Abstract
Recognition of nucleic acids by TLR9 requires its trafficking from the endoplasmic reticulum to endolysosomal compartments and its subsequent proteolytic processing. Both processes depend on interactions of TLR9 with the polytopic endoplasmic reticulum–resident protein UNC93B1. To examine the intracellular behavior of TLR9 in primary APCs, we generated transgenic mice expressing a TLR9-GFP fusion. The TLR9-GFP transgene is functional and is proteolytically processed in resting bone marrow–derived macrophages (BMDMs), dendritic cells, and B cells. Inhibition of cleavage impairs TLR9-dependent responses in all primary APCs analyzed. The kinetics of TLR9-GFP processing in BMDMs and B cells differs: in B cells, proteolysis occurs at a faster rate, consistent with an almost exclusive localization to endolysosomes at the resting state. In contrast to the joint requirement for cathepsins L and S for TLR9 cleavage in macrophages, TLR9-GFP cleavage depends on cathepsin L activity in B cells. As expected, in BMDMs and B cells from UNC93B1 (3d) mutant mice, cleavage of TLR9-GFP is essentially blocked, and the expression level of UNC93B1 appears tightly correlated with TLR9-GFP cleavage. We conclude that proteolysis is a universal requirement for TLR9 activation in the primary cell types tested, however the cathepsin requirement, rate of cleavage, and intracellular behavior of TLR9 varies. The observed differences in trafficking indicate the possibility of distinct modes of endosomal content sampling to facilitate initiation of TLR-driven responses in APCs.
Introduction
Toll-like receptors sense conserved microbial molecules (1). Recognition of microbial products by these receptors on APCs is essential for the onset of adaptive immune responses. TLRs are found on the surface of cells or in endosomal compartments. The surface-disposed TLRs sense molecular components exposed on microbes, whereas endosomal TLRs recognize pathogen-derived nucleic acids, such as dsRNA (TLR3), ssRNA (TLR7 and TLR8), and dsDNA (TLR9). Proper TLR localization within cells ensures the encounter of TLRs with their respective ligands, and therefore regulation of TLR trafficking is crucial for their function (2). The requirement for interaction partners to deliver TLRs to their destination varies among TLRs. Whereas multiple surface and intracellular TLRs require the endoplasmic reticulum (ER)–resident chaperones gp96 and PRAT4A for localization (3), delivery of nucleic acid–sensing TLRs to endolysosomal compartments requires an interaction with the ER-resident protein UNC93B1 (4). Endosomal TLRs undergo cleavage upon reaching their proper destination (5–9), a process required for their function. Because TLR7 and TLR9 can also respond to self nucleic acids, their proper localization within the endolysosomal compartment is crucial to avoid autoimmune responses (10).
Our current knowledge of TLR9 trafficking and cleavage relies, for the most part, on studies performed in cell lines and primary dendritic cells that express TLR9 upon retroviral transduction. Retroviral and lentiviral transduction efficiencies of primary APCs are relatively low, hampering biochemical analysis, complicated also by the dearth of anti-mouse TLR9 Abs that can be used for immunoprecipitation. The unwanted activation of primary APCs by the transduction process can be a further confounding factor. Therefore, whether TLR9 is processed in B cells and plasmacytoid dendritic cells (pDCs) is not known. Differential TLR9 activation within pDCs and B cells (11, 12) by CpG oligonucleotides (ODNs) as well as among other cell types (13, 14) suggests that TLR9 trafficking and processing vary in a cell type–specific manner. Thus, the types of compartments sampled by TLR9 and the ensuing responses evoked by TLR9 engagement may also depend on the type of APC involved.
To understand the trafficking behavior and posttranslational modifications of endosomal TLR9 in primary APCs, we generated transgenic mice that express a murine TLR9-GFP fusion protein to allow live cell imaging and to facilitate biochemical analysis. Our data not only reveal cell type–specific differences in the behavior of TLR9 but also serve to validate these mice as an accurate model to study TLR9 trafficking and its functional properties. It allows an examination of TLR9 behavior in primary APCs where transfection or retroviral transduction experiments are difficult and where no permanently established cell lines exist.
Materials and Methods
Generation of TLR9-GFP transgenic mice
TLR9-GFP was cloned by restriction digest of the TLR9-GFP pMSCVpuro construct (4) into the pgkATGfrt-CAGGS-RGBpolyA vector (15) (TLR9-GFP CAGGS). Cherry-KDEL was cloned by restriction digest of the mcherry-KDEL pMSCVneo construct (4) into the pgkATGfrt-CAGGS-RGBpolyA vector (mcherry-KDEL CAGGS). Both constructs were verified by sequencing. Fifty micrograms of TLR9-GFP CAGGS or mcherry-KDEL CAGGS was mixed with 25 μg FLPe recombinase, precipitated, and used for electroporation of V6.5 (C57BL/6 x129 F1) ES cells. Mice were genotyped with the following primers: colA 5′-GCACAGCATTGCGGACATGC-3′, colB 5′-CCCTCCATGTGTGACCAAGG-3′, and colC 5′-GCAGAAGCGCGGCCGTCTGG-3′.
Mice and cell lines
TLR9KO mice (16) were obtained from A. Marshak-Rothstein (University of Massachusetts, Worcester, MA). UNC93B1 H412R mutant (3d) mice (17) were obtained from Bruce Beutler (The Scripps Institute, La Jolla, CA). C57BL/6 wild-type (WT) mice were purchased from Taconic. All animals were maintained under specific pathogen-free conditions, and experiments were performed in accordance with institutional, state, and federal guidelines. Immortalized bone marrow–derived macrophages (BMDMs) derived from C57BL/6J WT mice (NR-9456; National Institutes of Health Biodefense and Emerging Infections Research Resources Repository, National Institute of Allergy and Infectious Diseases, National Institutes of Health) and HEK 293-T cells (CRL-11268; American Type Culture Collection) were maintained in high-glucose DMEM containing 10% heat-inactivated fetal calf serum (IFS) and penicillin/streptomycin (P/S).
Preparation of primary cells and cell culture
Untouched B and T cells were isolated from spleens using magnetic beads (Miltenyi) according to the manufacturer’s specifications. FLT3-ligand dendritic cells (FLDCs) were prepared as follows: femur and tibia were flushed with RPMI 1640 medium, RBCs lysed, and cells plated in RPMI 1640 containing 10% IFS, P/S, and 2-mercaptoethanol supplemented with 3.5% FLT3L-containing cell culture supernatant. At day 8, nonadherent cells were harvested, stained for expression of CD11b and B220 to determine myeloid dendritic cell (mDC) and pDC subsets (mDCs are CD11b+B220−, whereas pDCs are CD11b− and B220+), and plated for stimulation assays.
For BMDMs, femur and tibia were flushed, and cells were cultured in high-glucose DMEM, 10% IFS (Hyclone), P/S, and 5% MCSF (day 0). At day 1 of culture, nonadherent cells were transferred to a fresh culture dish. Medium was replaced at day 5, and cells were used for experiments at day 7 or 8 of MCSF culture. Cells were assayed by FACS for Cd11b expression to ensure homogeneity.
For the generation of mouse embryonic fibroblasts (MEFs), a TLR9-GFP+/+/WT transgenic mouse was crossed with a C57BL/6 mouse, and embryos were removed at day 14.5 d postcoitum. MEFs generated from C57BL/6 mice were used as negative control. MEFs were prepared according to standard protocols and used for experiments between passages 2 and 4.
