49.Sun FL, Dean WL, Kelsey G, Allen ND, Reik W. 1997. Transactivation of Igf2 in a mouse model of Beckwith–Wiedemann syndrome. Nature 389, 809–815 10.1038/39797 (doi:10.1038/39797) [DOI] [PubMed] [Google Scholar]
- 50.Hattori N, Davies TC, Anson-Cartwright L, Cross JC. 2000. Periodic expression of the cyclin-dependent kinase inhibitor p57(Kip2) in trophoblast giant cells defines a G2-like gap phase of the endocycle. Mol. Biol. Cell 11, 1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smits G, et al. 2008. Conservation of the H19 noncoding RNA and H19-IGF2 imprinting mechanism in therians. Nat Genet 40, 971–976 10.1038/ng.168 (doi:10.1038/ng.168) [DOI] [PubMed] [Google Scholar]
- 52.Bartolomei MS, Zemel S, Tilghman SM. 1991. Parental imprinting of the mouse H19 gene. Nature 351, 153–155 10.1038/351153a0 (doi:10.1038/351153a0) [DOI] [PubMed] [Google Scholar]
- 53.Yoshimizu T, et al. 2008. The H19 locus acts in vivo as a tumor suppressor. Proc. Natl Acad. Sci. USA 105, 12 417–12 422 10.1073/pnas.0801540105 (doi:10.1073/pnas.0801540105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawahara M, Morita S, Takahashi N, Kono T. 2009. Defining contributions of paternally methylated imprinted genes at the Igf2-H19 and Dlk1-Gtl2 domains to mouse placentation by transcriptomic analysis. J. Biol. Chem. 284, 17 751–17 765 10.1074/jbc.M109.000299 (doi:10.1074/jbc.M109.000299) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sekita Y, et al. 2008. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 40, 243–248 10.1038/ng.2007.51 (doi:10.1038/ng.2007.51) [DOI] [PubMed] [Google Scholar]
- 56.Moon YS, Smas CM, Lee K, Villena JA, Kim KH, Yun EJ, Sul HS. 2002. Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity. Mol. Cell Biol. 22, 5585–5592 10.1128/MCB.22.15.5585-5592.2002 (doi:10.1128/MCB.22.15.5585-5592.2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kagami M, et al. 2008. Deletions and epimutations affecting the human 14q32.2 imprinted region in individuals with paternal and maternal upd(14)-like phenotypes. Nat. Genet. 40, 237–242 10.1038/ng.2007.56 (doi:10.1038/ng.2007.56) [DOI] [PubMed] [Google Scholar]
- 58.Georgiades P, Watkins M, Surani MA, Ferguson-Smith AC. 2000. Parental origin-specific developmental defects in mice with uniparental disomy for chromosome 12. Development 127, 4719–4728 [DOI] [PubMed] [Google Scholar]
- 59.Tevendale M, Watkins M, Rasberry C, Cattanach B, Ferguson-Smith AC. 2006. Analysis of mouse conceptuses with uniparental duplication/deficiency for distal chromosome 12, comparison with chromosome 12 uniparental disomy and implications for genomic imprinting. Cytogenet. Genome Res. 113, 215–222 10.1159/000090835 (doi:10.1159/000090835) [DOI] [PubMed] [Google Scholar]
- 60.Haig D, Westoby M. 1989. Parent-specific gene-expression and the triploid endosperm. Am. Nat. 134, 147–155 10.1086/284971 (doi:10.1086/284971) [DOI] [Google Scholar]
- 61.Keverne EB, Curley JP. 2008. Epigenetics, brain evolution and behaviour. Front. Neuroendocrinol. 29, 398–412 10.1016/j.yfrne.2008.03.001 (doi:10.1016/j.yfrne.2008.03.001) [DOI] [PubMed] [Google Scholar]
