Summary
Background
Increased activity of metalloproteinases may play a role in the initiation and propagation of inflammation in sarcoidosis, and may also be one of the factors responsible for the development of lung fibrosis. The aim of this study was to verify whether polymorphisms of MMP2 C-735T, MMP7 A-181G, MMP9 T-1702A and tissue inhibitor of metalloproteinase (TIMP)2 G-418C predispose to sarcoidosis.
Material/Methods
The study included 139 patients with sarcoidosis and 100 healthy subjects. MMPs and TIMP2 mRNA were measured in peripheral blood lysate using real-time RT-PCR. DNA for genetic polymorphism was extracted from peripheral blood by GTC method. Protein concentrations in peripheral blood lysates were measured by ELISA, and MMP2 and 9 activities in BAL fluid were estimated by gel zymography.
Results
TT genotype in MMP9 T-1702A was more frequent in sarcoidosis (p<0.0001, OR=13.71, 95% CI 7.02–26.80) and resulted in higher expression of MMP9 mRNA (p<0.0001). No differences were found between TT and AT/AA patients in terms of radiological stage, lung function test parameters, activity markers and the presence/absence of Löfgren syndrome. There were no differences in the distribution of MMP2, MMP7 and TIMP2 polymorphisms. Messenger RNAs, as well as protein concentrations of MMP2, 7, 9, and TIMP2 were elevated in patients with sarcoidosis (p<0.0001 for each).
Conclusions
The TT homozygotes of MMP9 T-1702A genotype may be predisposed to sarcoidosis. Elevated MMP2, 7, 9, and TIMP2 mRNAs suggest their inducibility.
Keywords: sarcoidosis, metalloproteinases, genetic polymorphism
Background
Sarcoidosis is a granulomatous inflammatory disease most commonly affecting lungs and intrathoracic lymph nodes. The etiology of sarcoidosis remains unknown, but genetic factors implicate susceptibility to the illness and influence its clinical course and outcome. Although in the majority of cases the disease regresses spontaneously, up to 15% of patients develop a progressive lung disease with fibrosis [1].
The family of metalloproteinases is represented by 24 genes that collectively cleave all components of extracellular matrix (ECM). They are regulated by specific tissue inhibitors (TIMPs). The balance between MMPs and TIMPs is responsible for physiological tissue turnover, and excess protease activity may lead to tissue destruction, eventually initiating impaired healing and, in consequence, an uncontrolled production of connective tissue elements [2]. Additionally, metalloproteinases are one of the critical mediators of host defense and inflammation; therefore several other activities may be implicated in the pathogenesis of sarcoidosis. MMP2, 9 (gelatinases A and B) and 7 (matrilysin) degrade many ECM substrates. They activate cytokines, which may be responsible either for granuloma formation or tissue fibrosis, by their ability to cleave pro-TGF-β, pro-IL1β, pro-TNFα, other MMP propeptides and other cytokines [2–4]. On the other hand, TGF-β, IL1β and TNFα, key cytokines in the pathogenesis of sarcoidosis, are strong inducers of the MMP9 gene [5,6], which may result in formation of a self-perpetuating mechanism. Recently it was shown that Th1 lymphocytes, which are predominant in sarcoid inflammation, stimulate macrophages to increase production of MMP2 and MMP9 much more than Th2 or Th0 cells [7]. All these mechanisms may be involved in the pathogenesis of sarcoidosis. TIMPs 1 to 4 represent a family of tissue inhibitors responsible for post-translational regulation of MMPs activity.
Many experimental studies indicate the role of the imbalance between metalloproteinases and inhibitors in the pathogenesis of lung fibrosis [8,9]. Metalloproteinases, including MMP8 (collagenase), MMP7 and TIMP1 have been implicated in the pathogenesis of idiopathic pulmonary fibrosis (IPF) [10,11]. Previous studies on metalloproteinases and their inhibitors in sarcoidosis revealed increased collagenase activity of BAL fluid related to MMP8 [10]. According to other authors, increased concentrations of TIMP1 in BAL fluid of sarcoid patients are related to impaired lung function [12]. Another group reported increased levels of MMP9 in induced sputum of these patients, which correlated negatively with lung diffusion capacity [13].
In view of the above, there is a possibility that uninhibited activity of metalloproteinases in the lung may promote initial stages of granulomatous inflammation and influence the course of sarcoidosis. The genetic polymorphism could explain why some are predisposed to the disease and why only a small proportion of patients are predisposed to such an unfavorable outcome as lung fibrosis. Therefore, in our present study we looked for the selected genetic polymorphisms of MMP2, 7, 9 and TIMP2 in a population of patients from central Poland suffering from sarcoidosis. The selection of polymorphisms was based on the literature search. MMP2 C-735T, MMP7 A-181G and TIMP2 G-418C are common functional polymorphisms identified as risk factors for cancer development, progression and response to treatment [14,15], or vascular pathology [16]. MMP9 T-1702A is a less recognized polymorphism (1 of 4 identified in the promoter region of MMP9 gene), studied in the context of bronchial wall remodeling, but with negative results [17,18]. None of these polymorphisms have been studied in a population of patients with sarcoidosis or other interstitial lung diseases.
Material and Methods
Study group
From October 2007 to July 2010, 139 Caucasian residents of central Poland consulted with the Division of Pneumology and Allergy, Medical University of Lodz. Sarcoidosis was proven by biopsy or without biopsy, when clinical and radiological symptoms indicated stage I/II disease with Löfgren syndrome (27% of this subgroup did not have histo-pathological confirmation) [1,20]; 11 patients were on treatment during evaluation and blood sampling (maintenance dose of prednisone 5–10 mg/day). The sarcoidosis group consisted of never-smokers (n=96; 69%), ex-smokers (n=31; 22%; 13 pack-years) and active smokers (n=12; 9%; 10 pack-years). Other respiratory and non-respiratory chronic conditions were exclusion criteria. At the time of blood collection, bronchoscopy and other examinations, the patients were free of acute respiratory infection. The study was approved by the Ethics Committee of the Medical University of Lodz (Consent No. RNN/99/08/KE). All participants provided informed consent.
Lung function tests
Spirometry with flow-volume loop was performed according to ERS/ATS standards [21] on computer-based spirometer (Jaeger, Germany). Forced expiratory volume in 1st second (FEV1) and forced vital capacity (FVC) were provided as percent of predicted value. FEV1/FVC from best achieved values was calculated and presented as a percentage.