Abs and reagents
LysoTracker was purchased from Molecular Probes, and CpG-A (2336), CpG-B (1826), and CpG-B–Alexa 647 (1826) were synthesized by IDT. LPS (Escherichia coli 026:B6), polybrene, z-FA-FMK, and brefeldin A were purchased from Sigma, R848 and Pam3CysK4 from Invivogen, and complete protease inhibitors (PIs) from Roche. Specific cathepsin K, L, and S inhibitors were from Calbiochem. The polyclonal anti-UNC93B1 Ab recognizes the C terminus of murine UNC93B1 and was previously described (18). The Cd11b Ab clone M1/70 was from eBioscience, the Alexa Fluor 647–conjugated anti-mouse TNF-α Ab (clone MP6-XT22) from BD Pharmingen, and anti-GFP (ab290) from Abcam.
Histology
Organs of 3-mo-old mice (TLR9-GFP/WT−/+ and WT littermates) were fixed in 10% formalin and embedded in paraffin. Paraffin sections were deparaffinized in xylene and rehydrated in graded ethanol series. For Ag retrieval, slides were placed in a pressure cooker in citrate buffer (pH 7.6). Endogenous peroxidase was blocked with 3% H2O2. Subsequently, slides were blocked with ready to use blocking solution (Zytomed Systems).
Rabbit anti-GFP polyclonal A11222 (Invitrogen), diluted 1:1000 in Ab diluent from Zytomed Systems, was used as first Ab. As secondary Ab, goat anti-rabbit TR-060-BN (Thermo Scientific) was applied. Subsequently, streptavidin–HRP conjugate (Zytomed Systems), DAB chromogen, and substrate buffer (Zytomed Systems) were added. Counterstaining was done with hematoxylin (Merck).
Anti-nuclear Abs titers
Serum from 10- to 12-mo-old C57BL/6 (WT), TLR9-GFP heterozygous (one copy of the transgene) or homozygous (two copies of the transgene) was diluted and added to HEP-2 cells and IgG detected using anti-Ig-FITC Ab.
Cell lysate preparation
Organs were homogenized with a tissue disruptor in buffer containing 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA, and PIs. Nonidet P-40 (NP-40) was added to a final concentration of 1% (NP-40 lysis buffer). BMDMs, B cells, T cells, pDCs, and mDCs were lysed in RIPA buffer (see below). All cell lysates were centrifuged at 14,000 × g for 10 min, and the protein concentration of the supernatants was determined by bicinchoninic acid assay (Pierce).
Retrovirus production and transduction of MEFs and BMDMs
UNC93B1-hemagglutinin (HA) WT and UNC93B1-HA H412R or TLR9-GFP retroviruses were produced in 293-T cells as described earlier (18) and added to either MEFs generated from TLR9-GFP transgenic mice or immortalized BMDMs, respectively, in the presence of polybrene. Cells were spun for 1.5 h at 2200 rpm, medium was changed, and cells were selected with 1.2 mg/ml G418 (MEFs) or 5 μg/ml puromycin (BMDMs) (Invitrogen) 1 d after transduction.
Intracellular TNF FACS staining
BMDMs were seeded at day 7 of MCSF culture and stimulated with TLR agonists as indicated in the figure legends in the presence of 10 μg/ml brefeldin A for 4 h. Cells were trypsinized and fixed with 3.7% formaldehyde in PBS for 10 min at room temperature. Cells were permeabilized with 0.5% saponin in FACS buffer (PBS with 2% IFS) for 20 min at room temperature, stained with Alexa Fluor 647–conjugated anti-mouse TNF-α Ab for 30 min at room temperature (diluted 1:100), washed, and analyzed by FACS.
ELISA
IL-6 secretion was determined in media supernatants using the OptEIA kit (Becton Dickinson) according to the manufacturer’s specifications. IFN-α production was detected by ELISA using Abs specific for IFN-α (PBL).
Pulse-chase, immunoprecipitation, and endoglycosidase H/peptide-N-glycosidase F assays
In brief, cells were starved for methionine and cysteine in Met/Cys-free DMEM (starvation medium) for 30–60 min, pulsed for different time periods (see the figure legends) with 35S-labeled methionine/cysteine (PerkinElmer) in starvation medium supplemented with dialyzed FCS, and chased with an excess of non-radioactive amino acids in regular DMEM for various time periods (see the figure legends). Cells were lysed in either of the following lysis buffers supplemented with PIs as indicated in the figure legends: RIPA (20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 100 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS) or buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA with 1% digitonin as detergent. Lysates were equalized for incorporation of radioactive material with 35S counts in the trichloroacetic acid precipitate and immunoprecipitated with the indicated Abs. Washes were performed with the same buffers used for lysis except for digitonin lysates/immunoprecipitations, which were washed with 0.1–0.2% digitonin-containing buffer. Reimmunoprecipitations were carried out as follows: protein–Ab complexes were dissolved by denaturation with 1% SDS and 1% 2-mercaptoethanol for 1 h at 37°C (UNC93B-IPs) or 10 min at 95°C (TLR9-GFP-IPs). Subsequently, SDS and 2-mercaptoethanol were diluted to 0.1% by addition of 1% NP-40 lysis buffer, and reimmunoprecipitations were carried out with the indicated Abs. Immunoprecipitates were subjected to 10% SDS-PAGE and fluorography. Digestions with endoglycosidase H (Endo H) and peptide-N-glycosidase F (PNGase F) were performed, where indicated, in accordance with the manufacturer's instructions (New England BioLabs).
Confocal microscopy of B cells
For imaging primary B cells, 8-well-chambered coverglasses were coated with poly-L-lysine for 15 min at room temperature followed by a blocking step with RPMI 1640 supplemented with 20% IFS for 15 min at room temperature. Purified B cells were seeded at 0.75 × 106 to 1.5 × 106 cells/well and allowed to attach for 15 min at 37°C. During imaging, cells were maintained in RPMI 1640 supplemented with 2% IFS. Images were acquired using a PerkinElmer Ultraview spinning disk confocal microscope.
Quantification of TLR9-GFP localization
BMDMs generated from TLR9-GFP transgenic mice or immortalized BMDMs stably transduced with TLR9-GFP were seeded into 8-well-chambered coverglasses (Sarstedt). Cells were maintained in a climate chamber (EMBLEM, Heidelberg, Germany) at 37°C and 7.5% CO2 in phenol red–free DMEM supplemented with 10% IFS, 25 mM HEPES, and LysoTracker Red. Medium containing 1 μM CpG ODN 1826–Alexa 647 was used for stimulation experiments, and medium was exchanged after 1 h to avoid background fluorescence. Volocity software (Improvision) was used for image acquisition and quantification of TLR9-GFP localization to endosomes. To determine TLR9-GFP fluorescence values, all endosomes present in a given cell as well as the entire cell were encircled, and fluorescence intensities inside these regions were measured. Fluorescence counts of endosomes were divided by those of entire cells to obtain the percentage of TLR9-GFP fluorescence in endosomes.
