Abstract
PPARγ is a key regulator of glucose homeostasis and insulin sensitization. PPARγ must heterodimerize with its dimeric partner, the retinoid X receptor (RXR), to bind DNA and associated coactivators such as p160 family members or PGC-1α to regulate gene networks. To understand how coactivators are recognized by the functional heterodimer PPARγ/RXRα and to determine the topological organization of the complexes, we performed a structural study using small angle X-ray scattering of PPARγ/RXRα in complex with DNA from regulated gene and the TIF2 receptor interacting domain (RID). The solution structures reveal an asymmetry of the overall structure due to the crucial role of the DNA in positioning the heterodimer and indicate asymmetrical binding of TIF2 to the heterodimer.
1. Introduction
PPARγ, a member of the nuclear receptor family, is a key regulator of adipocyte differentiation and is involved in glucose homeostasis and insulin sensitization (reviewed in [1, 2]). PPARγ together with the CCAAT/enhancer-binding proteins had been identified as key transcription factors of driving fat cell differentiation (reviewed in [3]). PPARγ is absolutely required for both white and brown fat cell development. Several PPARγ coregulators have also been shown to affect positively or negatively this differentiation (reviewed in [4]).
PPARγ is activated through the binding of diverse ligands including natural fatty acid derivatives and nonsteroidal drugs and is the target of therapeutically active antidiabetics such as rosiglitazone (reviewed in [5]). Furthermore, cdk-5 phosphorylation of PPARγ leads to deregulation of some genes involved in metabolism [6].
The actions of PPARγ are mediated by 2 isoforms that result from alternative splicing. PPARγ2 is 28 amino acids longer at the N-terminal end (Figure 1(a)) and is mainly expressed in adipocyte cells, while PPARγ1 is ubiquitously expressed. Interestingly, PPARγ2 is ten times more active in ligand-independent transcriptional activation than PPARγ1 [7, 8].
Figure 1.
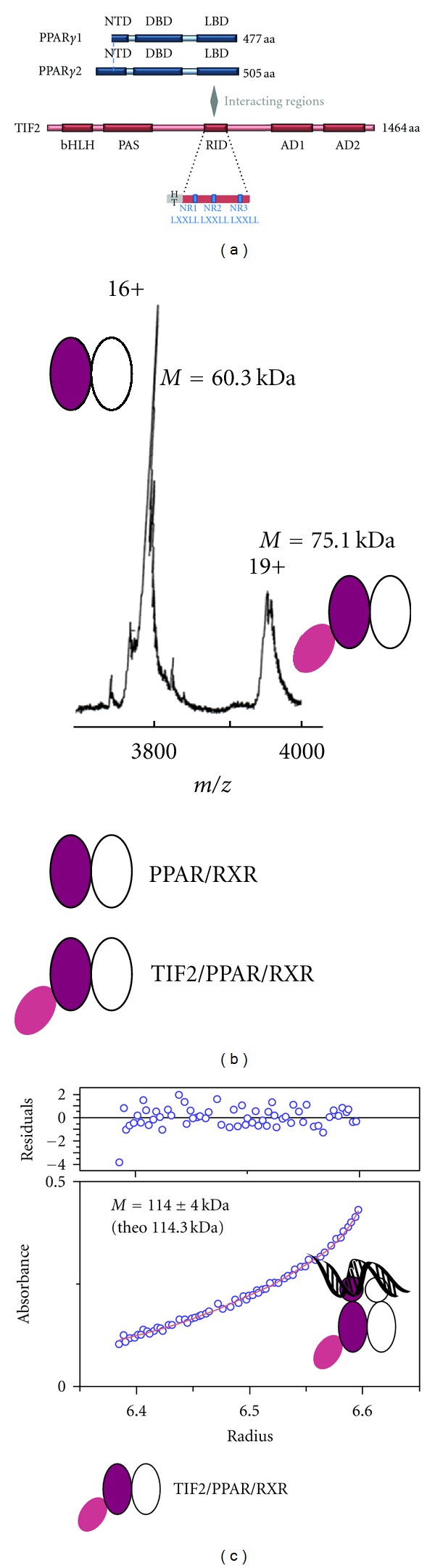
Biophysical characterization of the stoichiometry of the TIF2 RID/PPARγ/RXR complexes. (a) Structural organization of hPPARγ1, hPPARγ2, and hTIF2. (b) ESI mass spectra of TIF2 RID/PPARγ/RXR LBDs recorded under nondenaturing conditions in 200 mM ammonium acetate at pH = 7.4. The different charge states of the proteins are indicated above the peaks. The calculated molecular mass of the first peak corresponds to PPARγ/RXRα LBDs and the second one to the complex containing one PPARγ/RXRα LBDs dimer and one TIF2 RID. (c) Sedimentation equilibrium experiments. Best fits of experimental data for TIF2 RID/PPARγΔNTD/RXRΔNTD at 12,000 rpm with the self-association methods (SedPhat program). Sedimentation equilibrium data agrees with one TIF2 RID bound to PPARγΔNTD/RXRΔNTD.