Lung diffusion capacity for carbon monoxide (DLCO) was measured on Lungtest 1000 SB (MES, Poland) with a single breath method, according to ATS/ERS standards [21]. Values were corrected for hemoglobin concentration (DLCOc) and presented as% of predicted value.
Bronchoscopy was performed with a flexible bronchoscope (Pentax, Japan) according to British Thoracic Society guidelines [22].
Bronchoalveolar lavage fluid (BALF) was collected from the medial lobe or lingula, by administration and subsequent withdrawal of 4×50 ml of 0.9% NaCl. The fluid recovery was 57±2%. The crude BALF was filtered through gauze and centrifuged, and the pellet was suspended in a phosphate buffer. The total cell count (TCC) was presented as n×106. Cytospin slides were prepared and stained by May-Grünwald-Giemsa stain. Numbers of particular cell types were calculated under a light microscope and presented as a percent of TCC.
Laboratory activity markers
Serum C-reactive protein (CRP), serum angiotensin-converting enzyme (SACE), serum Ca2+ concentration and 24-hrs urine Ca2+ loss were measured.
The control group
The control group consisted of healthy non-smoking volunteers (n=100, 55 women, age 38.7 years). All had a negative personal and family history of sarcoidosis and their chest X-ray was normal. Other respiratory and non-respiratory chronic conditions were exclusion criteria. At the time of blood collection they were free of respiratory infection.
RNA extraction and reverse transcription
Total RNA was extracted from the blood samples using an RNA extraction reagent, TRIZOL (Invitrogen Life Technologies, USA), according to the standard acid-guanidinium-phenol-chloroform method [23]. The extracted RNA concentrations and purity were determined by spectrophotometer readings at 260 and 280 nm and analyzed by 1% agarose-2M formaldehyde gel electrophoresis. Total RNA was digested with Dnase I (Invitrogen Life Technologies, USA) at room temperature for 15 min. Five micrograms of digested RNA were reverse transcribed at 42°C for 60 min in a total 20 ml reaction volume using the ImProm-II™ Reverse Transcription System kit (Promega, USA). Obtained cDNA was used in real-time PCR reaction.
Detection of gene expression using real-time RT-PCR method
Real-time PCR based on TaqManTM technology was performed using master mix prepared according to the FastStart Universal Probe Master (ROX) from Roche Applied Science. Probes and primers were designed using the online Universal Probe Library (www.universalprobelibrary.com). Primer sequences and probe numbers are as follow: MMP2 (forward, 5′-ACTGTTGGTGGGAACTCAGAAG-3′, reverse, 5′-CAAGGTCAAT GTCAGGAGAGG-3′, probe: #1), MMP7 (forward, 5-CGGATGGTAAGCAGTCTAGGG-3′, reverse, 5′-AGGTTGGATACATCACTGCA3TTAG-3′, probe: #49), MMP9 (forward, 5-TGGGTGTACGACGGTGAAAA-3′, reverse, 5′-CATGGGTCTCTAGCCTGATA-3′ probe: #31), TIMP2(forward 5′-TCTGGAAACGACATTTATGG-3′, reverse 5′-GTTGGAGGCCTGCTTATGGG-3′ and 18sRNA (forward 5′ CCGATAACGAACGAGACTCTGG-3′, reverse 5′ TAGGGTAGGCACACGCTGAGCC-3′ probe: #29), which was used as internal control for real-time PCR.
Real-time PCR was carried out in a final volume of 50 μl, with 0.05 μg cDNA, 25 μl FastStart Universal Probe Master (ROX) 2×, 250 nM probe and 1 μM of each primer. Amplification was performed for 10 min at 95°C to activate FastStart Taq DNA polymerase and 40 rounds of 15 sec at 95°C and 1 min at 60°C for amplification and signal analysis. ABI Prism 7000 Sequence Detection System from Applied Biosystems was used to detect amplifications. Each sample was assayed in triplicate in independent reactions. RT PCR data were automatically calculated with the data analysis module. The results were analyzed according to the 2–ΔΔCt method [24]. Validation of PCR efficiency was performed with a standard curve. Standard curves were prepared for each gene by serial dilution.
Determination of peripheral cell lysate MMP2, 7, 9, TIMP2 levels using Enzyme-Linked Immunosorbent Assay (ELISA)
For the quantitative detection of MMP2, 7, 9 and TIMP2 in peripheral cells lysate, the RayBio® Human ELISA (Enzyme-Linked Immunosorbent Assay) from RayBiotech was used, according to the manufacturer’s recommendations.
Zymography
MMP2/MMP9 zymography from BAL fluid samples were carried out according to Gürkan et al. [25]. Electrophoresis was performed using 5% polyacrylamide stacking gel and 10% resolving polyacrylamide co-polymerized with 1 mg/mL gelatin. MMP-2/MMP-9, human zymography standards (Millipore, Billerica, MA, US), were simultaneously loaded onto the gel. Gels were run in standard Tris-glycine-SDS running buffer. Gelatin gels were washed overnight by gentle shaking at room temperature in rinse buffer [50 mmol/L Tris-HCl pH 8.0, 5 mmol/L CaCl2 and 2.5% (v/v) Triton X-100], incubated in 50 mmol/L Tris-HCl pH7.5, 5 mmol/L CaCl2 for 18 h. Parallel gels of gelatin zymography were incubated in buffers containing 10 mmol/L EDTA to inhibit metalloproteinase activity. Gelatinolytic activity appeared as a clear band over a blue background. Using Image Master VDS (Pharmacia Biotech) images taken at the same magnification were quantified by densitometry, on the basis of their contour quantity after background subtraction. The arbitrary densitometry units were correlated with a standard curve prepared by serial dilutions of human recombinant gelatinases across linear range (0.03–1.25 ng/ml). The final results were expressed as 103 arbitrary units/ml of BAL fluid. A total of 74 BAL samples were available for examination.