- 62.Curley JP, Barton S, Surani A, Keverne EB. 2004. Coadaptation in mother and infant regulated by a paternally expressed imprinted gene. Proc. Biol. Sci. 271, 1303–1309 10.1098/rspb.2004.2725 (doi:10.1098/rspb.2004.2725) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keverne EB. 2013. Importance of the matriline for genomic imprinting, brain development and behaviour. Phil. Trans. R. Soc. B 368, 20110327. 10.1098/rstb.2011.0327 (doi:10.1098/rstb.2011.0327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keverne EB, Fundele R, Narasimha M, Barton SC, Surani MA. 1996. Genomic imprinting and the differential roles of parental genomes in brain development. Brain Res. Dev. Brain Res. 92, 91–100 10.1016/0165-3806(95)00209-X (doi:10.1016/0165-3806(95)00209-X) [DOI] [PubMed] [Google Scholar]
- 65.Swaney WT, Curley JP, Champagne FA, Keverne EB. 2007. Genomic imprinting mediates sexual experience-dependent olfactory learning in male mice. Proc. Natl Acad. Sci. USA 104, 6084–6089 10.1073/pnas.0609471104 (doi:10.1073/pnas.0609471104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Constância M, Kelsey G, Reik W. 2004. Resourceful imprinting. Nature 432, 53–57 10.1038/432053a (doi:10.1038/432053a) [DOI] [PubMed] [Google Scholar]
- 67.Ivanova E, Kelsey G. 2011. Imprinted genes and hypothalamic function. J. Mol. Endocrinol. 47, R67–R74 10.1530/JME-11-0065 (doi:10.1530/JME-11-0065) [DOI] [PubMed] [Google Scholar]
- 68.Jiao B, et al. 2012. Paternal RLIM/Rnf12 is a survival factor for milk-producing alveolar cells. Cell 149, 630–641 10.1016/j.cell.2012.02.056 (doi:10.1016/j.cell.2012.02.056) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cooper DW, VandeBerg JL, Sharman GB, Poole WE. 1971. Phosphoglycerate kinase polymorphism in kangaroos provides further evidence for paternal X inactivation. Nat. New Biol. 230, 155–157 [DOI] [PubMed] [Google Scholar]
- 70.Gregg C, Zhang J, Butler JE, Haig D, Dulac C. 2010. Sex-specific parent-of-origin allelic expression in the mouse brain. Science 329, 682–685 10.1126/science.1190831 (doi:10.1126/science.1190831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang X, Soloway PD, Clark AG. 2010. Paternally biased X inactivation in mouse neonatal brain. Genome Biol. 11, R79. 10.1186/gb-2010-11-7-r79 (doi:10.1186/gb-2010-11-7-r79) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang Y, Tycko B. 1992. Monoallelic expression of the human H19 gene. Nat. Genet. 1, 40–44 10.1038/ng0492-40 (doi:10.1038/ng0492-40) [DOI] [PubMed] [Google Scholar]
- 73.Suzuki S, et al. 2007. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 3, e55. 10.1371/journal.pgen.0030055 (doi:10.1371/journal.pgen.0030055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.O'Neill MJ, Ingram RS, Vrana PB, Tilghman SM. 2000. Allelic expression of IGF2 in marsupials and birds. Dev. Genes Evol. 210, 18–20 10.1007/PL00008182 (doi:10.1007/PL00008182) [DOI] [PubMed] [Google Scholar]
- 75.Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N. 1991. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349, 84–87 10.1038/349084a0 (doi:10.1038/349084a0) [DOI] [PubMed] [Google Scholar]
- 76.Killian JK, Byrd JC, Jirtle JV, Munday BL, Stoskopf MK, MacDonald RG, Jirtle RL. 2000. M6P/IGF2R imprinting evolution in mammals. Mol. Cell 5, 707–716 10.1016/S1097-2765(00)80249-X (doi:10.1016/S1097-2765(00)80249-X) [DOI] [PubMed] [Google Scholar]
- 77.Moore GE, Abu-Amero SN, Bell G, Wakeling EL, Kingsnorth A, Stanier P, Jauniaux E, Bennett ST. 2001. Evidence that insulin is imprinted in the human yolk sac. Diabetes 50, 199–203 10.2337/diabetes.50.1.199 (doi:10.2337/diabetes.50.1.199) [DOI] [PubMed] [Google Scholar]