PPARγ as the other nuclear receptors is a modular protein with a DNA binding domain (DBD) and a ligand binding domain (LBD). It is active as a heterodimer with the retinoid X nuclear receptor (RXR). The PPAR/RXR heterodimer recognizes specific DNA sequences called PPAR response elements (PPRE), composed of imperfect direct repeats separated by one nucleotide (DR1) and an extended 5′ half-site hexanucleotide [8]. Both the nonsymmetrical DR1 and the 5′ flanking sequence contribute to the selective binding of PPAR. PPAR binds the 5′ hexanucleotide and the 5′ flanking region, while RXR binds the 3′ half-site. To be fully active, PPARg2/RXR must be associated with coactivators which include p300/CBP, p160 members, PGC-1α, as well as Med1 (reviewed in [9, 10]). These coactivators are themselves highly regulated at the transcriptional and posttranscriptional levels.
Structurally, most of our current knowledge on the molecular basis of the mechanism of action of NRs is based on the X-ray and NMR structures of isolated DNA and ligand binding domains. The crystal structures of various NR LBDs with coactivator peptides have revealed a conserved binding mode of the coactivators to NRs. Agonist ligands trigger a conformational change of the NR LBDs allowing a leucine-rich interacting motif (LXXLL motif) present in the coactivator sequence that forms an α-helix to interact through a charge clamp and hydrophobic interactions at the LBD surface [11–13]. Coactivators such as p160 or PGC-1α contain several LXXLL motifs (NR boxes) involved in specific binding to NRs and other transcription factors. For TIF2 (NCoA2, p160 member), 3 NR boxes are found in a central domain that interacts with NRs and is called the receptor interacting domain (RID) (Figure 1(a)). The third motif has been shown to preferentially bind to PPARγ [14].
Extensive studies on structure-function relationships have been done on PPARγ LBD monomers or dimers and have provided information on conformational change induced by various ligands. The structures of full-length PPARγ/RXR complexes have also been studied by X-ray crystallography [15] and by small-angle X-ray scattering (SAXS) [16, 17], although the N-terminal domain (NTD) was not visible in the electron density map. Remaining questions concern the mode of recognition of coactivator domains by PPARγ/RXRα complexes. To understand how TIF2 RID is recognized by PPARγ/RXRα and the topological organization of the complex, we performed a structural analysis in solution by SAXS of PPARγ/RXRα LBD in complexes with TIF2 RID and of PPARγ2/RXRα full-length or depleted of their N-terminal domain (ΔNTD) bound to PPRE and TIF2 RID protein.
2. Materials and Methods
2.1. Cloning, Protein Expression, and Purification
The HsPPARγ LBD (203–477), HsPPARγΔNTD (135–505) and HsTIF-2 (632–772) were expressed as hexahistidine fusion proteins. HsRXRα LBD (223–462) and HsRXRαΔNTD (130–462), were cloned into pACYC plasmid encoding nontagged proteins. The nuclear receptor heterodimers were coexpressed in E. coli BL21 (DE3) cells. Full-length HsPPARγ2 (1–505) and HsRXRα (1–462) were coexpressed in Sf9 insect cells as N-ter hexahistidine and Flag tagged fusion proteins, respectively. The PPAR/RXR dimers and the coactivator TIF-2 RID were purified by affinity chromatography and gel filtration as described [16, 18]. Ligands (CD3254 for RXR and rosiglitazone for PPAR) were added in a 2-fold excess to saturate the receptors. The DNA (GAAACTAGGGTAAAGGTCAG/CTTTGATCCCATTTCCAGTC) was added in a 1.2-fold excess to the dimers, and the complex was gel-filtrated on a Superdex S200 (16/60 or 10/300, GE Healthcare). For the PPAR/RXR/TIF2 RID complexes, TIF2 was added in a 2-fold excess and gel-filtrated on Superdex S200 (16/60 or 10/300, GE Healthcare). Protein samples were concentrated using Amicon Ultra centrifugal filter units (Millipore). Purity and homogeneity of the protein were assessed by SDS-PAGE, and complex formation was monitored by native PAGE. The final buffer for PPAR/RXR LBDs was Tris 20 mM pH 7.5, NaCl 200 mM, and DTT 5 mM and for PPARγ/RXRα/DNA complexes Tris 20 mM pH 7.5, NaCl 75 mM, KCl 75 mM, MgSO4 4 mM, Glycerol 5%, and Chaps 2 mM.
2.2. Ultracentrifugation Equilibrium Sedimentation
For sedimentation equilibrium experiments, samples were spun at 12,000 rpm and systems were first allowed to equilibrate for 12 hours before absorbance profiles were compared at different times to ensure that system had reached equilibrium. Using nonlinear least-squares analysis, these datasets were fitted using single component model and several equilibrium models with Sedphat program.
2.3. Electrospray Ionization Mass Spectrometry.
Prior to ESI-MS analysis, samples were desalted on Zeba Spin desalting columns (Pierce) in 150 mM ammonium acetate (pH 8.0). ESI-MS measurements were performed on an electrospray time-of-flight mass spectrometer (MicrOTOF, Bruker Daltonic, Germany). Purity and homogeneity of the proteins were verified by mass spectrometry in denaturing conditions (samples were diluted at 2 pmol/μL in a 1 : 1 water-acetonitrile mixture (v/v) acidified with 1% formic acid). The mass measurements of the noncovalent complexes were performed in ammonium acetate (200 mM; pH 8.0). Samples were diluted to 8 pmol/mL in the previous buffer and continuously infused into the ESI ion source at a flow rate of 3 mL/min through a Harvard syringe pump (Harvard Apparatus model 11). A careful optimization of the interface parameters was performed to obtain the best sensitivity and spectrum quality without affecting the noncovalent complexes stability. In particular, the capillary exit (CE) ranged from 60 to 150 V with a vacuum interface pressure of 2.3 mbar and was set to 80 V.