Analysis of sequence gene polymorphism
DNA was extracted from whole blood according to the GTC method [23]. The MMP2, 7, 9 and TIMP2 gene polymorphisms were analyzed using 0.1 μg genomic DNA, 200 μM each dNTP, 5× GoTaq buffer solution, 1u GoTaq polymerase (Promega, Madison WI, USA), 0.5 μM primers 5′ ATAGGGTAAACCTCCCCACATT 3′ 5′ GGTAAAATGAGGCTG AGACCTG 3′, 5′ TGGTAC CATAATGTCCTGAATG 3′ 5 ′TCGTTATTGGCAGGAA GCACACAATGAAT 3′, 5′ CAAGGTC ACATAGCTGGAA 3′ 5′ CACCACGCCTTG GCTAAAT-3′ and 5′ CGTCTCTTGTTGGCTGGTCA 3′ 5′-CCTTCAGCTCGACTCTG GAG-3′ specific for MMP2 C-735T, MMP7 A-181G, MMP9 T-1702A, TIMP2 G-418C, respectively [26–29]. After a 5 min denaturing step, amplification was performed according to the following cycling profile: 94°C for 30 sec, 59°C for 30 sec and 72°C for 30 sec (28 cycles). The final elongation step was 10 min at 72°C. Amplification product was digested with restriction enzyme MMP2 C-735T (300bp)/Hinf 1, MMP7 A-181G (150bp)/EcoRI, MMP9 T-1702A (353bp)/BseJ I and TIMP2 G-418C (327bp)/Eco88 I (New England BioLabs). The polymorphism was visualized by separating the digested amplification products on 6% PAA gel in TAE buffer. Bands were visualized by UV light, and the results were recorded photographically and analyzed densitometrically using an LKB Ultrascan XL Enhanced Laser Densitometer.
Statistical analysis
Data were expressed as mean ± standard error of means (SEM), with the exception of age, where standard deviation (SD) was provided. Chi-square test was used to estimate Hardy-Weinberg equilibrium within subgroups and differences in frequency distribution of various polymorphisms between sarcoidosis and control groups, and within the sarcoidosis group (radiological stages, LS vs. non-LS). Shapiro-Wilk’s W test was used to assess normality. Medians with 25th and 75th percentile were provided for non-normally distributed continuous data. Categorical data were compared using Yates’ corrected χ2 test or Pearson’s χ2 test. Distribution of alleles in the control group was tested against one expected from the Hardy-Weinberg (H-W) equilibrium using the χ2 test. Odds ratios were computed for recessive model of inheritance in all cases. Multivariate analysis of association between polymorphisms was performed using logistic regression incorporating assessment of interaction between the analyzed genotypes. Unpaired T-test (for normally distributed data) or Mann-Whitney test (for non-parametric data) was used to compare sarcoidosis with controls. When more than 2 groups were compared, one-way ANOVA and Tukey’s post-hoc test for unequal samples (for data with Gaussian distribution) or Kruskall-Wallis followed by Dunn’s Multiple Comparison Test (for data without normal distribution) was used. Spearman test was applied to assess correlations. The p value ≤0.05 was assumed as statistically significant.
Results
Table 1 shows the characteristics of the study group in relation to the presence/absence of Löfgren syndrome and Table 2 shows these characteristics in relation to radiological stages. The population was suitably matched for age (39.0±11.0 year compared to 38.7±6.7 for control, ns) but sex distribution was different (40% of women in sarcoidosis vs. 55% in controls, p=0.02 estimated by χ2 test). There were no interactions between sex and MMPs genotypes in the study group: p=0.92 for MMP2, p=0.94 for MMP7, p=0.19 for MMP9; however, the influence of sex on TIMP2 genotype may not be excluded (p=0.03).
Table 1.
The characteristics of the study group in relation to the presence or absence of Löfgren syndrome (LS).
| All | LS | Non-LS | LS vs. non-LS | |
|---|---|---|---|---|
| Females/males | 56/83 | 20/41 | 36/42 | NS |
| Age [mean ± SD] | 39.0±11.0 | 35.5±9.0 | 42.0±11.5 | p=0.0003 |
| Radiological stage 0/I/II/III/IV | 6/70/52/9/2 | 0/49/12/0/0 | 6/21/40/9/2 | p=0.0002 |
| FEV1% predicted [mean ± SEM] | 89.9±1.7 | 93.4±1.9 | 87.1±2.6 | NS |
| FVC% predicted [mean ± SEM] | 96.4±1.8 | 98.4±1.9 | 94.7±2.6 | NS |
| FEV1/FVC [mean ± SEM] | 0.78±0.01 | 0.79±0.01 | 0.78±0.01 | NS |
| DLCOc% predicted [mean ± SEM] | 91.5±1.8 | 97.9±2.6 | 86.9±2.5 | p=0.002 |
| SACE IU/L [median; 25–75 percentile] | 53.1 29.1–73.9 | 45.8 28.3–71.6 | 60.2 29.7–74.9 | NS |
| CRP mg/l [median; 25–75 percentile] | 4.0 1.9–13.1 | 9.2 1.9–69.5 | 3.0 1.8–7.9 | p=0.01 |
| Serum Ca2+ mmol/L [median; 25–75 percentile] | 2.49 2.40–2.53 | 2.46 2.37–2.53 | 2.50 2.43–2.55 | NS |
| Urine Ca2+ mmol/24 hrs [mean ±SEM] | 5.28±0.32 | 4.10±0.33 | 6.14±0.47 | p=0.002 |
| BALF lymphocytes% | 31.0±1.8 | 33.5±2.6 | 28.6±2.3 | NS |
| BALF neutrophils% | 1; 0–2 | 1; 0–2 | 1; 0–3 | NS |
| BALF eosinophils% | 0; 0–1 | 0; 0–1 | 1; 0–1 | NS |
BALF – bronchoalveolar lavage fluid; CRP – C-reactive protein; DLCOc – diffusion capacity corrected for hemoglobin; FEV1 – forced expiratory volume in 1st second of expiration; FVC – forced vital capacity; NS – not significant; SACE – serum angiotensin converting enzyme; SEM – standard error of means.
Table 2.
The characteristics of the study group in relation to radiological stages (0, I, II, III, IV).