- 78.Ager E, Suzuki S, Pask A, Shaw G, Ishino F, Renfree MB. 2007. Insulin is imprinted in the placenta of the marsupial, Macropus eugenii. Dev. Biol. 309, 317–328 10.1016/j.ydbio.2007.07.025 (doi:10.1016/j.ydbio.2007.07.025) [DOI] [PubMed] [Google Scholar]
- 79.Kaneko-Ishino T, et al. 1995. Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat. Genet. 11, 52–59 10.1038/ng0995-52 (doi:10.1038/ng0995-52) [DOI] [PubMed] [Google Scholar]
- 80.Suzuki S, Renfree MB, Pask AJ, Shaw G, Kobayashi S, Kohda T, Kaneko-Ishino T, Ishino F. 2005. Genomic imprinting of IGF2, p57(KIP2) and PEG1/MEST in a marsupial, the tammar wallaby. Mech. Dev. 122, 213–222 10.1016/j.mod.2004.10.003 (doi:10.1016/j.mod.2004.10.003) [DOI] [PubMed] [Google Scholar]
- 81.Qian N, et al. 1997. The IPL gene on chromosome 11p15.5 is imprinted in humans and mice and is similar to TDAG51, implicated in Fas expression and apoptosis. Hum. Mol. Genet. 6, 2021–2029 10.1093/hmg/6.12.2021 (doi:10.1093/hmg/6.12.2021) [DOI] [PubMed] [Google Scholar]
- 82.Suzuki S, Shaw G, Kaneko-Ishino T, Ishino F, Renfree MB. 2011. Characterisation of marsupial PHLDA2 reveals eutherian specific acquisition of imprinting. BMC Evol. Biol. 11, 244. 10.1186/1471-2148-11-244 (doi:10.1186/1471-2148-11-244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakabayashi K, et al. 2004. Genomic imprinting of PPP1R9A encoding neurabin I in skeletal muscle and extra-embryonic tissues. J. Med. Genet. 41, 601–608 10.1136/jmg.2003.014142 (doi:10.1136/jmg.2003.014142) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Müller B, et al. 2002. Evidence that paternal expression of the epsilon-sarcoglycan gene accounts for reduced penetrance in myoclonus-dystonia. Am. J. Hum. Genet. 71, 1303–1311 10.1086/344531 (doi:10.1086/344531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Leff SE, Brannan CI, Reed ML, Ozcelik T, Francke U, Copeland NG, Jenkins NA. 1992. Maternal imprinting of the mouse Snrpn gene and conserved linkage homology with the human Prader–Willi syndrome region. Nat. Genet. 2, 259–264 10.1038/ng1292-259 (doi:10.1038/ng1292-259) [DOI] [PubMed] [Google Scholar]
- 86.Rapkins RW, et al. 2006. Recent assembly of an imprinted domain from non-imprinted components. PLoS Genet. 2, e182. 10.1371/journal.pgen.0020182 (doi:10.1371/journal.pgen.0020182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Herzing LB, Cook EH, Jr, Ledbetter DH. 2002. Allele-specific expression analysis by RNA-FISH demonstrates preferential maternal expression of UBE3A and imprint maintenance within 15q11–q13 duplications. Hum. Mol. Genet. 11, 1707–1718 10.1093/hmg/11.15.1707 (doi:10.1093/hmg/11.15.1707) [DOI] [PubMed] [Google Scholar]
- 88.Wylie AA, Murphy SK, Orton TC, Jirtle RL. 2000. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 10, 1711–1718 10.1101/gr.161600 (doi:10.1101/gr.161600) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. 2008. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 24, 306–316 10.1016/j.tig.2008.03.011 (doi:10.1016/j.tig.2008.03.011) [DOI] [PubMed] [Google Scholar]
- 90.Weidman JR, Maloney KA, Jirtle RL. 2006. Comparative phylogenetic analysis reveals multiple non-imprinted isoforms of opossum Dlk1. Mamm. Genome 17, 157–167 10.1007/s00335-005-0116-x (doi:10.1007/s00335-005-0116-x) [DOI] [PubMed] [Google Scholar]