2.4. SAXS Experiments and Data Processing.
Synchrotron X-ray solution scattering data were collected at the X33 beamline (DESY, Hamburg) [19] using a PILATUS detector at a sample-detector distance of 2.7 m, covering the range of momentum transfer 0.01 < q < 0.6 Å−1 (q = 4πsin(θ)/λ, where 2θ is the scattering angle and λ = 0.15 nm is the X-ray wavelength) in eight frames (15 seconds each) to check for possible radiation damage. All scattering measurements were carried out at 10°C with automated filling, and samples were measured at several concentrations.
SAXS experiments were also conducted on the SWING beamline at SOLEIL Synchrotron (Gif-sur-Yvette, France), using a 17 × 17 cm2 low-noise Aviex CCD detector positioned at a distance of 2.107 m from the sample. Sample solutions were circulated in a thermostated Quartz capillary with a diameter of 1.5 mm and 10 μm wall thickness, positioned within a vacuum chamber. Fifty frames of 2 s each were collected, normalized to the transmitted intensity, and subsequently averaged using the image analysis software Foxtrot (SWING beamline at SOLEIL Synchrotron).
The SAXS data were averaged and processed by standard procedures using PRIMUS [20]. The forward scattering I(0) and the radii of gyration R g were evaluated using the Guinier approximation assuming that at very small angles (s < 1.3/R g) the intensity is represented as I(s) = I(0)exp(−(sR g)2/3). These parameters were also computed from the entire scattering pattern using the indirect transform package GNOM [21], which also provides the maximum dimension of the particle D max and the distance distribution function P(r). Low resolution shape analysis of the solutes was performed using the ab initio program DAMMIF [22]. The scattering from the atomic models was calculated using the program CRYSOL [23] which either predicts theoretical scattering patterns or fits the experimental data by adjusting the excluded volume and the contrast of the hydration layer. The program SASREF [24] was employed for molecular rigid body modeling of the PPARγΔNTD/RXRαΔNTD/DNA complex, based on the crystal structure of PPARγ/RXR (PDB ID: 3DZY). For both ab initio and rigid body analysis, multiple runs were performed to verify the stability of the solution, and the most typical reconstructions were selected using the programs DAMAVER [25] and SUPCOMB [26].
3. Results and Discussion
3.1. Characterization of PPARγ/RXRα Complexes
PPARγ/RXRα LBDs or full-length proteins were copurified in two steps as described [16, 18]. DNA from Cyp4A1 regulated gene was added in a 1.2-fold excess to the full-length heterodimer and the complex further purified by analytical gel-filtration. In order to investigate the binding of TIF2 RID on PPARγ/RXRα, a 15 kDa fragment of TIF2 (632–772) containing the three LXXLL motifs and interacting with NRs (Figure 1(a)) was added to the heterodimer and further purified by gel-filtration. After gel-filtration, the complexes were analyzed by SDS-PAGE and native electrophoresis confirming the formation and homogeneity of the complexes (see Supplementary Figure 1 available online at doi:10.1155/2012/701412). The stoichiometry of the TIF2 RID/PPARγ/RXRα complexes was determined using analytical ultracentrifugation (AUC) and electrospray ionization mass spectrometry (ESI-MS), a method that has proven its efficiency for the study of noncovalent complexes [27, 28]. ESI-MS experiments were performed under nodenaturing conditions on PPARγ/RXRα LBDs in the presence of TIF2 RID. The molecular mass obtained (Figure 1(b)) is consistent with a complex composed of one TIF2 RID molecule per heterodimer. The presence of mass peaks corresponding to free PPARγ/RXRα LBDs is due to partial dissociation of the complex in the ESI ion source. Equilibrium sedimentation AUC experiments were also carried out to characterize the association state of TIF2 RID with PPARγΔNTD/RXRαΔNTD/DNA in solution as well as its molecular mass (Figure 1(c)). The molecular mass of Mw of 114 kDa is in excellent agreement with the expected value of 114.3 kDa. Both measurements indicate that only one TIF2 RID molecule binds to PPARγ/RXRα heterodimer.
3.2. Topology of PPARγ/RXRα LBDs and TIF2 RID/PPARγ/RXRα LBDs Complexes
The observed stoichiometry of one TIF2 RID per heterodimer contrasts with the crystallographic structures of NRs bound to LXXLL peptides [11–13] which have revealed 2 peptides bound by each monomer of the dimer. This raises the question of the binding mode of TIF2 RID to PPARγ/RXRα either asymmetrically to one monomer or to the two subunits involving two NR boxes. To address this question we used SAXS, a structural method to characterize multidomain proteins and complexes. We analyzed by SAXS the recruitment of the TIF2 RID by the PPARγ/RXRα LBDs.