| Feature/radiological stage | 0/I | II | III/IV | p value |
|---|---|---|---|---|
| Females/males | 29/56 | 18/24 | 9/3 | 0.02 |
| Age [mean ± SD] | 38.0±10.5* | 38.7±11.8 | 47.1±2.9* | 0.03 |
| FEV1% predicted [mean ± SEM] | 94.3±1.8* | 83.4±3.2* | 84.9±7.8 | *0.01 |
| FVC% predicted [mean ± SEM] | 99.4±2.3 | 92.0±3.1 | 93.3±6.7 | NS |
| FEV1/FVC [mean ± SEM] | 0.97±0.02 | 0.77±0.02 | 0.77±0.04 | NS |
| DLCOc% predicted [mean ± SEM] | 97.1±2.1*# | 86.7±3.2# | 75.3±5.8* | *0.001; #0.05 |
| SACE IU/L [mean ± SEM] | 50.4±3.7 | 72.8±8.8 | 68.5±20.5 | NS |
| CRP [median; 25–75 percentile] | 3.7 (1.6–21.4) | 7.5 (3.0–12.1) | 2.8 (1.6–7.9) | NS |
| Serum Ca2+ mmol/L [median; 25–75 percentile] | 2.47 (2.40–2.53) | 2.51 (2.39–2.57) | 2.50 (2.48–2.57) | NS |
| Urine Ca2+ mmol/24 hrs [mean ±SEM] | 4.83±0.38 | 5.9±0.6 | 6.3±1.1 | NS |
| BALF lymphocytes% | 31.1±2.2 | 34.0±3.3 | 18.5±4.5 | NS |
| BALF neutrophils% | 1 (0–2) | 2 (0–3) | 0 (0–1) | NS |
| BALF eosinophils% | 0 (0–1) | 0.5 (0–1.5) | 0.5 (0–1) | NS |
BALF – bronchoalveolar lavage fluid; CRP – C-reactive protein; DLCOc – diffusion capacity corrected for hemoglobin; FEV1 – forced expiratory volume in 1st second of expiration; FVC – forced vital capacity; NS – not significant; SACE – serum angiotensin converting enzyme; SD – standard deviation; SEM – standard error of means. Signs * and # indicate compared values with statistical significance.
Differences in frequency distribution of various genotypes
MMP2 C-735T
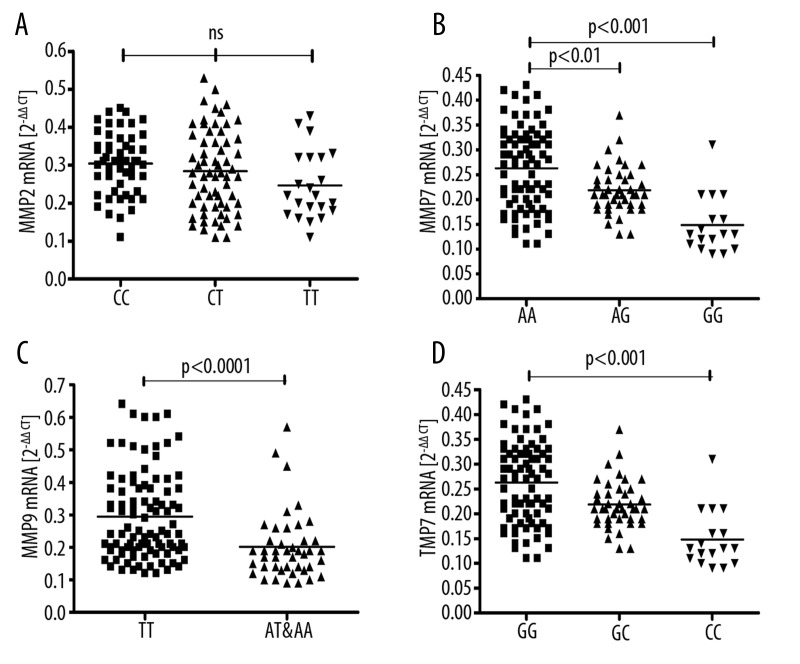
The distribution of alleles showed marginal deviation from the H-W equilibrium (p=0.06). There were no differences in genetic distribution between groups (Table 3). There were no differences in mRNA expression between genotypes in the study group (Figure 1A). Expression of proMMP2/MMP2 indicated by gel zymography did not show any differences between genotypes: proMMP2–4.07±0.33 vs. 3.79±0.17 vs. 3.82±0.38, and MMP2–4.48±0.34 vs. 4.27±0.21 vs. 4.35±0.49 103 units/ml BAL (for CC, CT and TT, respectively). CC genotype was more frequent in patients without LS (p=0.02). Patients with CC, CT and TT polymorphisms were not different in terms of parenchymal involvement (radiological stages), presence/absence of LS and LFT results.
Table 3.
Genotype distribution between sarcoidosis and control groups.
| χ2 | p | OR (alleles) | χ2 | 95%CI | p | ||||
|---|---|---|---|---|---|---|---|---|---|
| MMP-2 C735T | CC | CT | TT | C vs.T | |||||
| Control | 40 | 39 | 21 | ||||||
| Sarcoidosis | 54 | 63 | 22 | 1.43 | 0.49 | 1.09 | 0.20 | 0.89–1.20 | 0.66 |
| MMP-7 A181G | AA | AG | GG | A vs. G | |||||
| Control | 47 | 40 | 13 | ||||||
| Sarcoidosis | 52 | 58 | 29 | 3.38 | 0.18 | 0.69 | 3.76 | 0.47–1.01 | 0.053 |
| MMP-9 T1702A | TT | AT | AA | T vs. A | |||||
| Control | 14 | 40 | 46 | ||||||
| Sarcoidosis | 96 | 41 | 2 | 97.71 | <0.0001 | 5.74 | 62.71 | 3.67–8.97 | <0.0001 |
| TIMP-2 G418C | GG | GC | CC | G vs. C | |||||
| Control | 70 | 22 | 8 | ||||||
| Sarcoidosis | 80 | 42 | 17 | 3.90 | 0.14 | 0.74 | 1.72 | 0.48–1.16 | 0.19 |
OR – odds ratio; CI – confidence interval. Chi2 and p values characterize the distribution differences of various polymorphisms within a studied genotype between controls and sarcoidosis patients.
Figure 1.
Expression of mRNA in peripheral blood lysates of sarcoid patients with various MMPs/TIMP2 polymorphisms. Horizontal lines show mean values: (A) MMP2 C-735T (CC vs. CT vs. TT); (B) MMP7 A-181G (AA vs. AG vs. GG); (C) MMP9 T-1702A (TT vs. AT and AA); (D) TIMP2 G-418C (GG vs. GC vs. CC).
MMP7 A-181G
The distribution of MMP7 polymorphisms was in Hardy-Weinberg equilibrium (p=0.34). There were no differences in genetic distribution between groups (Table 3). Patients with AA genotype had higher mRNA expression compared to AG (0.26±0.01 vs. 0.22±0.01 2–ΔΔCt, p<0.01) and GG genotypes (vs. 0.15±0.01 2–ΔΔCt, p<0.001, Figure 1B). Patients with AA, AG and GG polymorphisms were not different in terms of parenchymal involvement (radiological stages), presence/absence of LS and LFT results.