Results
Cleavage of TLR9-GFP correlates with UNC93B1 expression levels
We had previously shown that retrovirally transduced TLR9-GFP trafficks together with UNC93B1 from the ER to the endolysosomal compartment upon stimulation with the TLR9 agonist CpG (4). Using the same TLR9-GFP DNA construct, we generated TLR9-GFP transgenic mice by FLPe-mediated site-specific integration into the collagen 1A (col1A) locus (15). Resulting mice express the TLR9-GFP transgene from the chicken β-actin (CAGGS) promoter (15). The colAI locus supports high levels of transgene expression even in cell types that do not normally express the type I collagen gene (19). Both heterozygous and homozygous TLR9-GFP mice were healthy and fertile. Immunostaining of paraffin-embedded tissue with an anti-GFP Ab showed expression of TLR9-GFP in spleen, liver, lymph node, lung, glandular stomach, small intestine, choroid plexus, and kidney (Supplemental Fig. 1). Histopathological evaluation of heterozygous TLR9-GFP mice revealed no alterations compared with WT littermates (data not shown). Aged mice did not develop high anti-nuclear Ab titers, and lymphocyte development was normal (Supplemental Fig. 2).
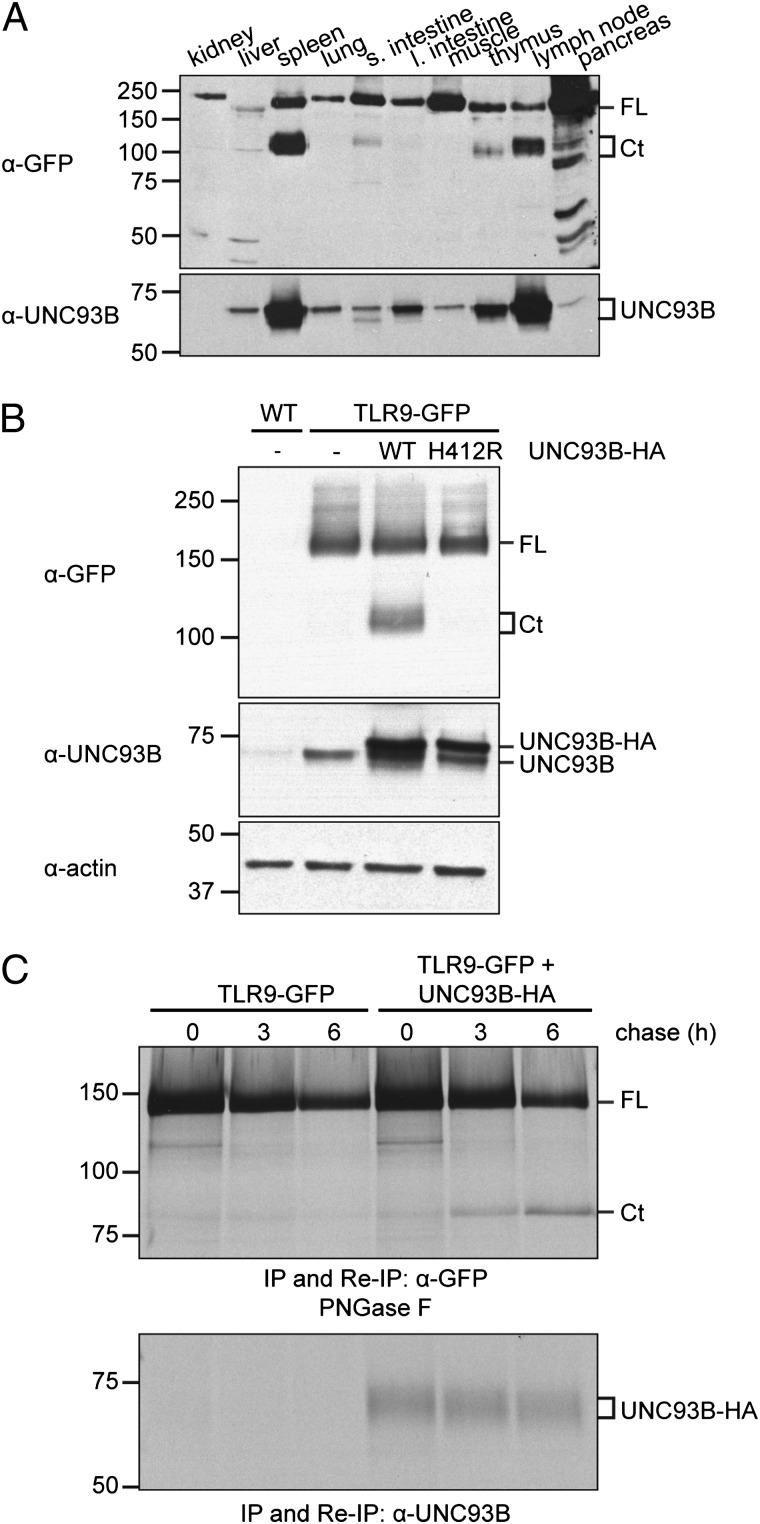
To ascertain the protein expression levels and molecular behavior of TLR9-GFP in different tissues, we analyzed homogenates of kidney, liver, spleen, lung, small and large intestine, muscle, thymus, lymph node, and pancreas by immunoblotting with an anti-GFP Ab. As expected, we observed TLR9-GFP expression in all samples, albeit at different levels (Fig. 1A, upper panel). Some tissues expressed TLR9 as a full-length protein (kidney, lung, large intestine, muscle), whereas others showed a band consistent with the C-terminal product of TLR9-GFP proteolysis (spleen and lymph node predominantly).
FIGURE 1.
Sufficient levels of UNC93B1 protein are required for TLR9-GFP cleavage. (A) Organs of TLR9-GFP/WT transgenic mice were lysed and equal protein concentrations analyzed by immunoblots with anti-GFP and anti-UNC93B1 Abs. (B) MEFs prepared from TLR9-GFP/WT mice were retrovirally transduced with either WT UNC93B1-HA (WT), the UNC93B1-HA H412R mutant (H412R), or not transduced (−). Non-transduced MEFs prepared from WT mice were used as negative control. Cells were lysed and equal protein concentrations analyzed by immunoblotting with anti-GFP (upper panel), anti-UNC93B1 (middle panel, detecting endogenous and HA-tagged UNC93B1), and anti–β-actin (lower panel) Abs. (C) MEFs prepared from TLR9-GFP/WT transgenic mice were retrovirally transduced with WT UNC93B1-HA or not transduced. Cells were metabolically labeled with [35S]methionine/cysteine for 1 h (pulse) and lysed after 0, 3, and 6 h of chase. Samples were split in half, and immunoprecipitation (IP) and reimmunoprecipitation (Re-IP) were carried out with either anti-GFP (upper panel) or anti-UNC93B1 (lower panel) Abs. The anti-GFP reimmunoprecipitations were digested with PNGase F. Shown are representative results from two independent experiments. Ct, C-terminal fragment of TLR9-GFP; FL, full-length TLR9-GFP.
Because UNC93B1 is required for trafficking to endolysosomes—the site of TLR9 cleavage—we examined UNC93B1 levels in lysates from the same tissue/organs by immunoblot (18) and found that UNC93B1 expression was highest in those samples where the C-terminal TLR9-GFP fragment was well detected (Fig. 1A, bottom panel). These results suggested that not only expression of UNC93B1 but also precise levels of this protein were necessary to promote TLR9 proteolysis. To test whether this was indeed the case, we used MEFs. MEFs express endogenous UNC93B1, but expression levels are apparently insufficient to promote TLR9 cleavage (Ref. 6 and Fig. 1B). To increase the levels of UNC93B1 protein, we retrovirally transduced MEFs generated from TLR9-GFP transgenic mice with WT UNC93B1-HA (4). In UNC93B1-HA transduced cells, cleavage of TLR9-GFP occurs (Fig. 1B). Ectopically expressed HA-tagged UNC93B1 was more abundant than endogenous UNC93B1 as measured by immunoblots using the anti-UNC93B1 antiserum (Fig. 1B). Cleavage of TLR9-GFP did not occur when we coexpressed the HA-tagged H412R mutant of UNC93B1, which does not bind TLR9 (18) (Fig. 1B). To follow the kinetics of TLR9-GFP conversion in the presence of appropriate levels of UNC93B1, we performed metabolic labeling of MEFs and chased for different time points to follow TLR9-GFP cleavage. Bands corresponding to full length and cleaved TLR9-GFP were retrieved after immunoprecipitation and reimmunoprecipitation with anti-GFP Abs, and digestion with PNGase F to remove all N-linked glycans and so improve electrophoretic resolution. Conversion of full-length TLR9-GFP to its cleaved product requires ∼3 h when adequate levels of UNC93B1 are provided (Fig. 1C). A threshold of UNC93B1 expression is thus required for TLR9-GFP cleavage to occur in MEFs, a result consistent with our findings in different tissues of TLR9-GFP mice.