As a reference, scattering profiles of PPARγ/RXRα LBDs alone were collected (Figure 2(a)). Monodisperse concentrated solutions of PPARγ/RXRα LBDs were measured, and the structural parameters including the radius of gyration (R g) and the maximum particle dimension (D max) were computed from the experimental scattering patterns from the heterodimer (Table 1). A symmetrical pair-distribution function is observed (Figure 2(b)) indicating a globular complex. The experimental SAXS data is well fitted by the scattering profile calculated from the crystal structure (PDB ID: 3H0A) using CRYSOL [23] and is in agreement with SAXS parameters measured for heterodimer LBDs [17, 18].
Figure 2.
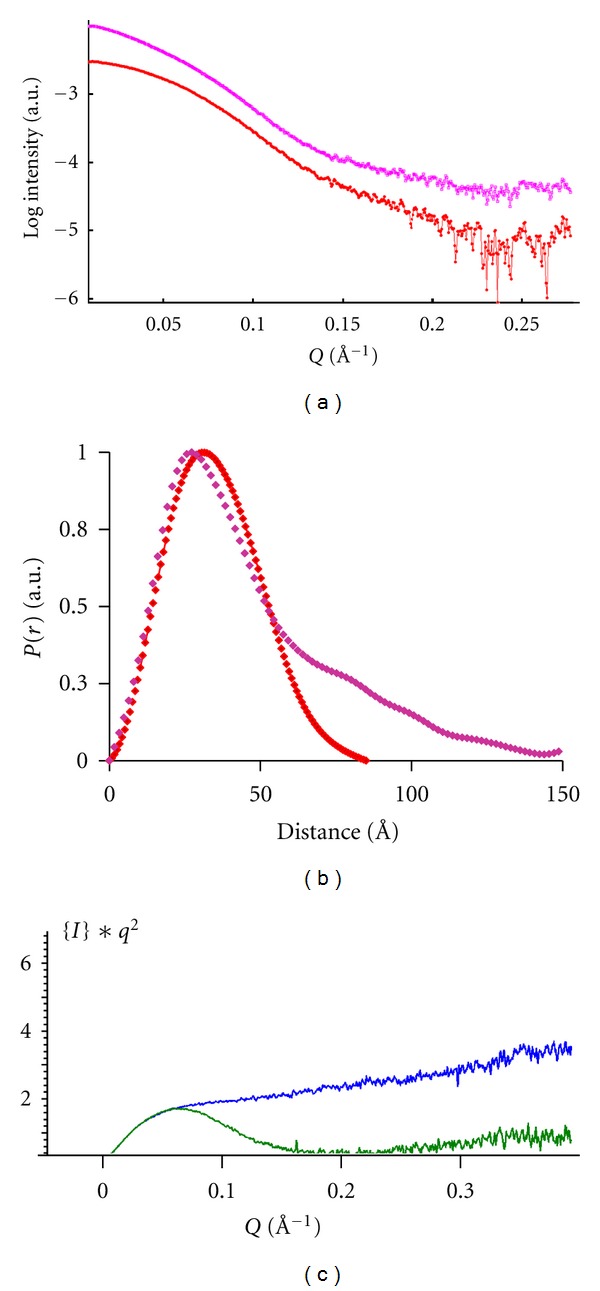
Experimental SAXS data of PPARγ/RXRα LBDs complexes. (a) Scattering profiles of PPARγ/RXRα LBDs (red) and TIF2 RID/PPARγ/RXRα LBDs (pink). (b) Distance distribution functions computed from the X-ray scattering patterns using the program GNOM. Same color code as in (a). (c) Kratky representations for TIF2 RID (blue) and TIF2 RID/PPARγ/RXRα LBDs (green).
Table 1.
Structural parameters from SAXS data.
| Complexes | R g (Å) | D max (Å) |
|---|---|---|
| PPARγ/RXRα LBD | 27.2 ± 0.1 | 85 ± 5 |
| TIF2 RID/PPARγ/RXRα LBD | 35.5 ± 0.1 | 150 ± 10 |
| TIF2 RID | 33 ± 1 | 130 ± 10 |
| PPARγ2ΔNTD/RXRαΔNTD/PPRE (CYP4A1) | 37.1 ± 0.4 | 140 ± 10 |
| PPARγ2/RXRαΔNTD/PPRE | 44 ± 0.5 | 160 ± 10 |
| PPARγ2/RXRα/PPRE | 52 ± 0.5 [16] | 180 ± 10 [16] |
| TIF2 RID/PPARγ2/RXRα/PPRE | 59 ± 0.5 | 200 ± 20 |
We next analyzed the TIF2 RID/PPARγ/RXRα LBDs complex. The values of R g and D max calculated from the distance pair-distribution function P(r) for TIF2 RID are larger (Table 1) than typical values expected for globular proteins suggesting that TIF2 RID is extended. To have an indication of TIF2 RID folding, we used the Kratky representations, which emphasize deviation from the high-q behavior of the scattering intensity I(q). The Kratky plot of TIF2 RID alone (Figure 2(c)) shows a continuously increasing curve that indicates an unfolded protein. For the TIF2 RID/PPARγ/RXRα LBDs complex, it corresponds to a partially folded protein with a bell-shaped curve in agreement with a folded macromolecule and thus suggesting an induced folding of the molecule upon complex formation. Binding of TIF2 RID to the heterodimer led to an increase of the R g (by 8 Å) and of the D max (by 65 Å) (Table 1). The large difference in R g and D max compared to those of PPARγ/RXRα LBDs suggests that the TIF2 RID is extended and partially disordered. This is also observed in the asymmetric profile of the pair-distribution function with a tail at high r values (Figure 2(b)) which corresponds to elongated molecules in contrast to the symmetric pair-distribution function of the globular PPAR/RXR LBDs.