MMP9 T-1702A
The distribution of MMP9 polymorphisms was in Hardy-Weinberg equilibrium (p=0.27). We found statistically significant differences in the distribution of MMP9 T-1702A polymorphisms, with more frequent TT genotype in sarcoidosis (Table 3). Odds ratio (OR) for TT genotype equaled 13.71 (95% CI 7.02–26.80, p<0.0001), suggesting a highly significant overabundance of this genotype in patients with sarcoidosis in comparison to the control group. Patients with TT genotype had higher expression of MMP9 mRNA compared to TA and AA taken together (Table 4, Figure 1C). Gel zymography did not show significant differences in MMP9 and pro-MMP9 activity in BAL between AT/AA and TT genotypes (Table 4). Moreover, there were no differences in proMMP9/MMP9 zymography results between patients with LS vs. non-LS and patients with and without parenchymal involvement. TT patients had significantly lower CRP concentrations (3.6; 1.8–10.5 vs. 7.9; 2.4–95.8 mg/dL, p=0.02) and BAL fluid lymphocyte percentage (28.0±2.1 vs. 36.3±2.9, p=0.02). Patients with TT and AT/AA polymorphisms were not different in terms of parenchymal involvement (radiological stages), presence/absence of LS and LFT results.
Table 4.
Peripheral blood lysate mRNA and protein concentrations, and pro-MMP9 and MMP-9 activity in BAL fluid (estimated by gel zymograpgy) in patients with TT vs. TA&AA T1702A MMP-9 genotypes.
| mRNA [2–ΔΔCt] | Protein concentration [ng/mL] | BAL pro-MMP9 activity [103 u/ml BAL] | BAL MMP9 activity [103 u/ml BAL] | |
|---|---|---|---|---|
| TT | 0.25 (0.19–0.38) | 286.1±9.7 | 2.40±0.18 | 7.79±0.46 |
| TA and AA | 0.19 (0.14–0.22) | 286.1±13.2 | 2.48±0.17 | 6.64±0.63 |
| p value | <0.0001 | 0.96 | 0.66 | 0.14 |
TIMP2 G-418C
The distribution of TIMP2 polymorphisms was not in Hardy-Weinberg equilibrium (p=0.004). There were no differences in genetic distribution between groups (Table 3). TIMP2 G-418C alleles subgroups were different in terms of mRNA expression, with the following values from the highest to the lowest: GG – 0.26±0.01, GC – 0.22±0.01, CC – 0.15±0.01 2–ΔCt (GG vs. CC p<0.001, Figure 1D). Patients with GG, GC and CC polymorphisms were not different in terms of parenchymal involvement (radiological stages), presence/absence of LS and LFT results.
Multivariate analysis of MMP-7 and MMP-9 polymorphisms confirmed univariate results, with MMP-9 TT homozygosity remaining a highly significant factor promoting the diagnosis of sarcoidosis. No interaction effect between genotypes at the analyzed sites was found (Table 5).
Table 5.
Multivariate analysis results of association between matrix metalloproteinase 7 (MMP-7) and 9 (MMP-9) gene polymorphisms and presence of sarcoidosis.
| Polymorphic variant | p-level | OR | 95% CI | |
|---|---|---|---|---|
| MMP-7 AA | 0.42 | 0.87 | 0.61 | 1.22 |
| MMP-9 TT | <0.0001 | 3.90 | 2.75 | 5.52 |
| Interaction between MMP-7 and MMP-9 genotypes | 0.16 | 1.28 | 0.90 | 1.82 |
OR – odds ratio; 95%CI – 95% Confidence interval.
There were no differences in mRNA expression between studied genotypes in the control group.
Expression of mRNA and concentration of proteins in sarcoid vs. non-sarcoid subjects
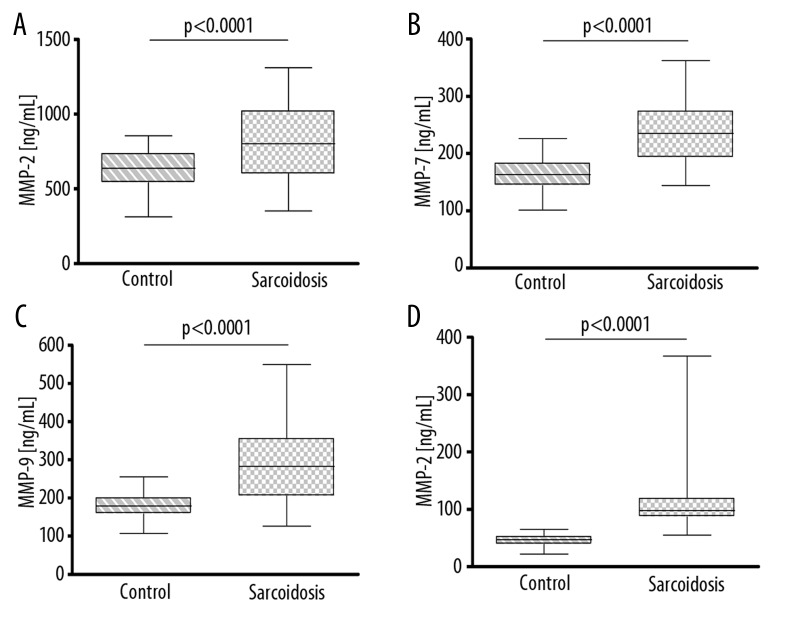
Messenger RNA expressions for MMP2, MMP7, MMP9 and TIMP2 were significantly higher in sarcoidosis (Figure 2A–D). Concentrations of proteins in whole peripheral blood lysates were also significantly higher (Figure 3A–D). In the study group, a correlation was found between MMP2 mRNA and MMP2 concentration (r=0.39; p<0.0001), and between MMP9 mRNA and MMP9 concentration in peripheral blood lysates (r=0.38; p<0.0001). There were no correlations between activity of MMP2 and MMP9 in BAL and mRNA concentrations or protein concentrations in peripheral blood lysates.
Figure 2.
Expression of mRNA in peripheral blood lysates in control and sarcoidosis groups for: (A) MMP2; (B) MMP7; (C) MMP9; (D) TIMP2. Data shown as median (horizontal line) and interquartile range (boxes). Whiskers show minimum and maximum values.
Figure 3.
Concentrations of proteins in peripheral blood lysates in control and sarcoidosis groups for: (A) MMP-2; (B) MMP-7; (C) MMP-9; (D) TIMP-2. Data shown as median (horizontal line) and interquartile range (boxes). Whiskers show minimum and maximum values.
