Abstract
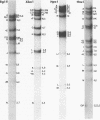
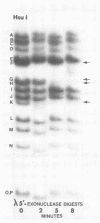
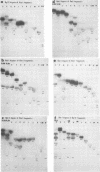
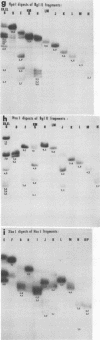
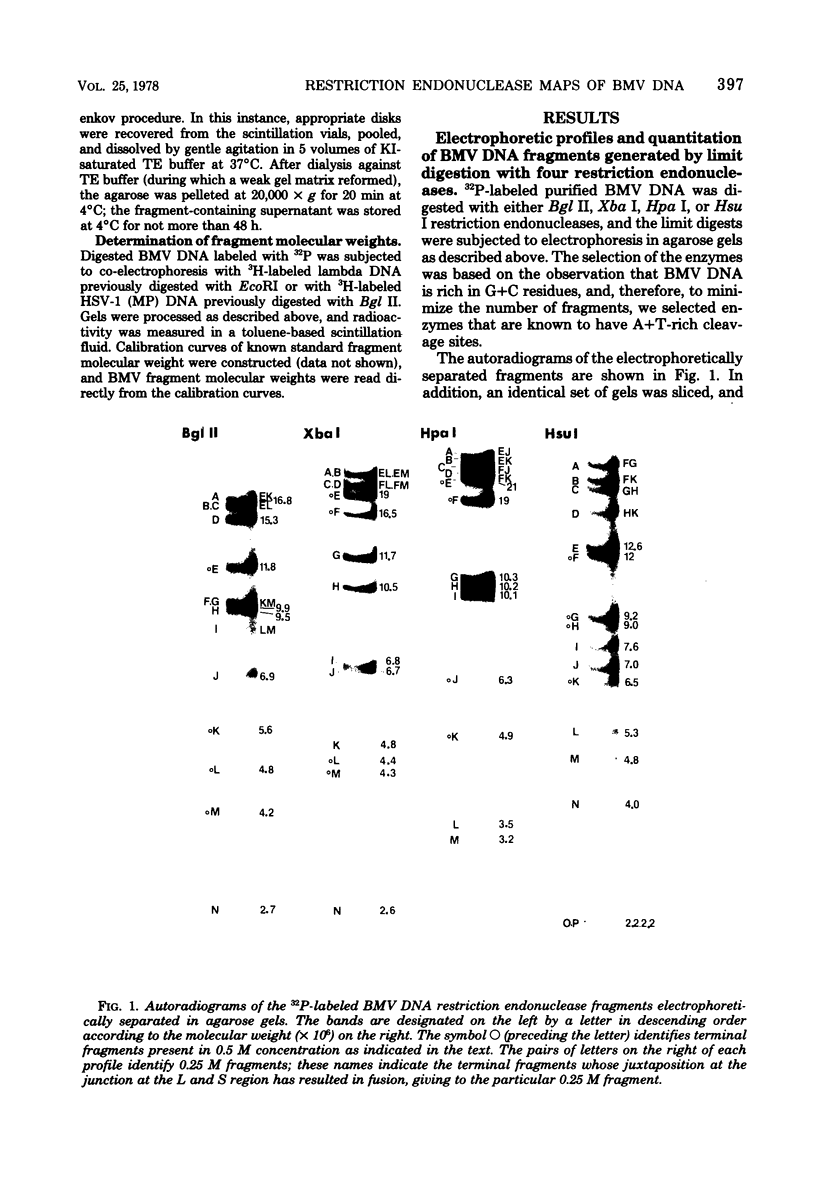
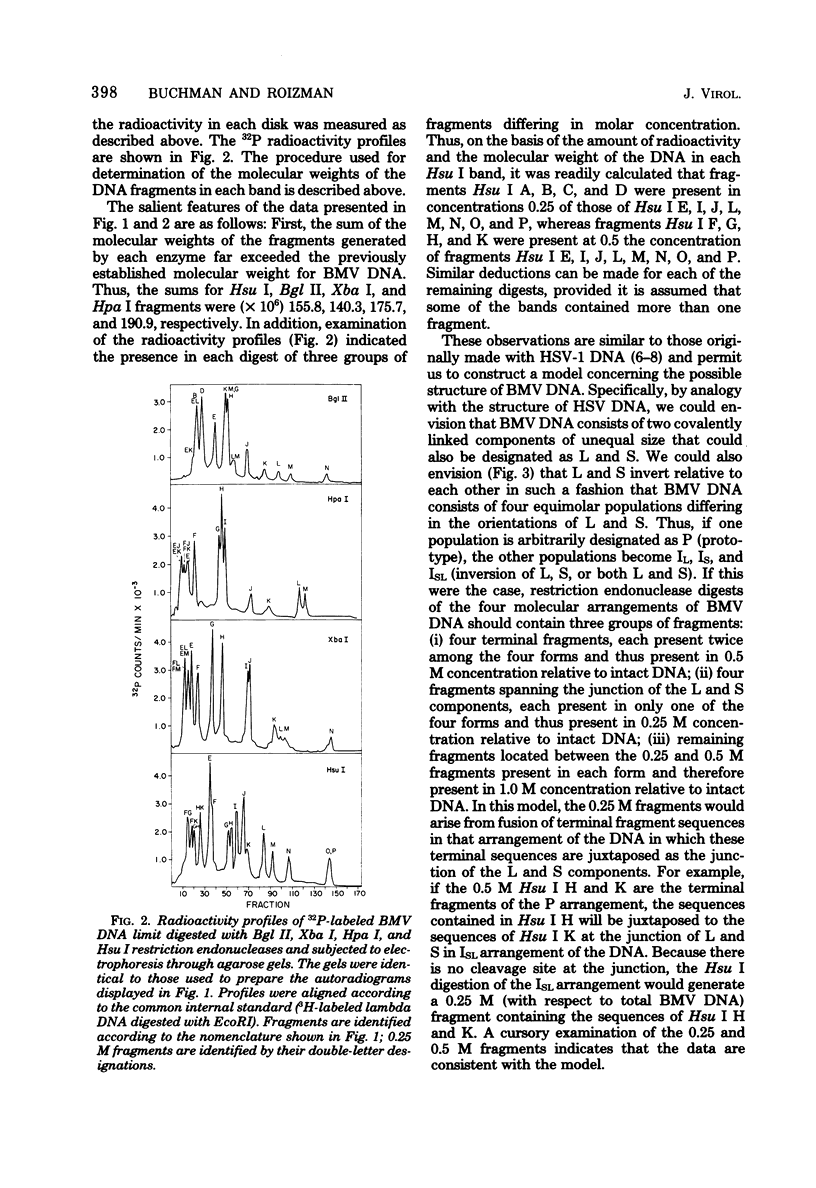
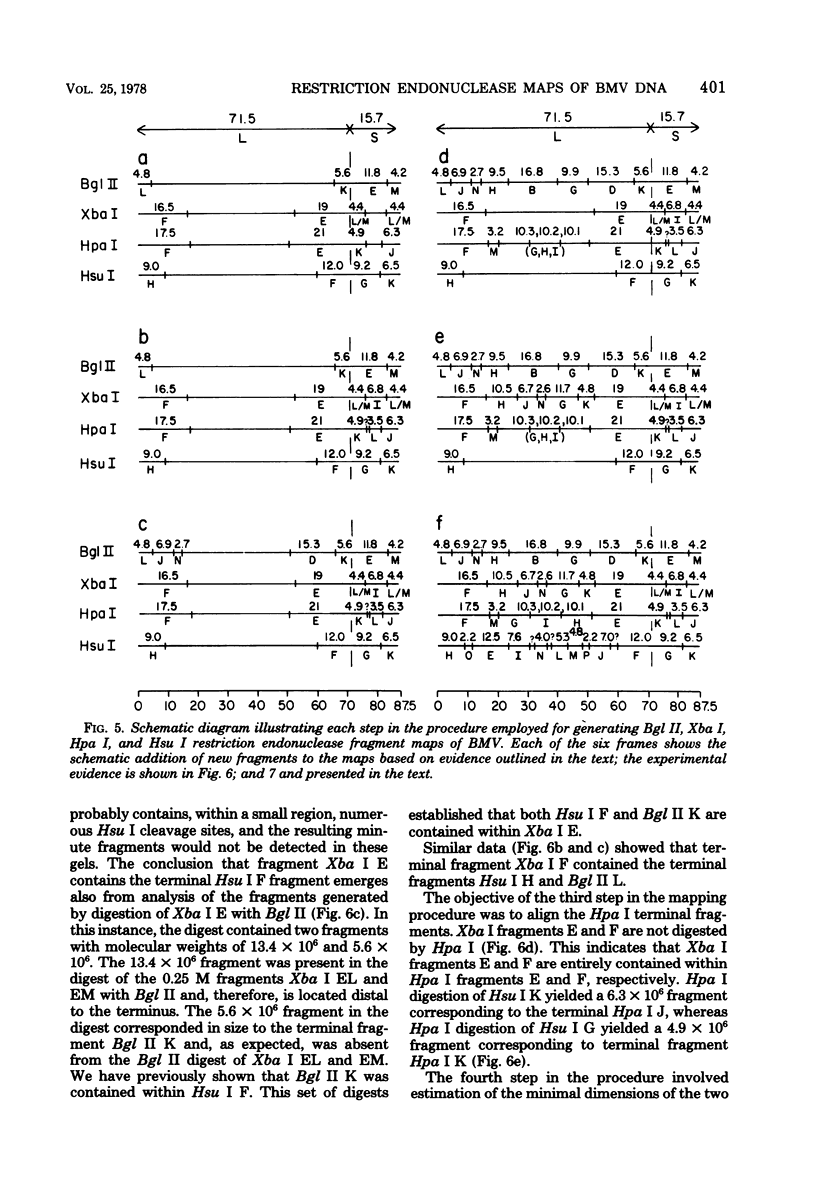
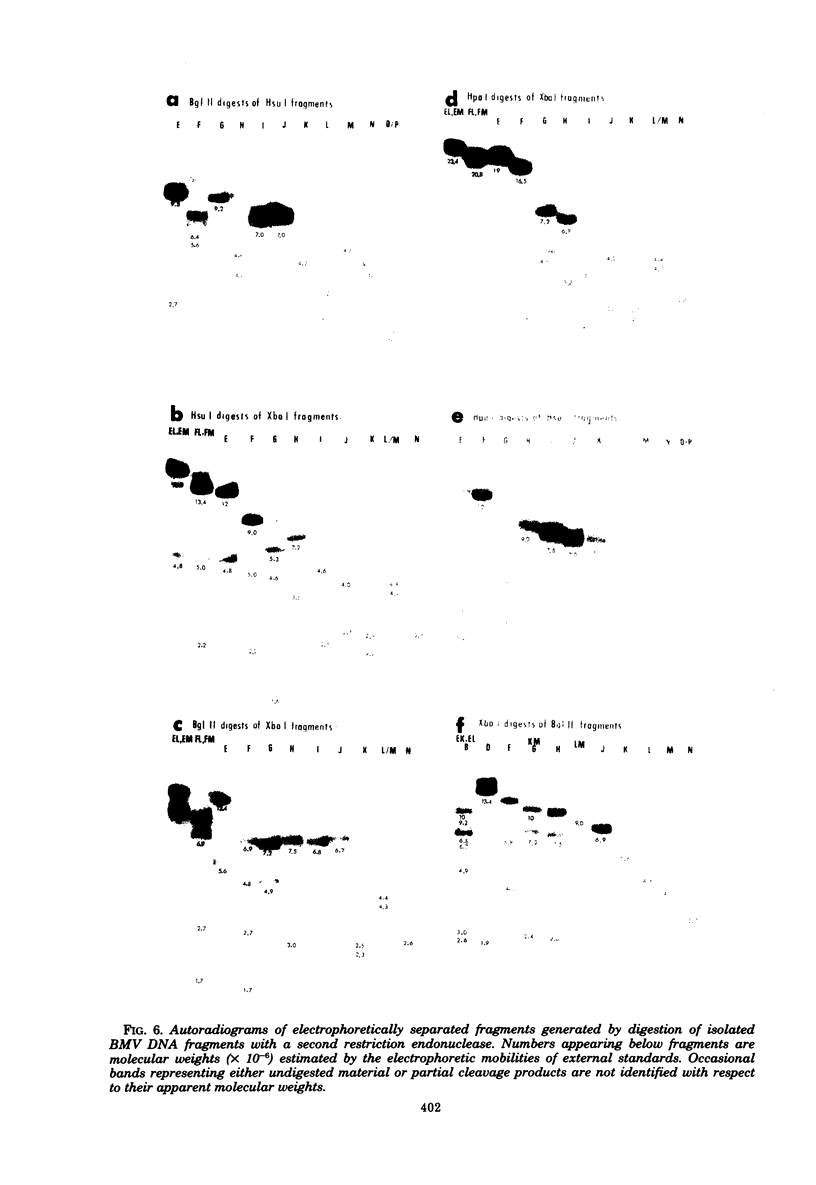
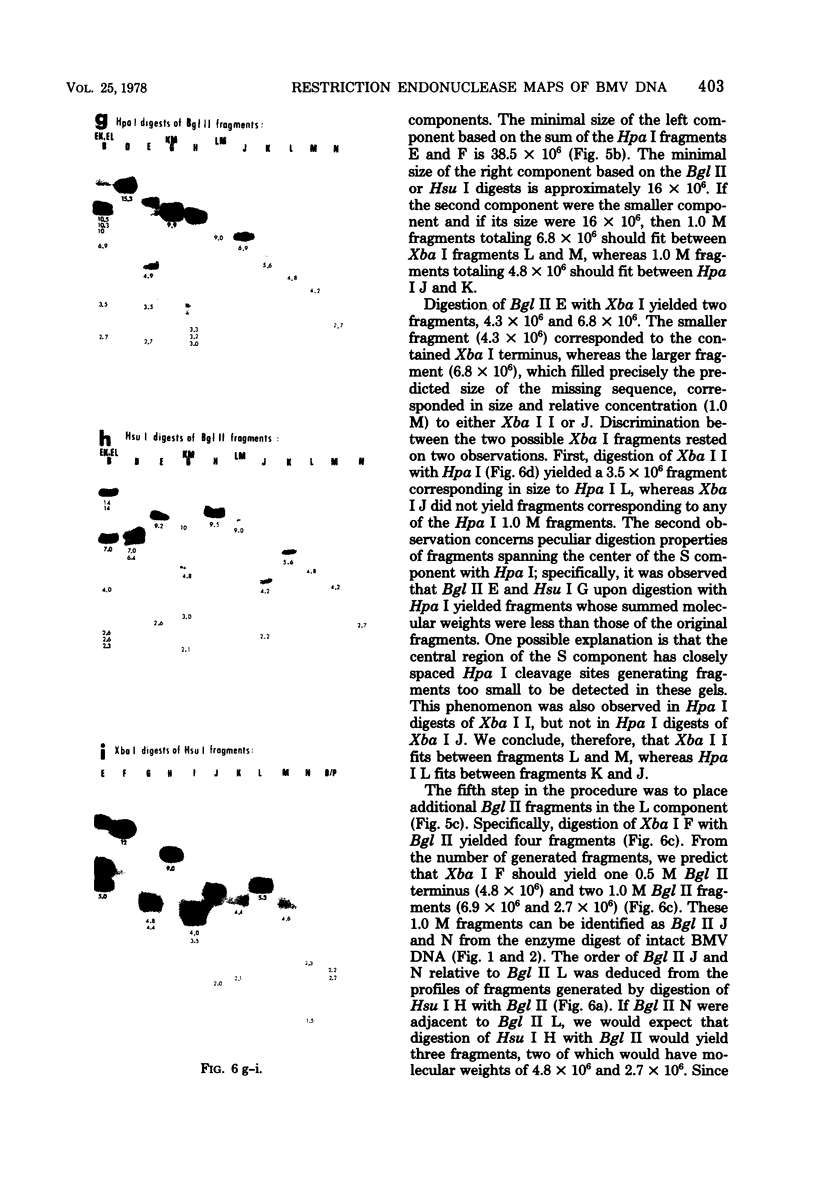
In this paper, we report that the DNA of bovine mammillitis virus (BMV) consists of two covalently linked components that are 71.5 × 106 and 15.7 × 106 in molecular weight and designated L and S, respectively. We further report that the BMV DNA consists of four equimolar populations differing only in the orientation of the L and S components relative to each other. This conclusion is based on the following: (i) The sum molecular weight of fragments generated by digestion of BMV DNA with Hsu I, Hpa I, Bgl II, or Xba I significantly exceeds the established