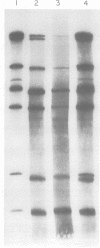
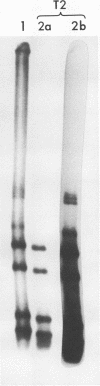
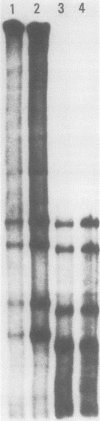
Abstract
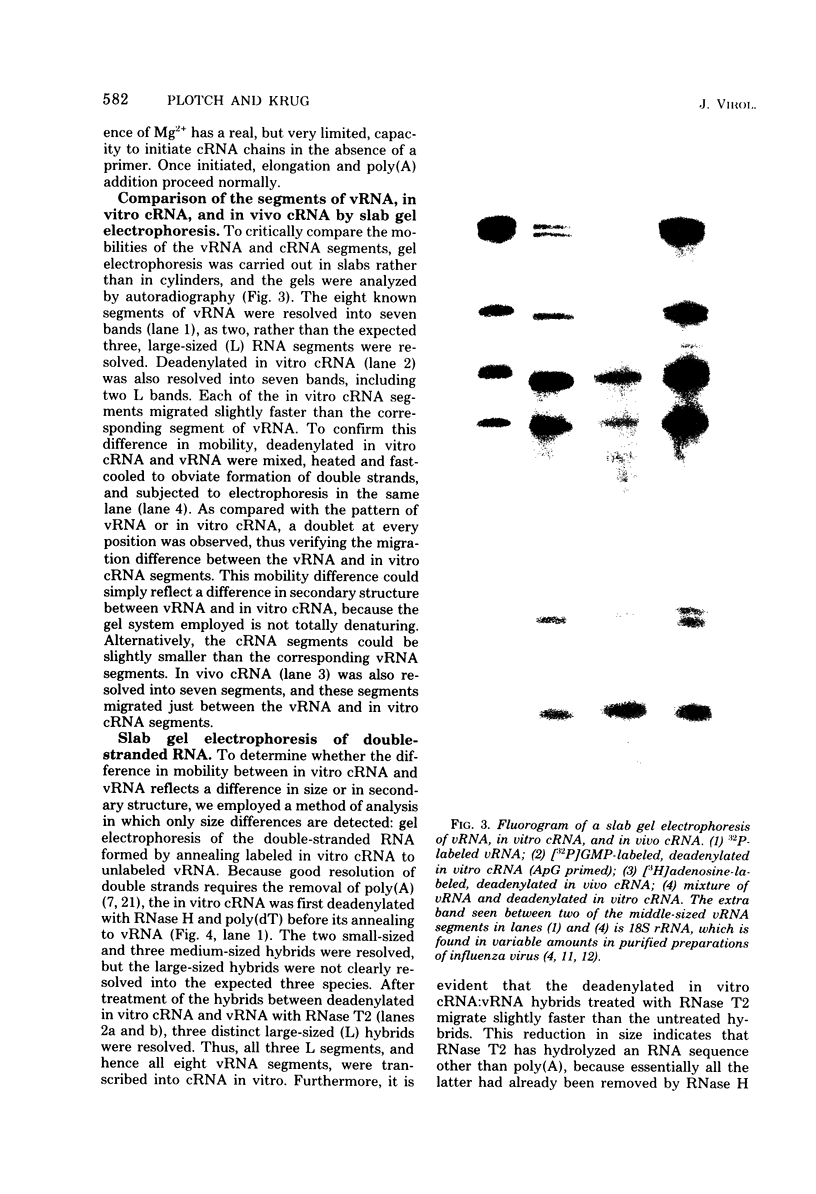
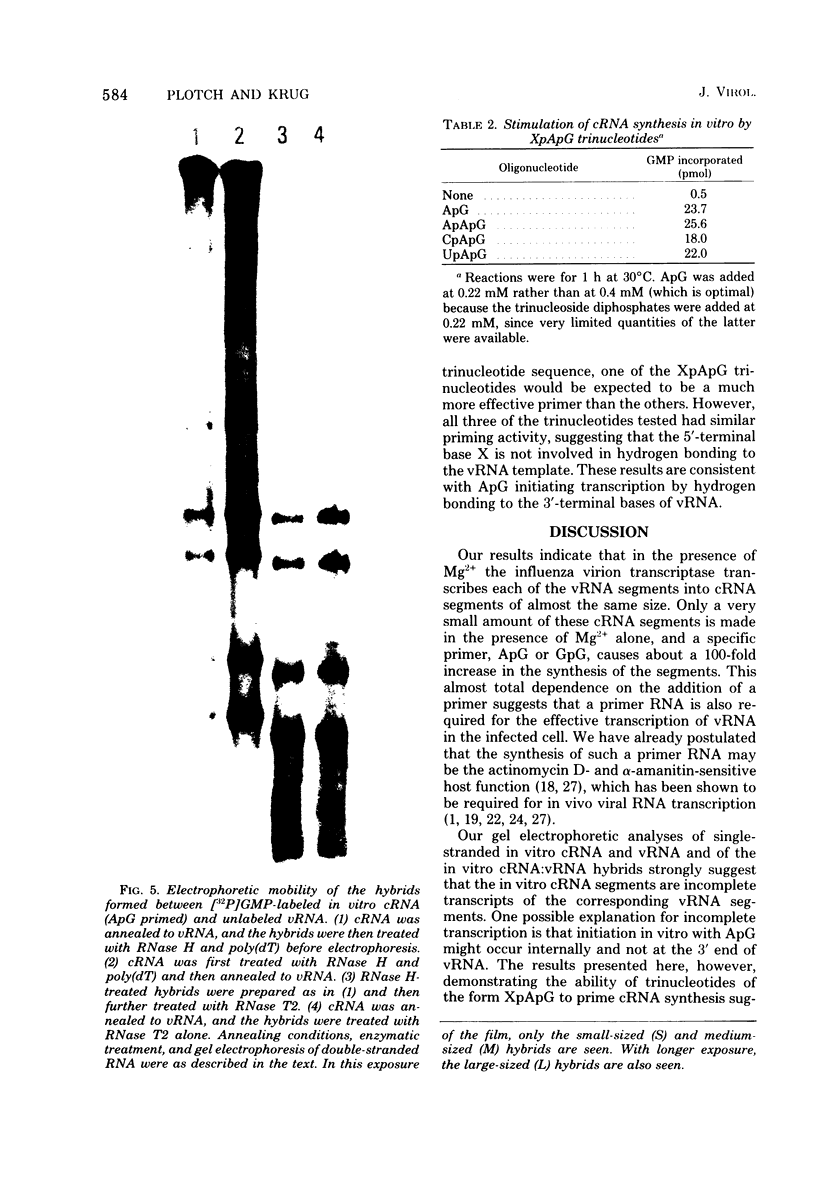
In the presence of Mg2+ and a specific primer, ApG or GpG, the influenza WSN virion transcriptase synthesizes large, polyadenylic acid-containing complementary RNA (cRNA) (Plotch and Krug, J. Virol., 21:24-34, 1977). After removal of its polyadenylic acid with RNase H in the presence of polydeoxythymidylic acid, the in vitro cRNA distributed into seven discrete bands during electrophoresis in acrylamide gels containing 6 M urea. The eight known segments of virion RNA (vRNA) also distributed into seven bands under these conditions as two, rather than the expected three, large-sized segments were resolved. Each of the in vitro cRNA segments migrated slightly faster than the corresponding vRNA segment. To determine whether this difference in mobility reflects a difference in size between cRNA and vRNA, the double-stranded RNA formed by annealing labeled in vitro cRNA to unlabeled vRNA was subjected to various nuclease treatments and was analyzed by gel electrophoresis. Hybrids treated with RNase T2 or a combination of RNase T2 and RNase H migrated slightly faster than those treated only with RNase H, indicating that RNase T2 removed an RNA sequence other than polyadenylic acid, most probably a short sequence of vRNA not hydrogen bonded to cRNA. These results suggest that the in vitro cRNA segments are shorter than, and thus incomplete transcripts of the corresponding vRNA segments. All eight hybrids were resolved by gel electrophoresis, indicating that all eight vRNA segments are transcribed into cRNA in vitro. We also present evidence suggesting that the ApG primer initiates in vitro transcription exactly at the 3′ end of vRNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bean W. J., Jr, Simpson R. W. Primary transcription of the influenza virus genome in permissive cells. Virology. 1973 Dec;56(2):646–651. doi: 10.1016/0042-6822(73)90067-6. [DOI] [PubMed] [Google Scholar]
- Bean W. J., Jr, Simpson R. W. Transcriptase activity and genome composition of defective influenza virus. J Virol. 1976 Apr;18(1):365–369. doi: 10.1128/jvi.18.1.365-369.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner W. M., Laskey R. A. A film detection method for tritium-labelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974 Jul 1;46(1):83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Cox N. J., Barry R. D. Hybridization studies of the relationship between influenza virus RNA and cellular DNA. Virology. 1976 Jan;69(1):304–313. doi: 10.1016/0042-6822(76)90217-8. [DOI] [PubMed] [Google Scholar]