Other correlations
Weak correlations were noticed between SACE and MMP2, and MMP7 mRNA (r=0.22; p=0.04 and r=0.23, p=0.01, respectively). MMP2 and MMP9 mRNA correlated with the number of episodes, where further episodes (>1) were defined as new relapses after spontaneous or drug-related remission had been achieved (r=0.28, p= 0.001 and r=0.23, p=0.02, respectively). Besides, MMP2 and MMP9 mRNA were higher in patients with >2 episodes (n=7, p=0.003 and p=0.02, respectively). MMP2 activity in BAL fluid estimated by zymography correlated negatively with DLCOc (r=–0.31, p=0.03).
Discussion
To our knowledge this is the first study on the genetic polymorphisms of MMP2, -7, -9 and TIMP2 in sarcoidosis. We have shown the higher prevalence of MMP9 TT1702 genotype and higher frequency of T allele in patients with sarcoidosis in the population under study. There were no differences in the distribution of MMP2, -7 and TIMP2 polymorphic genes between the sarcoidosis and control groups. The unexpected deviation of allele frequencies in MMP-2 and TIMP-2 polymorphic sites from the Hardy-Weinberg equilibrium made it impossible to include these variants into multivariate analysis. These polymorphisms should therefore be validated in other populations or study groups, as their association with sarcoidosis remains unclear.
Messenger RNAs for all studied MMP’s and TIMP were highly up-regulated in sarcoid patients, which strongly suggests that all of these molecules are inducible.
To date, the most extensively studied MMP9 polymorphism is the C-1562T, with the T allele reported to be linked to higher levels of gene expression, and in 1 study it was shown to influence the survival in non-small cell lung carcinoma [29]. T allele was also far more frequent in patients suffering from abdominal aortic aneurysms [30]. This polymorphism was not related to the susceptibility to idiopathic disseminated bronchiectasis [31] and decline of lung function in cigarette smokers [32]. It was shown to increase the susceptibility to COPD in the Korean population [33], but not in Brazilians [34]. Interestingly, an increased risk of multiple sclerosis, a Th1-related disease, was associated with the MMP9-1562 allele T [35].
There is far less published data on genetic MMP9 polymorphism of T-1702A. It was studied in the context of susceptibility to asthma and airway remodelling, but no associations were found, both for MMP9 C-1572T and T-1702A polymorphic genes [17,18]. Our results indicate that T-1702A TT homozygotes have significantly higher expression of MMP9 mRNA, which proves that this polymorphism is functional. It was shown to be much more frequent in patients suffering from sarcoidosis; therefore several links to the pathogenesis of sarcoidosis should be taken into consideration. First of all, patients with higher expression of MMP9 may be more susceptible to Th1 lymphocyte-mediated inflammation through increased activation of IL1β, TNFα and other cytokines [2–4]. These processes may result in further influx of lymphocytes, their differentiation, and stimulation of macrophages with their transformation to epithelioid and giant cells, forming a granuloma. Many cytokines involved in the initial steps of granuloma formation (ie, TNFα) may induce MMP9 genes, potentially increasing MMP9 burden in the lungs [5,6]. In the course of chronic, persistent inflammation, the increased activity of MMP9 and other matrix metalloproteinases may lead to increased ECM turnover, which may induce processes leading to fibrosis. This also releases growth factors from ECM. TGFβ and other growth factors, which may be important in the promotion of lung fibrosis, may also increase metalloproteinase expression [7], and conversely, MMPs may cleave proTGFβ, thus increasing its activity in the area of inflammation [3]. Messenger RNA of MMP2, 7 and TIMP2 and protein concentration in peripheral blood lysates were overexpressed in sarcoidosis patients, but the allelic distribution was not different between sarcoidosis patients and controls. This suggests that MMP genes are induced in the course of sarcoid inflammation. This observation is consistent with the results of Oviedo-Orta, who found that Th1 cells are more potent in the induction of MMP2 and MMP9 production by cocultured macrophages when compared to Th2 and Th0 cells [7]. Therefore, other metalloproteinases may also play a role in the pathogenesis of sarcoidosis.
There are limited functional data in the literature on the role of MMP9 in the pathogenesis of sarcoidosis. Fireman et al. found increased MMP9 concentrations in the induced sputum of sarcoidosis patients [13]. In one of our previous studies, we found the negative correlation between FEV1% predicted and the MMP9 expression in peripheral bronchial lung biopsies of sarcoid patients with parenchymal involvement [36]. MMP9 may be involved in airway remodeling in asthma [37,38].
Although on the basis of our results it seems highly probable that MMP9 TT1702 genotype predisposes to sarcoidosis, our work does not provide evidence for the negative prognostic value of this genotype. It was not linked to the more advanced radiological stage and worse lung function parameters. Also, mRNA expression was not negatively correlated with lung function parameters and there were no differences between radiological stages. However, MMP9 and MMP2 mRNA were correlated with the number of episodes. The significance of the difference between mRNA concentration in patients with 1 episode and ≥2 episodes was borderline, but the number of patients with recurrent sarcoidosis in our study group was small. Finally, we did not find a link between MMP9 polymorphism and enzyme activity in BAL. The most likely explanation of this apparent discrepancy is the influence of multi-level regulation of gene expression. Although the activity of MMP9 is regulated mostly at the transcriptional level [39], elevated mRNA for MMP9 in peripheral blood lysates of TT homozygotic patients without parallel increase in enzyme activity in BAL suggests the influence of posttranscriptional and posttranslational regulatory mechanisms. These may include regulation of mRNA stability, enzyme secretion and activation, and the influence of MMP inhibitors [2]. TIMP2 shows higher affinity towards MMP2, whereas MMP9 is strongly inhibited by TIMP1 [40]. Unfortunately, the latter inhibitor was not evaluated in this study. In addition, the MMP2 and 9 activities were measured in BAL fluid, whereas mRNA and protein concentrations were evaluated in peripheral blood. These limitations of our study prevent us from drawing final and definitive conclusions on possible posttranscriptional or posttranslational down-regulation of MMP activity in lower airways.
In view of the above, the presented results must be treated as preliminary. Significant differences in the expression of mRNA of all studied MMPs and TIMP2 between sarcoidosis and control groups strongly suggest the inducible character of these molecules. Also, significant differences in mRNA expression between various functional genotypes of MMP7, 9 and TIMP2 in sarcoidosis, but not in healthy subjects, suggest their inducibility.
In view of our results, further studies on the role of metalloproteinases in the pathogenesis of sarcoidosis may open new therapeutic possibilities. Matrix metalloproteinase inhibitors are a new class of drugs under development, which raises hopes of efficacy in the treatment of chronic inflammatory diseases [41].