TLR9-GFP transgene is functional
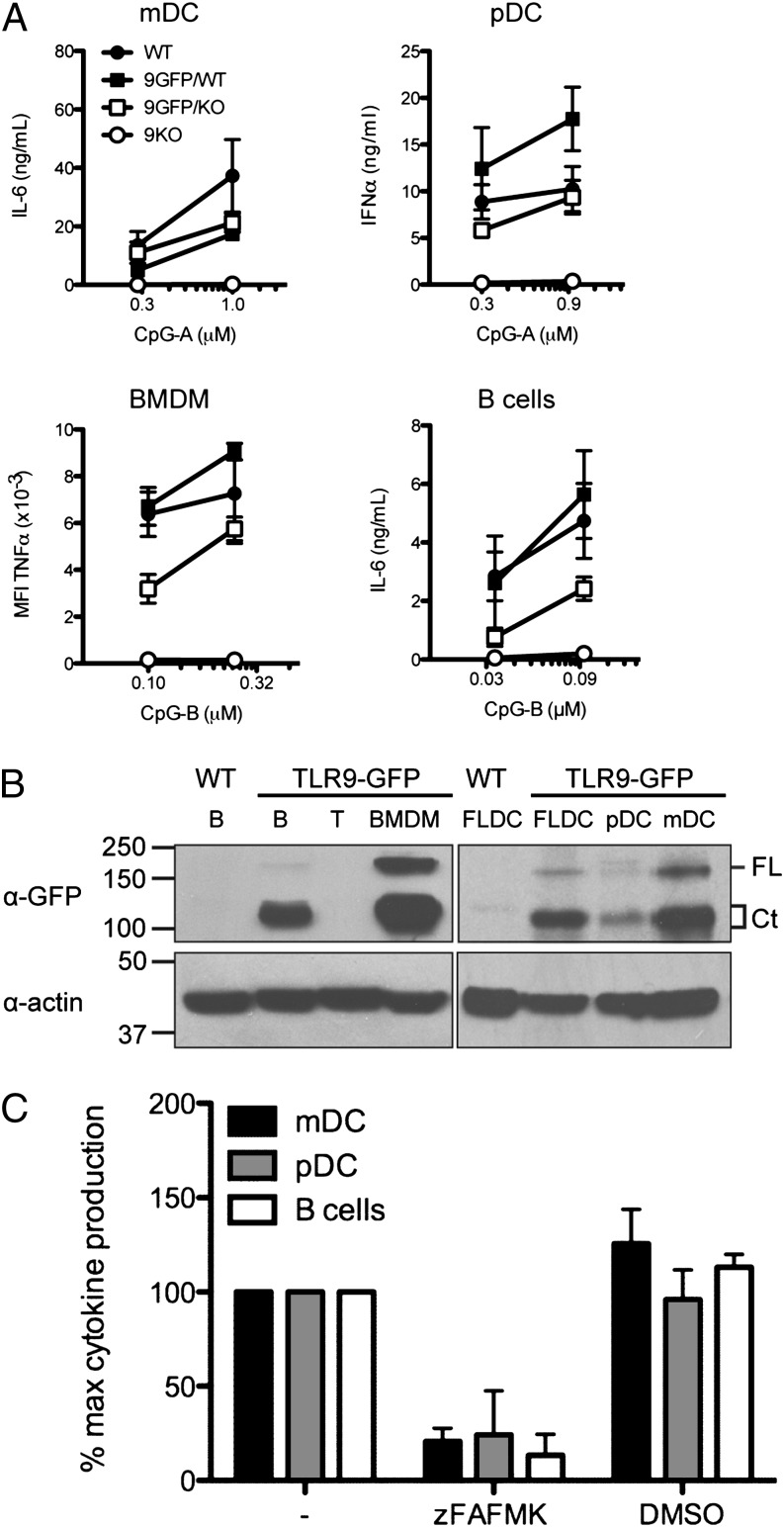
The transgenic TLR9-GFP mice express the TLR9-GFP transgene as well as endogenous copies of TLR9. We therefore first examined whether this had an effect on TLR9 responses to stimulation with its agonist CpG. We observed enhanced responses to CpG only in pDCs from TLR9-GFP/WT mice compared with WT pDCs. In BMDMs, mDCs, and B cells, cytokine production was comparable between TLR9-GFP/WT and WT mice (Fig. 2A). To determine whether the TLR9-GFP transgene was functional, we placed the TLR9-GFP transgene on a TLR9−/− background (TLR9-GFP/9KO). Although mDCs and pDCs from TLR9-GFP/9KO mice exhibited similar CpG responses compared with WT mice, responses to CpG were ∼50% lower in BMDMs and B cells from TLR9-GFP/9KO mice than in WT B cells (Fig. 2A). The reason for the incomplete restoration in BMDMs and B cells remains to be explored. Protein levels of full-length TLR9-GFP and the C-terminal fragment were comparable regardless of background (WT or 9KO) and zygosity (TLR9-GFP heterozygous or homozygous) (data not shown). Thus, additional copies of otherwise functional TLR9-GFP constructs do not affect endogenous TLR9 responses, except for pDCs, where the extra pool of TLR9-GFP led to increased IFN-α production.
FIGURE 2.
TLR9-GFP is functional, and cleavage of TLR9 is required for activation in primary APCs. (A) Primary mDCs (n = 3), pDCs (n = 2), BMDMs (n = 4), and B cells (n = 5) were isolated from TLR9KO, TLR9-GFP/WT, and TLR9-GFP/9KO mice. WT littermates of TLR9-GFP/WT transgenic mice served as control. BMDMs were stimulated with CpG-B, fixed cells were incubated with anti-TNF–A647, and intracellular TNF levels analyzed by FACS. Graph shows mean fluorescence intensity (MFI). IL-6 (mDCs and B cells) and IFN-α production (pDCs) in response to CpG-A and CpG-B stimulation was measured by ELISA. (B) Protein extracts from B cells, T cells, BMDMs, FLDCs, pDCs, and mDCs of TLR9-GFP/WT mice, or B cell and FLDC extracts from WT mice as control, bearing equal protein concentrations, were analyzed by immunoblot with an anti-GFP (top) or anti–β-actin (bottom) Ab. FL, full-length TLR9-GFP; Ct, C-terminal fragment of TLR9-GFP. (C) B cells (n = 3), mDCs (n = 4), and pDCs (n = 3) were preincubated with 3 μM z-FA-FMK in DMSO or equivalent volume of DMSO before addition of CpG. Secretion of IL-6 (B cells and mDCs) or IFN-α (pDCs) was measured by ELISA. Results shown are the mean + SEM of n (as specified) independent experiments (A, C) and a representative result of two independent experiments (B).