- Etkind P. R., Buchhagen D. L., Herz C., Broni B. B., Krug R. M. The segments of influenza viral mRNA. J Virol. 1977 May;22(2):346–352. doi: 10.1128/jvi.22.2.346-352.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etkind P. R., Krug R. M. Purification of influenza viral complementary RNA: its genetic content and activity in wheat germ cell-free extracts. J Virol. 1975 Dec;16(6):1464–1475. doi: 10.1128/jvi.16.6.1464-1475.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman G. J., Rose J. K., Clinton G. M., Huang A. S. RNA synthesis of vesicular stomatitis virus. VII. Complete separation of the mRNA's of vesicular stomatitis virus by duplex formation. J Virol. 1977 Mar;21(3):1094–1104. doi: 10.1128/jvi.21.3.1094-1104.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug R. M. Cytoplasmic and nucleoplasmic viral RNPs in influenza virus-infected MDCK cells. Virology. 1972 Oct;50(1):103–113. doi: 10.1016/0042-6822(72)90350-9. [DOI] [PubMed] [Google Scholar]
- Krug R. M., Etkind P. R. Cytoplasmic and nuclear virus-specific proteins in influenza virus-infected MDCK cells. Virology. 1973 Nov;56(1):334–348. doi: 10.1016/0042-6822(73)90310-3. [DOI] [PubMed] [Google Scholar]
- Krug R. M. Influenza viral RNPs newly synthesized during the latent period of viral growth in MDCK cells. Virology. 1971 Apr;44(1):125–136. doi: 10.1016/0042-6822(71)90159-0. [DOI] [PubMed] [Google Scholar]
- Krug R. M., Morgan M. A., Shatkin A. J. Influenza viral mRNA contains internal N6-methyladenosine and 5'-terminal 7-methylguanosine in cap structures. J Virol. 1976 Oct;20(1):45–53. doi: 10.1128/jvi.20.1.45-53.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewandowski L. J., Content J., Leppla S. H. Characterization of the subunit structure of the ribonucleic acid genome of influenza virus. J Virol. 1971 Nov;8(5):701–707. doi: 10.1128/jvi.8.5.701-707.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. B., Brass L. F., Doty P. A comparison of the binding to polynucleotides of complementary and noncomplementary oligonucleotides. Biochemistry. 1975 Jul 15;14(14):3164–3171. doi: 10.1021/bi00685a020. [DOI] [PubMed] [Google Scholar]