molecular weight of the intact DNA. (ii) In each digest, the fragments form three groups differing in molar concentration. In reference to the molar concentration of intact DNA, each enzyme digest contained a set of four fragments 0.25 M in concentration, a set of four fragments 0.5 M in concentration, and a variable size set, unique for each enzyme digest, 1.0 M in concentration. (iii) Experiments involving digestion of intact DNA by lambda exonuclease followed by restriction endonuclease digestion established that each of four 0.5 M fragments were positioned at the termini of the BMV DNA. (iv) Complete maps for the fragments generated by each enzyme established that the 0.25 M fragments arise by fusion of the sequences of the terminal fragments when these are juxtaposed as a consequence of the inversion of L and S components. The maps also established the dimensions of the L and S components. We conclude that the structure of BMV DNA is similar to that of HSV DNA previously shown to consist of two unequal size components that invert relative to each other.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clements J. B., Cortini R., Wilkie N. M. Analysis of herpesvirus DNA substructure by means of restriction endonucleases. J Gen Virol. 1976 Feb;30(2):243–256. doi: 10.1099/0022-1317-30-2-243. [DOI] [PubMed] [Google Scholar]
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Fleckenstein B., Bornkamm G. W., Ludwig H. Repetitive sequences in complete and defective genomes of Herpesvirus saimiri. J Virol. 1975 Feb;15(2):398–406. doi: 10.1128/jvi.15.2.398-406.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward G. S., Jacob R. J., Wadsworth S. C., Roizman B. Anatomy of herpes simplex virus DNA: evidence for four populations of molecules that differ in the relative orientations of their long and short components. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4243–4247. doi: 10.1073/pnas.72.11.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieff E., Hoyer B., Bachenheimer S., Roizman B. Genetic relatedness of type 1 and type 2 herpes simplex viruses. J Virol. 1972 May;9(5):738–745. doi: 10.1128/jvi.9.5.738-745.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W. B., Hay D., Crawford L. V., Bouvier G. L., Crawford E. M. Characteristics of bovine mammillitis virus. J Gen Microbiol. 1966 Nov;45(2):325–332. doi: 10.1099/00221287-45-2-325. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Regulation of lambda exonuclease. I. Properties of lambda exonuclease purified from lysogens of lambda T11 and wild type. J Mol Biol. 1966 Jul;18(2):235–250. doi: 10.1016/s0022-2836(66)80243-7. [DOI] [PubMed] [Google Scholar]
- Roizman B., Hayward G., Jacob R., Wadsworth S., Frenkel N., Honess R. W., Kozak M. Human herpersviruses I: a model for molecular organization and regulation of herpesviruses-a review. IARC Sci Publ. 1975;(11 Pt 1):3–38. [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Sterz H., Ludwig H., Rott R. Immunologic and genetic relationship between herpes simplex virus and bovine herpes mammillitis virus. Intervirology. 1974;2(1):1–13. doi: 10.1159/000149398. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M., Cortini R. Sequence arrangement in herpes simplex virus type 1 DNA: identification of terminal fragments in restriction endonuclease digests and evidence for inversions in redundant and unique sequences. J Virol. 1976 Oct;20(1):211–221. doi: 10.1128/jvi.20.1.211-221.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie N. M. Physical maps for Herpes simplex virus type 1 DNA for restriction endonucleases Hind III, Hpa-1, and X. bad. J Virol. 1976 Oct;20(1):222–233. doi: 10.1128/jvi.20.1.222-233.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]