TLR9 cleavage is required for functional TLR9 signaling in primary APCs
To assess the expression level and cleavage state of TLR9-GFP in cells of the immune system, we analyzed lysates prepared from purified primary B cells, T cells, BMDMs, FLDCs (these include both pDCs and mDCs), and sorted pDC and mDC populations from total FLDCs by immunoblot using an anti-GFP Ab. Surprisingly, neither full-length nor cleaved TLR9-GFP was detected in T cells. All other cell types (pDCs, mDCs, B cells, BMDMs) expressed significantly higher levels of cleaved TLR9-GFP than full-length TLR9-GFP (Fig. 2B). Because relocalization to an endolysosomal compartment is required for cleavage, this result indicates TLR9 had trafficked to an endosomal compartment in unstimulated, primary immune cells.
Cleavage of TLR9 is crucial for its function in the macrophage cell line RAW264.7 (5). We explored whether this holds also true for primary APCs derived from C57BL/6 mice. In the presence of z-FA-FMK, a broadly specific thiol protease inhibitor that blocks TLR9 cleavage, the response to stimulation with CpG ODN, as measured by IL-6 production, was significantly reduced in B cells (Fig. 2C). We also examined CpG-evoked responses in total FLDCs by measuring IL-6 production and IFN-α secretion, indicative of stimulation of mDCs and pDCs, respectively (20). z-FA-FMK blocked FLDC responses (Fig. 2C). Hence, cleavage of TLR9-GFP is necessary not only for the activation of TLR9 in macrophages, but also is a more general requirement for TLR9 signaling in cells of the immune system, including primary mDCs, pDCs, and B cells.
Proteolytic conversion of TLR9-GFP in BMDMs
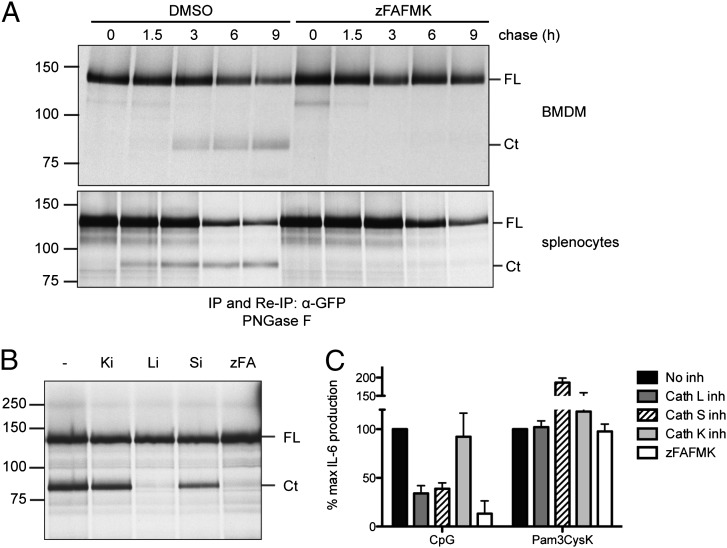
Because we found TLR9 was cleaved in unstimulated APCs, we explored the kinetics of TLR9-GFP cleavage in primary BMDMs and B cells by performing pulse-chase analysis on BMDMs and splenocytes from TLR9-GFP/WT mice. To inhibit cleavage of TLR9, z-FA-FMK was included. TLR9-GFP was recovered by immunoprecipitation and reimmunoprecipitation using anti-GFP, and the immunoprecipitates were digested with PNGase F to improve electrophoretic resolution. Material recovered using anti-GFP Ab in total splenocytes represented the TLR9-GFP content in B cells as T cells do not express the transgene. Cleavage of TLR9-GFP was detected after 3 h of chase in BMDMs, whereas a C-terminal TLR9 fragment was found as early as 1.5 h of chase in B cells (Fig. 3A). As expected, z-FA-FMK blocked cleavage (Fig. 3A). These results indicate that TLR9-GFP can reach endolysosomal compartments also in the absence of added CpG in B cells and BMDMs. The conversion of TLR9-GFP to the cleaved form occurred with similar kinetics in BMDMs derived from TLR9-GFP/WT and TLR9-GFP/9KO mice (data not shown).
FIGURE 3.
High rate of TLR9-GFP processing and cathepsin L requirement in B cells. (A) Primary BMDMs (upper panel) and primary splenocytes (lower panel) derived from TLR9-GFP/WT transgenic mice were metabolically labeled with [35S]methionine/cysteine for 1 h (pulse) and lysed after 0, 1.5, 3, 6, and 9 h of incubation in normal medium (chase). z-FA-FMK (10 μM for BMDMs, 3 μM for splenocytes) or DMSO (negative vehicle control) was added throughout the pulse and chase times. TLR9-GFP was recovered by immunoprecipitation (IP) with an anti-GFP Ab. Reimmunoprecipitation (Re-IP) was performed after denaturation of the initial immunoprecipitation with an anti-GFP Ab. Precipitates were subjected to digestion with PNGase F and resolved by SDS-PAGE. (B) Total splenocytes from TLR9-GFP/WT mice were metabolically labeled and chased for 4 h in the presence of DMSO (−), inhibitors of cathepsin K (Ki), cathepsin L (Li), or cathepsin S (Si), or z-FA-FMK (zFA). Cells were lysed and treated as in (A). (C) Purified B cells from C57BL/6 mice were incubated with 1 μM CpG or 1 μg/ml Pam3CysK in the absence (no inhibitor) or presence of cathepsin K, L, or S inhibitors or z-FA-FMK. IL-6 production was measured by ELISA. Shown are representative results from two (A) and three (B) independent experiments and the mean + SEM of three independent experiments (C). Ct, C-terminal fragment of TLR9-GFP; FL, full-length TLR9-GFP.
We then explored the cathepsin requirement for TLR9-GFP cleavage in B cells. In the RAW 264.7 macrophage cell line, TLR9 is cleaved by the combined activity of cathepsins L and S (5). Using selective inhibitors of cathepsin K, L, and S, we recovered TLR9-GFP by anti-GFP immunoprecipitation and reimmunoprecipitation from metabolically labeled splenocytes. We observed that inhibitors of cathepsin L and cathepsin S impaired TLR9-GFP cleavage (Fig. 3B), and both cathepsin L and cathepsin S inhibited responses to CpG (Fig. 3C). Cathepsin K inhibitors did not affect cleavage or activity (Fig, 3B, 3C). Treatment of splenocytes with cathepsin inhibitors did not produce a pre-C-terminal TLR9 (partial proteolysis) product as was observed in macrophages (5) but a fragment of size consistent with the functional C-terminal TLR9 fragment. The observed differences between B cells and macrophages in cathepsin requirement and cleavage products indicate dissimilar mechanisms responsible for TLR9 cleavage and activation in these cells.