Conclusions
On the basis of the above, we conclude that the MMP9 TT1702 genotype may confer considerable predisposition to sarcoidosis. Our results did not show any relationship with prognostic factors such as presence/absence of Löfgren syndrome, radiological stages or lung function parameters. A separate follow-up study should be conducted to evaluate such associations and verify the effect of MMP-2 and TIMP-2 polymorphic loci. MMP2, 7, 9, and TIMP2 genes are induced in the course of sarcoidosis, as shown by elevated mRNA and protein concentrations in peripheral blood lysates.
Acknowledgements
Many thanks to David Fam, a student of the Medical University of Lodz, for linguistic corrections.
Footnotes
Source of support: Medical University of Lodz (grant No 502–16-807) and Polish Ministry of Science (grant NN 402 350838), both granted to JS. WF received financial support from the Innovative Economy Operational Program – Activity 1.2 (the TEAM Program coordinated by the Foundation for Polish Science)
References
- 1.American Thoracic Society. European Respiratory Society: World Association of Sarcoidosis and Other Granulomatous Disorders. Statement on sarcoidosis. Am J Respir Crit Care Med. 1999;160:736–55. doi: 10.1164/ajrccm.160.2.ats4-99. [DOI] [PubMed] [Google Scholar]
- 2.Greenlee KJ, Werb Z, Kheradmand F. Matrix metalloproteinases in lung: Multiple, multifarious, and multifaceted. Physiol Rev. 2007;87:69–98. doi: 10.1152/physrev.00022.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu Q, Stemenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolitically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14:163–76. [PMC free article] [PubMed] [Google Scholar]
- 4.Schonbeck U, Mach F, Libby P. Generation of biologically active IL-1 beta by matrix metalloproteinases: a novel caspase-1-independent pathway of IL-1 beta processing. J Immunol. 1998;161:3340–46. [PubMed] [Google Scholar]
- 5.Yao PM, Maitre B, Delacourt C, et al. Divergent regulation of 92-kDa gelatinase and TIMP-1 by HBECs in response to IL-1 beta and TNF-alpha. Am J Physiol Lung Cell Mol Physiol. 1997;17:L866–74. doi: 10.1152/ajplung.1997.273.4.L866. [DOI] [PubMed] [Google Scholar]
- 6.Tobar N, Viller V, Sentibenez JF. ROS-NFkappaB mediates TGF-beta1-induced expression of urokinase-type plasminogen activator, matrix metalloproteinase-9 and cell invasion. Mol Cell Biochem. 2010;340:195–202. doi: 10.1007/s11010-010-0418-5. [DOI] [PubMed] [Google Scholar]
- 7.Oviedo-Orta E, Bermudez-Fajardo A, Karanam S, et al. Comparison of MMP-2 and MMP-9 secretion from T helper 0, 1 and 2 lymphocytes alone or in coculture with macrophages. Immunology. 2008;124:42–50. doi: 10.1111/j.1365-2567.2007.02728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swiderski RE, Dencoff JE, Floerchinger CS, et al. Defferential expression of extracellular matrix remodelling genes in a murine model of bleomycin-induced pulmonary fibrosis. Am J Pathol. 1998;152:821–28. [PMC free article] [PubMed] [Google Scholar]
- 9.Ruiz V, Ordonez RM, Berumen J, et al. Unbalanced collagenase/TIMP1 expression and epithelial apoptosis in experimental lung fibrosis. Am J Physiol Lung Cell Mol Physiol. 2003;285:L1026–36. doi: 10.1152/ajplung.00183.2003. [DOI] [PubMed] [Google Scholar]
- 10.Henry MT, McMahon K, Mackarel AJ, et al. Matrix metalloproteinases and tissue inhibitor of metalloproteinase-1 in sarcoidosis and IPF. Eur Respi J. 2002;20:1220–27. doi: 10.1183/09031936.02.00022302. [DOI] [PubMed] [Google Scholar]
- 11.Huh JW, Kim DS, Oh Y-M, et al. Is metalloproteinase-7 specific for idiopathic pulmonary fibrosis ? Chest. 2008;133:1101–16. doi: 10.1378/chest.07-2116. [DOI] [PubMed] [Google Scholar]
- 12.Shimada A, Koga T, Oshita Y, et al. Reduced pulmonary function is associated with enhanced inflammation and tissue inhibitor of metalloproteinase 1 concentration in the broncholaveolar lavage fluid of patients with lung parenchymal sarcoidosis. Kurume Med J. 2008;55:13–17. doi: 10.2739/kurumemedj.55.13. [DOI] [PubMed] [Google Scholar]
- 13.Fireman E, Kraiem Z, Sade O, et al. Induced sputum-retrieved matrix metalloproteinase 9 and tissue metalloproteinase inhibitor 1 in granulomatous diseases. Clin Exp Immunol. 2002;130:331–37. doi: 10.1046/j.1365-2249.2002.t01-1-02001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scherf DB, Dally H, Müller P, et al. Single nucleotide polymorphisms in matrix metalloproteinase genes and lung cancer chemotherapy response and prognosis. Eur Respir J. 2010;35:381–90. doi: 10.1183/09031936.00125608. [DOI] [PubMed] [Google Scholar]
- 15.Park KS, Kim SJ, Kim KH, Kim JC. Clinical characteristics of TIMP2, MMP2, and MMP9 gene polymorphisms in colorectal cancer. J Gastroenterol Hepatol. 2011;26:391–97. doi: 10.1111/j.1440-1746.2010.06504.x. [DOI] [PubMed] [Google Scholar]
- 16.Lamblin N, Bauters C, Hermant X, et al. Polymorphisms in the promoter regions of MMP-2, MMP-3, MMP-9 and MMP-12 genes as determinants of aneurismal coronary artery disease. J Am Coll Cardiol. 2002;40:43–48. doi: 10.1016/s0735-1097(02)01909-5. [DOI] [PubMed] [Google Scholar]
- 17.Ganter K, Deichmann KA, Heinzmann A. Association study of polymorphisms within matrix metalloproteinase 9 with bronchial asthma. Int J Immunogenet. 2005;32:233–36. doi: 10.1111/j.1744-313X.2005.00516.x. [DOI] [PubMed] [Google Scholar]
- 18.Lose F, Thompson PJ, Duffy D, et al. A novel tissue inhibitor of metalloproteinase-1 (TIMP1) polymorphism associated with asthma in Australian women. Thorax. 2005;60:623–28. doi: 10.1136/thx.2004.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wells AU, Hirani N, et al. Interstitial lung disease guideline: the British Thoracic Society in collaboration with the Thoracic Society of Australia and New Zealand and the Irish Thoracic Society. Thorax. 2008;63:v1–v58. doi: 10.1136/thx.2008.101691. [DOI] [PubMed] [Google Scholar]