- 91.Edwards CA, et al. 2008. The evolution of the DLK1-DIO3 imprinted domain in mammals. PLoS Biol. 6, e135. 10.1371/journal.pbio.0060135 (doi:10.1371/journal.pbio.0060135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsai CE, Lin SP, Ito M, Takagi N, Takada S, Ferguson-Smith AC. 2002. Genomic imprinting contributes to thyroid hormone metabolism in the mouse embryo. Curr. Biol. 12, 1221–1226 10.1016/S0960-9822(02)00951-X (doi:10.1016/S0960-9822(02)00951-X) [DOI] [PubMed] [Google Scholar]
- 93.Seitz H, Youngson N, Lin SP, Dalbert S, Paulsen M, Bachellerie JP, Ferguson-Smith AC, Cavaille J. 2003. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat. Genet. 34, 261–262 10.1038/ng1171 (doi:10.1038/ng1171) [DOI] [PubMed] [Google Scholar]
- 94.Lyle R, et al. 2000. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat. Genet. 25, 19–21 10.1038/75546 (doi:10.1038/75546) [DOI] [PubMed] [Google Scholar]
- 95.Weidman JR, Dolinoy DC, Maloney KA, Cheng JF, Jirtle RL. 2006. Imprinting of opossum Igf2r in the absence of differential methylation and Air. Epigenetics 1, 49–54 10.4161/epi.1.1.2592 (doi:10.4161/epi.1.1.2592) [DOI] [PubMed] [Google Scholar]
- 96.Miyoshi N, et al. 2000. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 5, 211–220 10.1046/j.1365-2443.2000.00320.x (doi:10.1046/j.1365-2443.2000.00320.x) [DOI] [PubMed] [Google Scholar]
- 97.Kagitani F, et al. 1997. Peg5/Neuronatin is an imprinted gene located on sub-distal chromosome 2 in the mouse. Nucleic Acids Res. 25, 3428–3432 10.1093/nar/25.17.3428 (doi:10.1093/nar/25.17.3428) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Evans HK, Weidman JR, Cowley DO, Jirtle RL. 2005. Comparative phylogenetic analysis of blcap/nnat reveals eutherian-specific imprinted gene. Mol. Biol. Evol. 22, 1740–1748 10.1093/molbev/msi165 (doi:10.1093/molbev/msi165) [DOI] [PubMed] [Google Scholar]
- 99.Suzuki S, Shaw G, Kaneko-Ishino T, Ishino F, Renfree MB. 2011. The evolution of mammalian genomic imprinting was accompanied by the acquisition of novel CpG islands. Genome Biol. Evol. 3, 1276–1283 10.1093/gbe/evr104 (doi:10.1093/gbe/evr104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ager EI, Pask AJ, Shaw G, Renfree MB. 2008. Expression and protein localisation of IGF2 in the marsupial placenta. BMC Dev. Biol. 8, 17. 10.1186/1471-213X-8-17 (doi:10.1186/1471-213X-8-17) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Renfree MB, et al. 2011. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 12, 414. 10.1186/gb-2011-12-12-414 (doi:10.1186/gb-2011-12-12-414) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Trott JF, Simpson KJ, Moyle R, Hearn CM, Shaw G, Nicholas KR, Renfree MB. 2003. Maternal regulation of milk composition, milk production and pouch young development during lactation in the tammar wallaby (Macropus eugenii). Biol. Reprod. 68, 929–936 10.1095/biolreprod.102.005934 (doi:10.1095/biolreprod.102.005934) [DOI] [PubMed] [Google Scholar]
- 103.Menzies BR, Shaw G, Fletcher TP, Renfree MB. 2007. Perturbed growth and development in marsupial young after reciprocal cross-fostering between species. Reprod. Fertil. Dev. 19, 976–983 10.1071/RD07142 (doi:10.1071/RD07142) [DOI] [PubMed] [Google Scholar]