Differences in TLR9-GFP maturation and stability in BMDMs and B cells
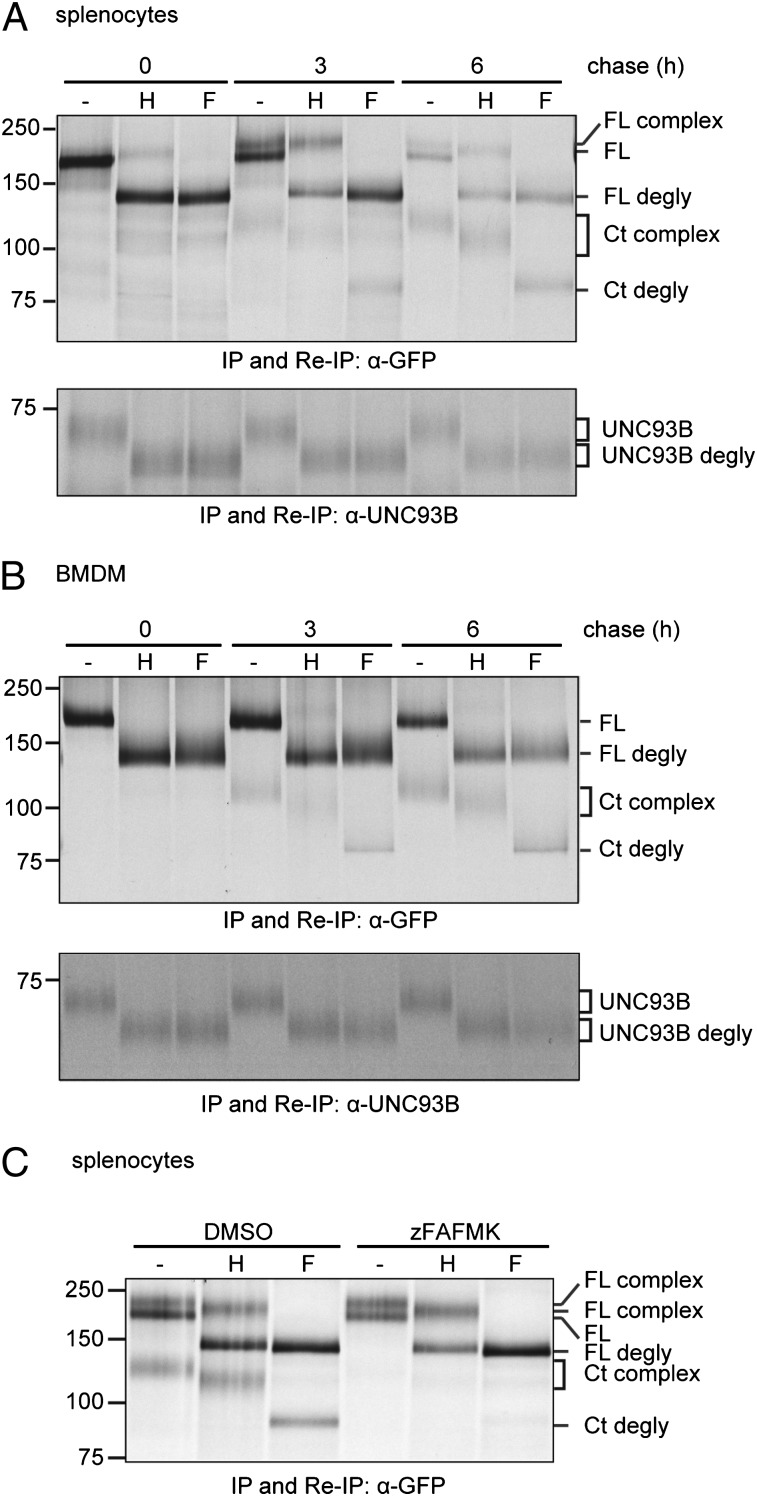
Because we observed differences in the rate of cleavage of TLR9-GFP in B cells and BMDMs, trafficking and maturation of TLR9-GFP and its accessory molecule UNC93B1 were examined. We metabolically labeled B cells and BMDMs, chased for different periods, and recovered TLR9-GFP and UNC93B1 from lysates by immunoprecipitation and reimmunoprecipitation using anti-GFP and anti-UNC93B1 Abs, respectively. Immunoprecipitates were digested with Endo H and PNGase F. Proteins that undergo complex N-linked glycan modification in the Golgi compartment are refractory to Endo H digestion, whereas ER-resident proteins are Endo H sensitive. Endo H–resistant material (“complex”) corresponding to full-length and cleaved TLR9-GFP was recovered from B cell lysates from TLR9-GFP/WT and TLR9-GFP/9KO mice, demonstrating TLR9-GFP trafficked through the Golgi compartment in these cells (Fig. 4A, “H” lanes). In BMDMs, TLR9-GFP also trafficks through the Golgi compartment, but full-length Endo H–resistant TLR9-GFP was barely detectable (Fig. 4B, upper panel). In contrast, B cells produce an Endo H–resistant full-length polypeptide (Fig. 4A). We were unable to detect Endo H–resistant forms of UNC93B1 in either BMDMs or B cells (Fig. 4A, 4B, lower panels). When cleavage was impeded by inclusion of z-FA-FMK, total levels of full-length TLR9-GFP were decreased in B cells (Fig. 4C). These results indicate that trafficking and cleavage lead to the presence of TLR9-GFP forms of different stability in BMDMs and B cells.
FIGURE 4.
Maturation of TLR9-GFP and UNC93B1 in primary APCs. Primary splenocytes (A) or BMDMs (B) derived from TLR9-GFP/WT transgenic mice were metabolically labeled with [35S]methionine/cysteine for 1 h and lysed after 0, 3, or 6 h of chase. TLR9-GFP was recovered by immunoprecipitation and reimmunoprecipitation with an anti-GFP Ab [(A, B) upper panels]. Endogenous UNC93B1 was recovered by immunoprecipitation and reimmunoprecipitation with an anti-UNC93B1 Ab [(A, B) lower panels]. Anti-GFP and anti-UNC93B1 immunoprecipitates were subjected to digestion with Endo H, PNGase F, or not digested (−) and resolved by SDS-PAGE. (C) Primary splenocytes derived from TLR9-GFP/WT transgenic mice were metabolically labeled with [35S]methionine/cysteine for 1 h and chased for 6 h in the presence of z-FA-FMK or DMSO. Cells were lysed and subjected to immunoprecipitation and reimmunoprecipitation with an anti-GFP Ab. Precipitated proteins were subjected to digestion with Endo H, PNGase F, or not digested (−) and resolved by SDS-PAGE. Shown are representative results of three independent experiments. Ct, C-terminal fragment of TLR9-GFP; degly, deglycosylated; F, PNGase F; FL, full-length TLR9-GFP; H, Endo H; IP, immunoprecipitation; Re-IP, reimmunoprecipitation.