- 20.Pellegrino R, Viegi G, Brusasco V, et al. ATS/ERS Task Force: Standardisation of lung function testing. Interpretative strategies for lung function tests. Eur Respir J. 2005;26:948–68. doi: 10.1183/09031936.05.00035205. [DOI] [PubMed] [Google Scholar]
- 21.British Thoracic Society guidelines on diagnostic flexible bronchoscopy. British Thoracic Society Bronchoscopy Guidelines Committee, a Subcommittee of the Standards of Care Committee of the British Thoracic Society. Thorax. 2001;56(Suppl I):i1–i21. doi: 10.1136/thorax.56.suppl_1.i1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chomczynski P, Sacchi N. Single – step method of RNA isolation by acid guanidinium thiocyanate- phenol- chloroform extraction. Anal Biochem. 1987;162:156–59. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 23.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 24.Luz Sampieri C, Pena S, Ochoa-Lara M, et al. Expression of matrix metalloproteinases 2 and 9 in human gastic cancer and superficial gastritis. World J Gastroenterol. 2010;16:1500–5. doi: 10.3748/wjg.v16.i12.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gürkan A, Emingil G, Saygan B, et al. Gene polymorphisms of matrix metalloproteinase-2, -9 and -12 in periodontal health and severe chronic periodontitis. Arch Oral Biol. 1997;53(4):337–45. doi: 10.1016/j.archoralbio.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 26.Lu Z, Wang Y, Zhang Q, et al. Association between the functional polymorphism in the matrix metalloproteinase-7 promoter and susceptibility to adult astrocytoma. Brain Res. 2006;1118:6–12. doi: 10.1016/j.brainres.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Ganter K, Deichmann A, Heinzmann A. Association study of polymorphisms within matrix metalloproteinase 9 with bronchial asthma. Int J Immunogenet. 2005;32:233–36. doi: 10.1111/j.1744-313X.2005.00516.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhou Y, Yu C, Miao X, et al. Substantial reduction in risk of breast cancer associated with genetic polymorphisms in the promoters of the matrix metalloproteinase-2 and tissue inhibitor of metalloproteinase-2 genes. Carcinogenesis. 2004;25:399–404. doi: 10.1093/carcin/bgh020. [DOI] [PubMed] [Google Scholar]
- 29.Jin G, Miao R, Hu Z, et al. Putative functional polymorphisms of MMP9 predict survival of NSCLC in a Chinese population. Int J Cancer. 2009;124:2172–78. doi: 10.1002/ijc.24190. [DOI] [PubMed] [Google Scholar]
- 30.Jones GT, Phillips VL, Harris EL, et al. Functional matrix metalloproteinase-9 polymorphism (C-1562T) associated with abdominal aortic aneurysm. J Vasc Surg. 2003;38:1363–67. doi: 10.1016/s0741-5214(03)01027-9. [DOI] [PubMed] [Google Scholar]
- 31.Stankovic M, Nikolic A, Divac A, et al. Matrix metalloproteinases gene variants in idiopathic disseminated bronchiectasis. J Investig Med. 2009;57:500–3. doi: 10.2310/JIM.0b013e318198277c. [DOI] [PubMed] [Google Scholar]
- 32.Joos L, He JQ, Shepherdson MB, et al. The role of matrix metalloproteinase polymorphisms in the rate of decline in lung function. Hum Mol Gen. 2002;1:569–76. doi: 10.1093/hmg/11.5.569. [DOI] [PubMed] [Google Scholar]
- 33.Lee SY, Kim MJ, Kang HG, et al. Polymorphisms in Matrix Metalloproteinase-1, -9 and -12 Genes and the Risk of Chronic Obstructive Pulmonary Disease in a Korean Population. Respiration. 2010;80:133–38. doi: 10.1159/000284926. [DOI] [PubMed] [Google Scholar]
- 34.Schirmer H, Basso da Silva L, et al. Matrix metalloproteinase gene polymorphisms: lack of association with chronic obstructive pulmonary disease in a Brazilian population. Genet Mol Res. 2009;8:1028–34. doi: 10.4238/vol8-3gmr596. [DOI] [PubMed] [Google Scholar]
- 35.Mirowska-Guzel D, Gromadzka G, Członkowski A, Członkowska A. Association of MMP1, MMP3, MMP9, and MMP12 polymorphisms with risk and clinical course of multiple sclerosis in a Polish population. J Neuroimmunology. 2009;214:113–17. doi: 10.1016/j.jneuroim.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 36.Piotrowski WJ, Nawrocka-Kunecka A, Antczak A, et al. Metalloproteinases MMP9, MMP2 and their tissue inhibitors TIMP1, TIMP2 in peripheral transbronchial lung biopsies of patients with sarcoidosis. Pol Arch Med Wewn. 2009;119:628–35. [PubMed] [Google Scholar]
- 37.Tonnel AB, Gosset P, Tillie-Leblond I. Characteristics of the inflammatory response in bronchoalveolar lavage fluid from patients with status asthmaticus. Int Arch Allergy Immunol. 2001;124:267–71. doi: 10.1159/000053729. [DOI] [PubMed] [Google Scholar]
- 38.Vermeer PD, Dunker J, Estin M, et al. MMP9 modulates tight junction integrity and cell viability in human airway epithelium. Am J Physiol Lung Cell Mol Physiol. 2009;296:L751–62. doi: 10.1152/ajplung.90578.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberhardt W, Huwiler A, Beck HF, et al. Amplification of IL-1beta-induced matrix metalloproteinase-9 expression by superoxide in rat glomerular mesangial cells is mediated by increased activities of NF-kappaB and activating protein-1 and involves activation of the MAPK pathways. J Immunol. 2000;165:5788–97. doi: 10.4049/jimmunol.165.10.5788. [DOI] [PubMed] [Google Scholar]
- 40.Vermeer PD, Denker J, Estin M, et al. MMP9 modulates tight junction integrity and cell viability in human airway epithelia. Am J Physiol Lung Cell Mol Physiol. 2009;196:L751–62. doi: 10.1152/ajplung.90578.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dormán G, Cseh S, Hajdu I, et al. Matrix metalloproteinase inhibitors: A critical appraisal of design principles and proposed therapeutic utility. Drugs. 2010;70:949–64. doi: 10.2165/11318390-000000000-00000. [DOI] [PubMed] [Google Scholar]