- 104.Blackburn DG, Hayssen V, Murphy CJ. 1989. The origins of lactation and the evolution of milk—a review with new hypotheses. Mamm. Rev. 19, 1–26 10.1111/j.1365-2907.1989.tb00398.x (doi:10.1111/j.1365-2907.1989.tb00398.x) [DOI] [Google Scholar]
- 105.Vorbach C, Capecchi MR, Penninger JM. 2006. Evolution of the mammary gland from the innate immune system? BioEssays 28, 606–616 10.1002/bies.20423 (doi:10.1002/bies.20423) [DOI] [PubMed] [Google Scholar]
- 106.Yballe CM, Vu TH, Hoffman AR. 1996. Imprinting and expression of insulin-like growth factor-II and H19 in normal breast tissue and breast tumor. J. Clin. Endocrinol. Metab. 81, 1607–1612 10.1210/jc.81.4.1607 (doi:10.1210/jc.81.4.1607) [DOI] [PubMed] [Google Scholar]
- 107.Pedersen IS, et al. 1999. Frequent loss of imprinting of PEG1/MEST in invasive breast cancer. Cancer Res. 59, 5449–5451 [PubMed] [Google Scholar]
- 108.Stringer J, Suzuki S, Pask A, Shaw G, Renfree MB. 2012. Selected imprinting of INS in the marsupial. Epigenetics Chromatin 5, 14. 10.1186/1756-8935-5-14 (doi:10.1186/1756-8935-5-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morrione A, Valentinis B, Li S, Ooi JY, Margolis B, Baserga R. 1996. Grb10: a new substrate of the insulin-like growth factor I receptor. Cancer Res. 56, 3165–3167 [PubMed] [Google Scholar]
- 110.Charalambous M, Smith FM, Bennett WR, Crew TE, Mackenzie F, Ward A. 2003. Disruption of the imprinted Grb10 gene leads to disproportionate overgrowth by an Igf2-independent mechanism. Proc. Natl Acad. Sci. USA 100, 8292–8297 10.1073/pnas.1532175100 (doi:10.1073/pnas.1532175100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Charalambous M, Cowley M, Geoghegan F, Smith FM, Radford EJ, Marlow BP, Graham CF, Hurst LD, Ward A. 2010. Maternally-inherited Grb10 reduces placental size and efficiency. Dev. Biol. 337, 1–8 10.1016/j.ydbio.2009.10.011 (doi:10.1016/j.ydbio.2009.10.011) [DOI] [PubMed] [Google Scholar]
- 112.Smith FM, et al. 2007. Mice with a disruption of the imprinted Grb10 gene exhibit altered body composition, glucose homeostasis, and insulin signaling during postnatal life. Mol. Cell Biol. 27, 5871–5886 10.1128/MCB.02087-06 (doi:10.1128/MCB.02087-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bartolomei MS, Ferguson-Smith AC. 2011. Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 3, pii: a002592. 10.1101/cshperspect.a002592 (doi:10.1101/cshperspect.a002592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kobayashi H, Sakurai T, Sato S, Nakabayashi K, Hata K, Kono T. 2012. Imprinted DNA methylation reprogramming during early mouse embryogenesis at the Gpr1-Zdbf2 locus is linked to long cis-intergenic transcription. FEBS Lett. 586, 827–833 10.1016/j.febslet.2012.01.059 (doi:10.1016/j.febslet.2012.01.059) [DOI] [PubMed] [Google Scholar]
- 115.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322, 1717–1720 10.1126/science.1163802 (doi:10.1126/science.1163802) [DOI] [PubMed] [Google Scholar]
- 116.Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 32, 232–246 10.1016/j.molcel.2008.08.022 (doi:10.1016/j.molcel.2008.08.022) [DOI] [PubMed] [Google Scholar]
- 117.Kaneko-Ishino T, Kohda T, Ono R, Ishino F. 2006. Complementation hypothesis: the necessity of a monoallelic gene expression mechanism in mammalian development. Cytogenet. Genome Res. 113, 24–30 10.1159/000090811 (doi:10.1159/000090811) [DOI] [PubMed] [Google Scholar]