Steady flow of TLR9-GFP from the ER to endolysosomes
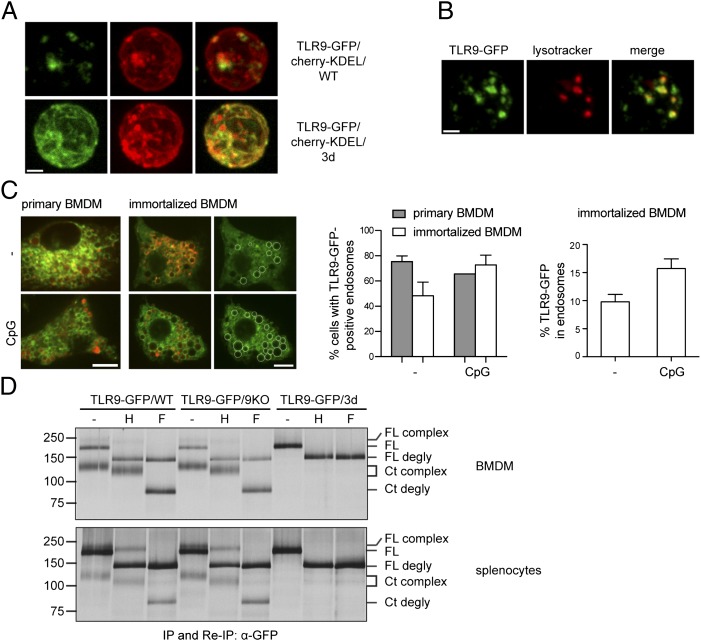
The observed cleavage of TLR9 in resting/unstimulated primary APCs is notable, as this result indicates that a major pool of TLR9 already localizes to the endosomal compartment where proteolysis occurs. Earlier reports rather suggested the major pool of TLR9 to localize to the ER (4, 13, 21). In B cells at rest, TLR9 appears to reside in early endosomal compartments (14). To analyze the distribution of TLR9-GFP in resting/unstimulated B cells relative to the ER, we crossed TLR9-GFP mice with mice transgenic for mCherry equipped with a C-terminal KDEL sequence (mCherry-KDEL). This reporter protein is retrieved from the cis-Golgi and redistributed to the ER via the KDEL receptor (22). We found a small portion of TLR9-GFP colocalizing with cherry KDEL in resting B cells (Fig. 5A, upper panel), whereas most TLR9-GFP was found in LysoTracker-positive compartments (Fig. 5B). To determine the distribution of TLR9-GFP in resting and stimulated BMDMs, we quantified TLR9-GFP–positive endolysosomes by confocal microscopy in unstimulated and CpG-stimulated BMDMs using LysoTracker as an endolysosome marker (Fig. 5C). Such quantifications were not possible in earlier studies because retroviral transduction levels were too low. Because all cells express the TLR9-GFP transgene, we were able to quantify the distribution of TLR9-GFP in primary BMDMs and compared it with immortalized BMDMs stably transduced with TLR9-GFP. In the absence of CpG, TLR9-GFP localized to endolysosomes in ∼75% of primary and ∼50% of immortalized cells. After 1 h of CpG stimulation, ∼65% of primary and ∼70% of immortalized cells showed TLR9-GFP colocalization with LysoTracker in endolysosomes (Fig 5C, middle panel). We next compared the distribution of TLR9-GFP in endosomal compartments and the ER in unstimulated and CpG-stimulated immortalized cells. In unstimulated immortalized WT BMDMs, ∼10% of TLR9-GFP localized to endosomes and ∼90% to the ER (Fig. 5C, right panel). After 1 h of CpG stimulation, ∼15% of TLR9-GFP localized to endosomes (Fig. 5C, right panel). These data show that at least in vitro, a significant proportion of TLR9-GFP already localizes to the endolysosomal compartment in unstimulated APCs, and a smaller percentage relocates to endolysosomes upon CpG stimulation.
FIGURE 5.
Functional UNC93B1 is required for TLR9-GFP cleavage, trafficking, and maturation in BMDMs and B cells. (A) Purified resting B cells of TLR9-GFP/cherry-KDEL/WT or TLR9-GFP/cherry-KDEL/3d mice were isolated and imaged by confocal microscopy. (B) Purified resting B cells of TLR9-GFP/WT mice were incubated with LysoTracker and imaged by confocal microscopy. (C) BMDMs derived from TLR9-GFP/WT transgenic mice or immortalized BMDMs stably transduced with TLR9-GFP were incubated with LysoTracker and either left unstimulated (−) or stimulated for 1 h with 1 μM of CpG and analyzed by confocal microscopy. Left panel, Representative image of the quantifications in the middle and right panels. TLR9-GFP–positive endolysosomes are marked with white circles. Middle panel, Shown is the percentage of primary and immortalized cells expressing TLR9-GFP in the endolysosome with or without CpG stimulation, with cells considered positive expressing TLR9-GFP in at least one endosome. Right panel, Shown is the percentage of TLR9-GFP fluorescence in endolysosomes versus the entire cell with or without CpG stimulation in immortalized BMDMs. Quantification of TLR9-GFP expression in endolysosomes was accomplished by measuring the fluorescence intensities of all TLR9-GFP–positive endosomes (encircled regions in left panel) and dividing the values by the fluorescence intensity of the entire cell. (D) BMDMs (upper panel) and total splenocytes (lower panel) from TLR9-GFP/WT, TLR9-GFP/9KO, or TLR9-GFP/3d mice were metabolically labeled with [35S]methionine/cysteine for 1 h and chased for six (BMDMs) or three (splenocytes) hours. TLR9-GFP was immunoprecipitated and reimmunoprecipitated with an anti-GFP Ab. Immunoprecipitates were left untreated (−) or digested with Endo H or PNGase F and separated by SDS-PAGE. IP, Immunoprecipitation; Re-IP, reimmunoprecipitation; H, Endo H; F, PNGase F; FL, full-length TLR9-GFP; Ct, C-terminal fragment of TLR9-GFP; degly, deglycosylated. Scale bar, 1.5 μm (A, B) and 10 μm (C). For (C), a minimum of 68 cells was analyzed for each condition (unstimulated and CpG stimulated). Shown are representative results from two (A) and three (B, D) independent experiments, and a representative image and mean + SEM of two (primary BMDMs) and three (immortalized BMDMs) independent experiments (C).
Trafficking of TLR9 depends on interaction with the ER protein UNC93B1, as a H412R point mutation in UNC93B1 in 3d mutant mice (17) leads to retention of TLR9-GFP in the ER (4). To investigate possible differences in the UNC93B1 requirement for TLR9-GFP trafficking in primary APCs from 3d mice, we crossed TLR9-GFP to 3d mice to obtain TLR9-GFP/3d mice and analyzed TLR9-GFP cleavage and maturation by pulse-chase analysis. The C-terminal cleaved fragment of TLR9-GFP was not detectable in BMDMs or B cells of TLR9-GFP/3d mice compared with TLR9-GFP/WT and TLR9-GFP/9KO cells (Fig. 5D). In B cells from TLR9-GFP/cherry-KDEL mice in the 3d context, TLR9-GFP colocalizes with cherry-KDEL (Fig. 5A, lower panel). Together, these results establish retention of TLR9-GFP in the ER of BMDMs and B cells of TLR9-GFP/3d mice. Importantly, these data demonstrate that cleavage of TLR9-GFP in TLR9-GFP transgenic mice is not due to overexpression of the transgene that could lead to aberrant trafficking.
Discussion
Transgenic mice created by site-specific integration of a TLR9-GFP fusion construct into the collagen locus allow the analysis of TLR9 receptor trafficking and processing in primary cell types previously not readily accessible to this type of analysis. The TLR9-GFP transgene is expressed at levels sufficient for detection by immunoprecipitation and immunoblotting with an anti-GFP Ab, as well as by live cell imaging. We demonstrate a steady flow of TLR9-GFP from the ER to the endolysosomes in BMDMs and B cells. This flow occurred more rapidly in B cells than in BMDMs, with the C-terminal TLR9 fragment appearing as early as 1.5 h of chase in B cells and only after 3 h in BMDMs. On the time frame usually observed for glycoprotein maturation, these differences are substantial. Moreover, cathepsin requirement varied between macrophages and B cells. It is likely that other endosomal TLRs traffic in a similar fashion, as cleavage has been demonstrated for TLR7 and TLR3 (7–9), with TLR3 cleavage dependent on activity of cathepsins H and B (7–9). Whether cell-specific differences exist for TLR3 and TLR7 cleavage remains to be established.