The molecular envelopes as bead models, derived from the SAXS data, were computed for PPARγ/RXRα LBDs and TIF2 RID/PPARγ/RXRα LBDs (Figure 3). It corresponds to a symmetrical globular shape for the LBD dimer (Figure 3(c)). In contrast, for the TIF2 RID complex, a marked asymmetry as compared to the LBD dimer is observed (Figure 3(d)). In analogy with the TIF2 RID and SRC-1 RID complexes with RAR/RXR and RAR homodimer [16, 29], we can speculate that the globular region of the molecular envelope corresponds to the LBD dimer and the asymmetric extended tail to TIF2 RID. The large difference in the structural parameters of the TIF2 RID complex compared to the LBD dimer and the molecular envelope of the complex suggests that TIF2 RID is flexibly attached asymmetrically to the PPARγ/RXRα through only one LXXLL motif. Importantly, the data preclude a model in which two LXXLL motifs bind to each subunit of the PPARγ/RXR heterodimer. A preliminary SAXS study of PPARγ/RXRα bound to PGC-1α RID provides similar results with similar R g and D max values suggesting that in both cases the architecture of the two different complexes is similar with the CoA RID asymmetrically bound to PPAR subunit through only one LXXLL motif. Interestingly, it has been shown that NR2 motif of PGC-1α is sufficient to have a full transcriptional response by PPARγ/RXRα [14]. While for TIF2, it has been shown that the third LXXLL motif binds preferentially to PPARγ [14] and a combination of LXXLL motifs is required for a full transcriptional response [30]. However the SAXS data are in agreement with the crystal structure of PPARγ/RXRα LBDs (PDB ID: 3H0A) that reveals one coactivator peptide tightly bound to PPARγ and one coactivator peptide loosely bound to RXRα as indicated by the poor electron density and high B factor for the peptide bound to RXR (averaged B factor for the peptide bound to RXR is 116.2 compared to the averaged B factor for the CoA peptide bound to PPARγ, 31.6). These observations demonstrate that TIF2 RID binds asymmetrically to PPARγ subunit within the heterodimer through one LXXLL.
Figure 3.
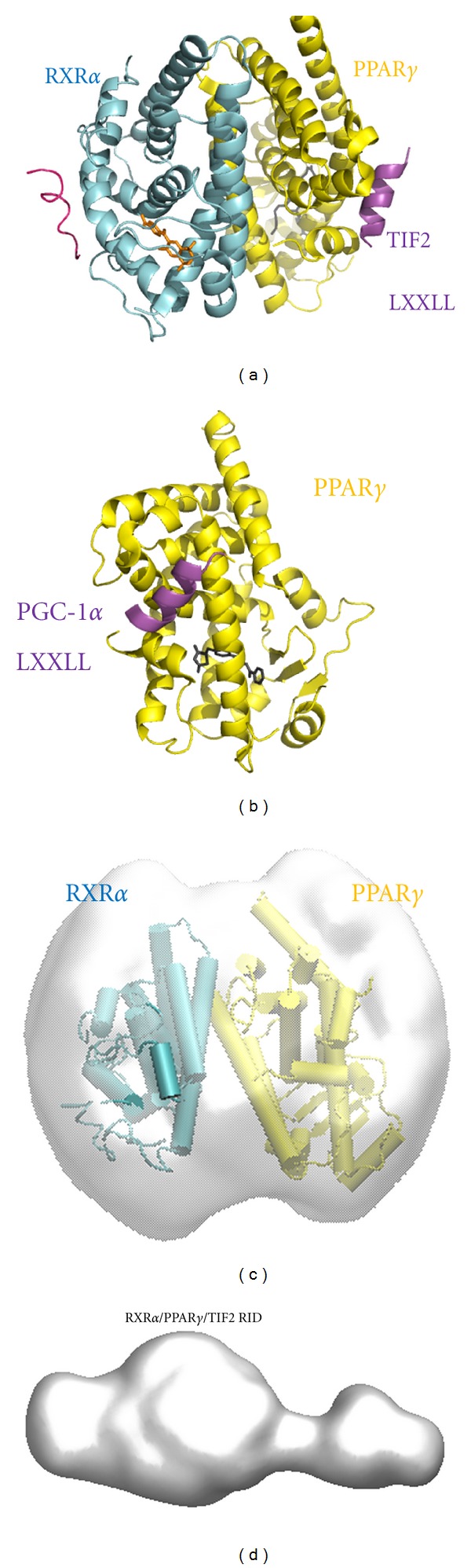
Molecular envelope of PPARγ/RXRα LBDs complexes and comparison with the crystal structure of PPARγ/RXRα LBDs. (a) Crystal structure of PPARγ/RXRα LBDs in complex with TIF2 coactivator peptide (PDB ID: 1H0A) shown in schematic cartoon representations with PPARγ in yellow, RXRα in cyan, and the coactivator peptides in pink. (b) Crystal structure of PPARγ LBD bound to PGC-1α NR2 motif (PDB ID: 3CS8). (c) Molecular envelopes of the complexes PPARγ/RXRα LBDs (grey surface) together with the crystal structure of the complex. (d) Molecular envelope of the complexes TIF2 RID/PPARγ/RXRα LBDs (grey surface).