- 118.Ekram MB, Kang K, Kim H, Kim J. 2012. Retrotransposons as a major source of epigenetic variations in the mammalian genome. Epigenetics 7, 370–382 10.4161/epi.19462 (doi:10.4161/epi.19462) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Barlow DP. 1993. Methylation and imprinting: from host defense to gene regulation? Science 260, 309–310 10.1126/science.8469984 (doi:10.1126/science.8469984) [DOI] [PubMed] [Google Scholar]
- 120.Surani MA. 1998. Imprinting and the initiation of gene silencing in the germ line. Cell 93, 309–312 10.1016/S0092-8674(00)81156-3 (doi:10.1016/S0092-8674(00)81156-3) [DOI] [PubMed] [Google Scholar]
- 121.Matzke MA, Mette MF, Matzke AJ. 2000. Transgene silencing by the host genome defense: implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol. Biol. 43, 401–415 10.1023/A:1006484806925 (doi:10.1023/A:1006484806925) [DOI] [PubMed] [Google Scholar]
- 122.McDonald JF, Matzke MA, Matzke AJ. 2005. Host defenses to transposable elements and the evolution of genomic imprinting. Cytogenet. Genome Res. 110, 242–249 10.1159/000084958 (doi:10.1159/000084958) [DOI] [PubMed] [Google Scholar]
- 123.Brown JD, Piccuillo V, O'Neill RJ. 2012. Retroelement demethylation associated with abnormal placentation in Mus musculus×Mus caroli hybrids. Biol. Reprod. 86, 88. 10.1095/biolreprod.111.095273 (doi:10.1095/biolreprod.111.095273) [DOI] [PubMed] [Google Scholar]
- 124.Wood AJ, Oakey RJ. 2006. Genomic imprinting in mammals: emerging themes and established theories. PLoS Genet. 2, e147. 10.1371/journal.pgen.0020147 (doi:10.1371/journal.pgen.0020147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Renfree MB, Hore TA, Shaw G, Graves JA, Pask AJ. 2009. Evolution of genomic imprinting: insights from marsupials and monotremes. Annu. Rev. Genom. Hum. Genet. 10, 241–262 10.1146/annurev-genom-082908-150026 (doi:10.1146/annurev-genom-082908-150026) [DOI] [PubMed] [Google Scholar]
- 126.Pask AJ, Papenfuss AT, Ager EI, McColl KA, Speed TP, Renfree MB. 2009. Analysis of the platypus genome suggests a transposon origin for mammalian imprinting. Genome Biol. 10, R1. 10.1186/gb-2009-10-1-r1 (doi:10.1186/gb-2009-10-1-r1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Dunzinger U, Haaf T, Zechner U. 2007. Conserved synteny of mammalian imprinted genes in chicken, frog, and fish genomes. Cytogenet. Genome Res. 117, 78–85 10.1159/000103167 (doi:10.1159/000103167) [DOI] [PubMed] [Google Scholar]
- 128.Wood AJ, Roberts RG, Monk D, Moore GE, Schulz R, Oakey RJ. 2007. A screen for retrotransposed imprinted genes reveals an association between X chromosome homology and maternal germ-line methylation. PLoS Genet. 3, e20. 10.1371/journal.pgen.0030020 (doi:10.1371/journal.pgen.0030020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, James K, Dean W, Kelsey G. 2009. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev. 23, 105–117 10.1101/gad.495809 (doi:10.1101/gad.495809) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Smith EY, Futtner CR, Chamberlain SJ, Johnstone KA, Resnick JL. 2011. Transcription is required to establish maternal imprinting at the Prader–Willi syndrome and Angelman syndrome locus. PLoS Genet. 7, e1002422. 10.1371/journal.pgen.1002422 (doi:10.1371/journal.pgen.1002422) [DOI] [PMC free article] [PubMed] [Google Scholar]