Only a small proportion of TLR9-GFP trafficks to the endolysosome upon CpG stimulation in BMDMs, and this result contrasts with findings previously reported, where the majority of TLR9 resided in the ER at rest and only traveled to endolysosome upon CpG stimulation (4, 13, 21). In B cells, no increase of cleavage upon stimulation with anti-IgM or CpG was found, even though TLR9 was reported to traffic to autophagosome-like compartments in anti-IgM–stimulated cells (14). We do not think TLR9-GFP trafficking properties derive from unphysiologically high levels of transgene expression, as UNC93B1 was required to execute cleavage (Fig. 5D). Our results show that a sizeable pool of cleaved TLR9 is localized to endolysosomes in resting primary APCs. Because CpG reaches endosomes rapidly (11), a ready source of cleaved TLR9-GFP would ensure rapid responses to ligand, with subsequent further recruitment from the ER. We also found differences in protein stability of full-length and cleaved TLR9-GFP in BMDMs and B cells. The lower stability of the full-length Endo H–resistant form of TLR9-GFP in BMDMs may explain why early reports suggested that TLR9 trafficking might bypass the Golgi compartment, an admittedly unlikely route for membrane proteins to reach the endocytic pathway (4, 13, 18, 21). Our inability to detect Endo H–resistant UNC93B1 indicates that the mature form may be rapidly degraded, as it seems unlikely, given the tight interaction between them, that UNC93B1 would separate from TLR9. As TLR pathways are examined toward the design of prophylactics and therapeutics, our results highlight the importance of studying molecular trafficking in cell-specific manner as TLRs may behave differently depending on the target cell. It is thus important to be cautious when generalizing TLR trafficking patterns based on effects observed in cells amenable to transduction. How trafficking occurs in response to more physiological TLR9 ligands, which require phagocytosis and processing in the endolysosome, remains to be established. These studies should be approachable with the TLR9-GFP mouse in hand.
Supplementary Material
Acknowledgments
We thank Brigitte Denker and Anna Rinkel (Helmholtz Centre for Infection Research) for excellent technical assistance and the Flow Cytometry and Keck Microscopy Facility at the Whitehead Institute for Biomedical Research.
This work was supported by National Institutes of Health Grant R01 GM100518 (to H.L.P.), a Merck–MIT postdoctoral fellowship (to A.M.A.), the Charles A. King Trust, Bank of America, Co-Trustee (to M.M.B.), the Initiative and Networking Fund of the Helmholtz Association (to M.M.B. and J.M.O.), and the Deutsche Forschungsgemeinschaft (SFB900 to M.M.B.).
The online version of this article contains supplemental material.
- BMDM
- bone marrow–derived macrophage
- Endo H
- endoglycosidase H
- ER
- endoplasmic reticulum
- FLDC
- FLT3-ligand dendritic cell
- HA
- hemagglutinin
- IFS
- inactivated fetal calf serum
- mDC
- myeloid dendritic cell
- MEF
- mouse embryonic fibroblast
- NP-40
- Nonidet P-40
- ODN
- oligonucleotide
- pDC
- plasmacytoid dendritic cell
- PI
- protease inhibitor
- PNGase F
- peptide-N-glycosidase F
- P/S
- penicillin/streptomycin
- WT
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Kawai T., Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11: 373–384 [DOI] [PubMed] [Google Scholar]
- 2.Blasius A. L., Beutler B. 2010. Intracellular toll-like receptors. Immunity 32: 305–315 [DOI] [PubMed] [Google Scholar]
- 3.Lee C. C., Avalos A. M., Ploegh H. L. 2012. Accessory molecules for Toll-like receptors and their function. Nat. Rev. Immunol. 12: 168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim Y.-M., Brinkmann M. M., Paquet M.-E., Ploegh H. L. 2008. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 452: 234–238 [DOI] [PubMed] [Google Scholar]
- 5.Park B., Brinkmann M. M., Spooner E., Lee C. C., Kim Y.-M., Ploegh H. L. 2008. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat. Immunol. 9: 1407–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ewald S. E., Lee B. L., Lau L., Wickliffe K. E., Shi G.-P., Chapman H. A., Barton G. M. 2008. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 456: 658–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewald S. E., Engel A., Lee J., Wang M., Bogyo M., Barton G. M. 2011. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J. Exp. Med. 208: 643–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia-Cattaneo A., Gobert F. X., Müller M., Toscano F., Flores M., Lescure A., Del Nery E., Benaroch P. 2012. Cleavage of Toll-like receptor 3 by cathepsins B and H is essential for signaling. Proc. Natl. Acad. Sci. USA 109: 9053–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qi R., Singh D., Kao C. C. 2012. Proteolytic processing regulates Toll-like receptor 3 stability and endosomal localization. J. Biol. Chem. 287: 32617–32629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton G. M., Kagan J. C. 2009. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat. Rev. Immunol. 9: 535–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., Taniguchi T. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 434: 772–777 [DOI] [PubMed] [Google Scholar]
- 12.Avalos A. M., Latz E., Mousseau B., Christensen S. R., Shlomchik M. J., Lund F., Marshak-Rothstein A. 2009. Differential cytokine production and bystander activation of autoreactive B cells in response to CpG-A and CpG-B oligonucleotides. J. Immunol. 183: 6262–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latz E., Schoenemeyer A., Visintin A., Fitzgerald K. A., Monks B. G., Knetter C. F., Lien E., Nilsen N. J., Espevik T., Golenbock D. T. 2004. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5: 190–198 [DOI] [PubMed] [Google Scholar]
- 14.Chaturvedi A., Dorward D., Pierce S. K. 2008. The B cell receptor governs the subcellular location of Toll-like receptor 9 leading to hyperresponses to DNA-containing antigens. Immunity 28: 799–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beard C., Hochedlinger K., Plath K., Wutz A., Jaenisch R. 2006. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44: 23–28 [DOI] [PubMed] [Google Scholar]
- 16.Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., Akira S. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408: 740–745 [DOI] [PubMed] [Google Scholar]
- 17.Tabeta K., Hoebe K., Janssen E. M., Du X., Georgel P., Crozat K., Mudd S., Mann N., Sovath S., Goode J., et al. 2006. The Unc93b1 mutation 3d disrupts exogenous antigen presentation and signaling via Toll-like receptors 3, 7 and 9. Nat. Immunol. 7: 156–164 [DOI] [PubMed] [Google Scholar]
- 18.Brinkmann M. M., Spooner E., Hoebe K., Beutler B., Ploegh H. L., Kim Y. M. 2007. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J. Cell Biol. 177: 265–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCreath K. J., Howcroft J., Campbell K. H. S., Colman A., Schnieke A. E., Kind A. J. 2000. Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature 405: 1066–1069 [DOI] [PubMed] [Google Scholar]
- 20.Yasuda K., Richez C., Uccellini M. B., Richards R. J., Bonegio R. G., Akira S., Monestier M., Corley R. B., Viglianti G. A., Marshak-Rothstein A., Rifkin I. R. 2009. Requirement for DNA CpG content in TLR9-dependent dendritic cell activation induced by DNA-containing immune complexes. J. Immunol. 183: 3109–3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leifer C. A., Kennedy M. N., Mazzoni A., Lee C., Kruhlak M. J., Segal D. M. 2004. TLR9 is localized in the endoplasmic reticulum prior to stimulation. J. Immunol. 173: 1179–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munro S., Pelham H. R. 1987. A C-terminal signal prevents secretion of luminal ER proteins. Cell 48: 899–907 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.