3.3. Solution Structure of PPARγ2/RXRα/Cyp4A1 DR1 Complexes
PPARγ/RXRα without DNA has been shown to be elongated with no interdomain interactions [17]. In complex with idealized DNA, the first atomic resolution structure of integral PPARγ/RXRα obtained by X-ray crystallography [15] confirmed the structural information concerning isolated domains [11, 31, 32] and also revealed an interdomain contact between the LBD of PPARγ and the DBD of RXR [13], although the functional correlation is limited to a single point mutation. The N-terminal domain, unfolded, was not visible in the electron density map. We previously measured the SAXS profile of PPARγ2/RXRα bound to an idealized DR1 [16] and have shown that the conformation in solution is different to that observed in the crystal structure with the heterodimer exhibiting an extended asymmetric shape without additional interdomain contacts between the DBDs and LBDs beyond the connection through the hinge regions. We have now characterized the solution structure of PPARγ2/RXRα full length or truncated of their NTDs and bound to a natural PPRE from CYP4A1. The structural parameters calculated from the experimental scattering patterns (Figures 4(a) and 4(b)) given in Table 1 reveal an extended shape of the complex. With the full length PPARγ complex, the structural parameters are even larger (R g = 44 Å and D max = 160 Å), suggesting a distinct dissociated NTD. Low resolution models were reconstructed ab initio from the corresponding experimental scattering patterns. The most typical ab initio model of PPARγ2ΔNTD/RXRαΔNTD/CYP4A1 PPRE presented in Figure 4(c) clearly displays separate DBD and LDB domains similar to the architecture observed for the complex with the idealized DR1 and those observed for other NR heterodimers. For PPARγ2ΔNTD/RXRαΔNTD/PPRE, its atomic structure was refined against SAXS experimental data (Figure 4(a)). The position of the domains was adjusted by rigid body modeling using the available high resolution crystal structure of the complex (PDB ID: 3DZY). The model obtained by rigid body refinement agrees well with the ab initio models as seen from the superposition in Figure 4(c). The refined structure reveals an asymmetric shape with the LBD dimer positioned at the 5′ end of the DNA (Figure 4(d)), an asymmetry already observed for other heterodimers studied by SAXS [16].
Figure 4.
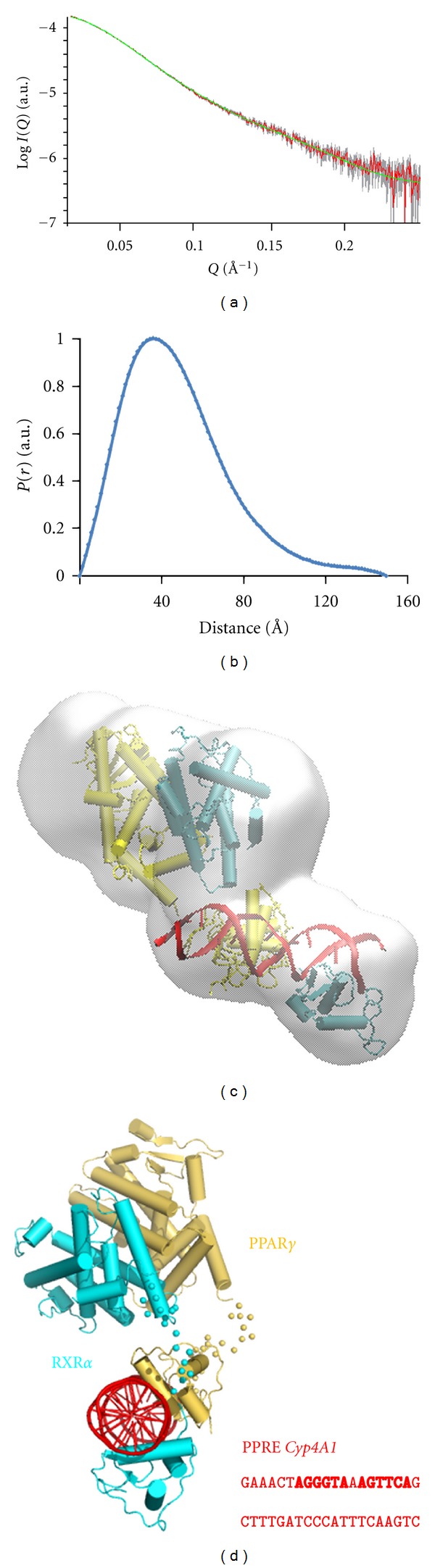
Solution structure of PPARγ2/RXRα bound to Cyp4A1 PPRE. (a) Scattering profiles of PPARγ2ΔNTD/RXRαΔNTD/PPRE. Experimental data are shown as red dots. Green fit is computed from the solution structure of the complex. (b) Distance distribution function computed from the X-ray scattering pattern using the program GNOM. (c) Most typical molecular envelope of PPARγ2ΔNTD/RXRαΔNTD/PPRE generated by DAMMIF (beads model shown as a grey surface) together with the refined model by rigid body refinement using the program SASREF (fit to the experimental data with χR = 0.98). (d) Pseudoatomic solution structure of PPARγ2ΔNTD/RXRαΔNTD/Cyp4A1 PPRE shown in schematic cartoon representation together with the sequence of the DNA.