- McGeoch D., Fellner P., Newton C. Influenza virus genome consists of eight distinct RNA species. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3045–3049. doi: 10.1073/pnas.73.9.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palese P., Ritchey M. B., Schulman J. L. P1 and P3 proteins of influenza virus are required for complementary RNA synthesis. J Virol. 1977 Mar;21(3):1187–1195. doi: 10.1128/jvi.21.3.1187-1195.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotch S. J., Krug R. M. Influenza virion transcriptase: synthesis in vitro of large, polyadenylic acid-containing complementary RNA. J Virol. 1977 Jan;21(1):24–34. doi: 10.1128/jvi.21.1.24-34.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pons M. W. A reexamination of influenza single-and double-stranded RNAs by gel electrophoresis. Virology. 1976 Feb;69(2):789–792. doi: 10.1016/0042-6822(76)90508-0. [DOI] [PubMed] [Google Scholar]
- Pons M. W. The inhibition of influenza virus RNA synthesis by actinomycin D and cycloheximide. Virology. 1973 Jan;51(1):120–128. doi: 10.1016/0042-6822(73)90372-3. [DOI] [PubMed] [Google Scholar]
- Rhodes D. P., Abraham G., Colonno R. J., Jelinek W., Banerjee A. K. Characterization of vesicular stomatitis virus mRNA species synthesized in vitro. J Virol. 1977 Mar;21(3):1105–1112. doi: 10.1128/jvi.21.3.1105-1112.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rott R., Scholtissek C. Specific inhibition of influenza replication by alpha-amanitin. Nature. 1970 Oct 3;228(5266):56–56. doi: 10.1038/228056a0. [DOI] [PubMed] [Google Scholar]
- Scholtissek C., Harms E., Rohde W., Orlich M., Rott R. Correlation between RNA fragments of fowl plague virus and their corresponding gene functions. Virology. 1976 Oct 15;74(2):332–344. doi: 10.1016/0042-6822(76)90340-8. [DOI] [PubMed] [Google Scholar]
- Scholtissek C., Rott R. Synthesis in vivo of influenza virus plus and minus strand RNA and its preferential inhibition by antibiotics. Virology. 1970 Apr;40(4):989–996. doi: 10.1016/0042-6822(70)90145-5. [DOI] [PubMed] [Google Scholar]
- Stavrianopoulos J. G., Chargaff E. Purification and properties of ribonuclease H of calf thymus. Proc Natl Acad Sci U S A. 1973 Jul;70(7):1959–1963. doi: 10.1073/pnas.70.7.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrianopoulos J. G., Gambino-Giuffrida A., Chargaff E. Ribonuclease H of calf thymus: substrate specificity, activation, inhibition. Proc Natl Acad Sci U S A. 1976 Apr;73(4):1087–1091. doi: 10.1073/pnas.73.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J. M., Illmensee R., Litwin S., Herring L., Broni B., Krug R. M. Use of specific radioactive probes to study transcription and replication of the influenza virus genome. J Virol. 1977 Feb;21(2):530–540. doi: 10.1128/jvi.21.2.530-540.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]