The scattering patterns of the full-length PPARγ2/RXRα/PPRE in complex with TIF2 RID were also monitored. The increase of the R g and of the D max (Table 1) upon TIF2 RID interaction to the full-length heterodimer is similar to that observed for the complex with the LBD dimer, suggesting a similar mode of binding as the LBD dimer with no additional interactions of the TIF2 RID with the full-length complex. The solution studies of DNA complexes with integral receptors provided also evidence for a stoichiometry of 1 coactivator per receptor dimer. Each heterodimer binds only one coactivator protein via PPAR, this preferential binding being controlled by affinity, rather than steric exclusion. However, other domains of TIF2 may be implicated in additional interactions to the receptor as TIF2 has been shown to bridge the N-terminal and C-terminal receptor domains of estrogen receptor [33].
4. Conclusion
The solution structures of the functional heterodimer PPARγ2/RXRα bound to natural DR1 from regulated gene and TIF2 RID reveal the asymmetry induced by the nonsymmetric DNA target of the NR dimer with the position of the LBDs at the 5′ end of the target DNA and the asymmetric mode of recruitment of TIF2. The extended conformation in solution of integral PPARγ/RXRα bound to DNA is recognized and maintained during coactivator binding. The TIF2 RID, which is intrinsically disordered, partially folds on binding to PPARγ but retains an extended conformation in the complex. The functional consequence imposed by the DNA is the asymmetric binding of the cofactor with the complex that orients the coactivator on one side of the DNA, allowing interactions with other regulatory proteins and thus leading to specific and controlled NR-mediated gene transcription. As other domains outside the TIF2 RID may interact with the heterodimer, further studies should be carried out to characterize the topology of such large complexes. A better understanding at the molecular level of the regulation of PPARγ/RXRα by their coactivators would benefit new therapeutic approaches for the treatment of metabolic diseases.
Supplementary Material
Supplementary Figure 1 with the coomassie-stained SDS-PAGE and Native-PAGE of the PPARg2/RXRa complexes after the final purification step. (a) 12% SDS gel for PPARg2/RXRa complex, (b) 12% SDS gel (left) and 8-25% Native gel for PPARg2/RXRaDeltaNTD/DNA, (c) 12% SDS gel for PPARg2DeltaNTD/RXRaDeltaNTD/DNA/TIF2 RID and (d) 12% SDS gel for PPARg LBD/TIF2 RID complex.
Conflict of Interests
The authors declare no competing financial interests.
Acknowledgments
The authors thank Carole Peluso-Iltis for technical assistance, Isabelle Kolb-Cheynel (IGBMC Baculovirus Facility) for productions in insect cells, Catherine Birck (IGBMC Structural Biology and Genomics Platform) for help in the analytical ultracentrifugation experiment, Adeline Page (IGBMC Proteomics Platform) for the mass spectrometry analysis, and Alastair McEwen for critical reading of the paper. They thank Pierre Roblin and the staff of the SOLEIL SWING beamline (Gif-sur-Yvette, France) for assistance. This work was supported by the Centre National pour la Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM), the French Infrastructure for Integrated Structural Biology (FRISBI), the Association pour la Recherche sur le Cancer (ARC), the Fédération pour la Recherche Médicale (FRM), and the IGBMC facilities.
References
- 1.Michalik L, Wahli W. Peroxisome proliferator-activated receptors (PPARs) in skin health, repair and disease. Biochimica et Biophysica Acta. 2007;1771(8):991–998. doi: 10.1016/j.bbalip.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Tontonoz P, Spiegelman BM. Fat and beyond: the diverse biology of PPARγ . Annual Review of Biochemistry. 2008;77:289–312. doi: 10.1146/annurev.biochem.77.061307.091829. [DOI] [PubMed] [Google Scholar]
- 3.Farce A, Renault N, Chavatte P. Structural insight into PPARγ ligands binding. Current Medicinal Chemistry. 2009;16(14):1768–1789. doi: 10.2174/092986709788186165. [DOI] [PubMed] [Google Scholar]
- 4.Farmer SR. Transcriptional control of adipocyte formation. Cell Metabolism. 2006;4(4):263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seale P, Kajimura S, Spiegelman BM. Transcriptional control of brown adipocyte development and physiological function-of mice and men. Genes and Development. 2009;23(7):788–797. doi: 10.1101/gad.1779209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi JH, Banks AS, Estall JL, et al. Anti-diabetic drugs inhibit obesity-linked phosphorylation of PPARγ 3 by Cdk5. Nature. 2010;466(7305):451–456. doi: 10.1038/nature09291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen R, Grøntved L, Stunnenberg HG, Mandrup S. Peroxisome proliferator-activated receptor subtype- and cell-type-specific activation of genomic target genes upon adenoviral transgene delivery. Molecular and Cellular Biology. 2006;26(15):5698–5714. doi: 10.1128/MCB.02266-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ijpenberg A, Tan NS, Gelman L, et al. In vivo activation of PPAR target genes by RXR homodimers. EMBO Journal. 2004;23(10):2083–2091. doi: 10.1038/sj.emboj.7600209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu S, Reddy JK. Transcription coactivators for peroxisome proliferator-activated receptors. Biochimica et Biophysica Acta. 2007;1771(8):936–951. doi: 10.1016/j.bbalip.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 10.Sugii S, Evans RM. Epigenetic codes of PPARγ in metabolic disease. FEBS Letters. 2011;585(13):2121–2128. doi: 10.1016/j.febslet.2011.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nolte RT, Wisely GB, Westin S, et al. Ligand binding and co-activator assembly of the peroxisome proliferator- activated receptor-γ . Nature. 1998;395(6698):137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 12.Shiau AK, Barstad D, Loria PM, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95(7):927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 13.Darimont BD, Wagner RL, Apriletti JW, et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes and Development. 1998;12(21):3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Kovach A, Suino-Powell K, Martynowski D, Xu HE. Structural and biochemical basis for the binding selectivity of peroxisome proliferator-activated receptor γ to PGC-1α . Journal of Biological Chemistry. 2008;283(27):19132–19139. doi: 10.1074/jbc.M802040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chandra V, Huang P, Hamuro Y, et al. Structure of the intact PPAR-γ-RXR-α nuclear receptor complex on DNA. Nature. 2008;456(7220):350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rochel N, Ciesielski F, Godet J, et al. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nature Structural & Molecular Biology. 2011;18(5):564–570. doi: 10.1038/nsmb.2054. [DOI] [PubMed] [Google Scholar]
- 17.Bernardes A, Batista FA, de Oliveira Neto M, et al. Low-resolution molecular models reveal the oligomeric state of the PPAR and the conformational organization of its domains in solution. PLoS One. 2012;7(2) doi: 10.1371/journal.pone.0031852.e31852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sato Y, Ramalanjaona N, Huet T, et al. The, “Phantom Effect” of the rexinoid LG100754: structural and functional insights. PLoS One. 2010;5(11) doi: 10.1371/journal.pone.0015119.e15119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roessle MW, Klaering R, Ristau U, et al. Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory Hamburg. Journal of Applied Crystallography. 2007;40:S190–S194. [Google Scholar]
- 20.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. Journal of Applied Crystallography. 2003;36(5):1277–1282. [Google Scholar]
- 21.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. Journal of Applied Crystallography. 1992;25(4):495–503. [Google Scholar]
- 22.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2009;42(2):342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Svergun D, Barberato C, Koch MH. CRYSOL-a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. Journal of Applied Crystallography. 1995;28(6):768–773. [Google Scholar]
- 24.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophysical Journal. 2005;89(2):1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. Journal of Applied Crystallography. 2003;36(3):860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. Journal of Applied Crystallography. 2001;34(1):33–41. [Google Scholar]
- 27.Sanglier S, Bourguet W, Germain P, et al. Monitoring ligand-mediated nuclear receptor-coregulator interactions by noncovalent mass spectrometry. European Journal of Biochemistry. 2004;271(23-24):4958–4967. doi: 10.1111/j.1432-1033.2004.04466.x. [DOI] [PubMed] [Google Scholar]
- 28.Bich C, Bovet C, Rochel N, et al. Detection of nucleic acid-nuclear hormone receptor complexes with mass spectrometry. Journal of the American Society for Mass Spectrometry. 2010;21(4):635–645. doi: 10.1016/j.jasms.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Osz J, Brlivet Y, Peluso-Iltis C, et al. Structural basis for a molecular allosteric control mechanism of cofactor binding to nuclear receptors. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(10):E588–E594. doi: 10.1073/pnas.1118192109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McInerney EM, Rose DW, Flynn SE, et al. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes and Development. 1998;12(21):3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gampe RT, Jr, Montana VG, Lambert MH, et al. Asymmetry in the PPARγ/RXRα crystal structure reveals the molecular basis of heterodimerization among nuclear receptors. Molecular Cell. 2000;5(3):545–555. doi: 10.1016/s1097-2765(00)80448-7. [DOI] [PubMed] [Google Scholar]
- 32.Rastinejad F, Wagner T, Zhao Q, Khorasanizadeh S. Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. EMBO Journal. 2000;19(5):1045–1054. doi: 10.1093/emboj/19.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benecke A, Chambon P, Gronemeyer H. Synergy between estrogen receptor a activation functions AF1 and AF2 mediated by transcription intermediary factor TIF2. EMBO Reports. 2000;1(2):151–157. doi: 10.1093/embo-reports/kvd028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1 with the coomassie-stained SDS-PAGE and Native-PAGE of the PPARg2/RXRa complexes after the final purification step. (a) 12% SDS gel for PPARg2/RXRa complex, (b) 12% SDS gel (left) and 8-25% Native gel for PPARg2/RXRaDeltaNTD/DNA, (c) 12% SDS gel for PPARg2DeltaNTD/RXRaDeltaNTD/DNA/TIF2 RID and (d) 12% SDS gel for PPARg LBD/TIF2 RID complex.


