Abstract
Transcription factors are proteins that regulate gene expression by modulating the synthesis of messenger RNA. Since this process is frequently one dominant control point in the production of many proteins, transcription factors represent the key regulators of numerous cellular functions, including proliferation, differentiation, and apoptosis. Pancreatic cancer progression is characterized by the activation of inflammatory signaling pathways converging on a limited set of transcription factors that fine-tune gene expression patterns contributing to the growth and maintenance of these tumors. Thus, strategies targeting these transcriptional networks activated in pancreatic cancer cells could block the effects of upstream inflammatory responses participating in pancreatic tumorigenesis. In this article we review this field of research and summarize current strategies to target oncogenic transcription factors and their activating signaling networks in the treatment of pancreatic cancer.
Keywords: transcription factors, pancreatic cancer, inflammation, therapy
Oncogenic transcription factors are final effectors of inflammatory signalling pathways during pancreatic carcinogenesis
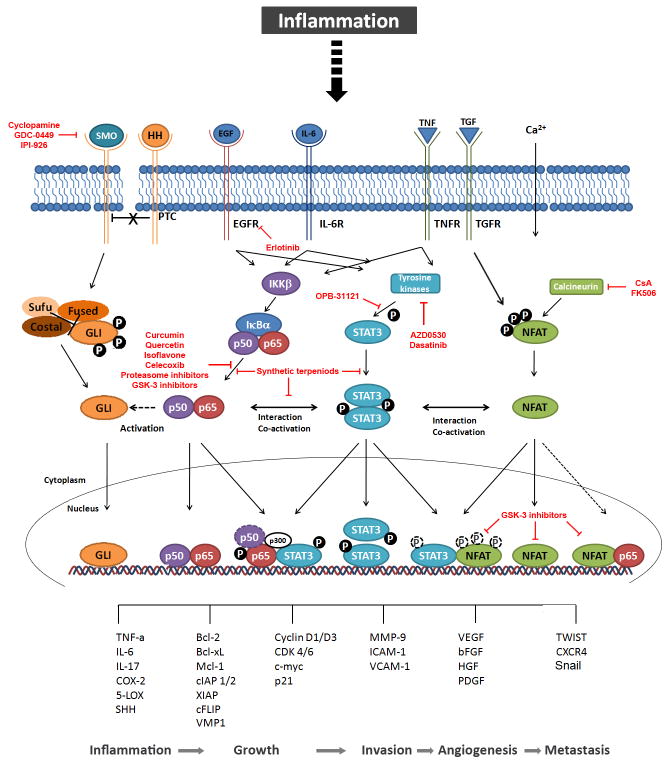
Pancreatic cancer, one of the most devastating malignancies, is triggered by genetic and epigenetic alterations leading to the aberrant activation of key oncogenes and inactivation of tumour suppressor pathways, and consequently confer transforming cells with a growth and survival advantage over normal cells. The most common genetic abnormality in pancreatic cancer is the activating mutation of KRAS, which is an initial key event in pancreatic carcinogenesis and found in virtually all invasively growing tumors1. However, mutation of KRAS alone is not sufficient for full neoplastic progression, and instead additional abnormalities (e.g. deletions of tumor suppressor genes) and signals that originate from the tumor microenvironment are required for tumor promotion and progression2,3. The tumor-associated microenvironment is comprised of stromal cells (mainly fibroblast and stellate cells), endothelial cells as well as cells from the innate (e.g. macrophages, neutrophils, dendritic cells and natural killer (NK) cells) and adaptive immune system (T and B lymphocytes), which can act in an autocrine and/or paracrine manner to trigger tumor progression. It is now apparent from epidemiological, pharmacological, and genetic studies that chronic inflammation can stimulate pancreatic cancer progression through extensive crosstalk interactions between malignant epithelial cells and the surrounding microenvironment4–6. Accordingly, recent pathological studies suggest that the establishment of pancreatic precursor lesions (PanIN) and the progression to frank adenocarcinoma occurs in most if not all cases in the presence of chronic inflammation and is associated with secretion of large amounts of proinflammatory cytokines, growth factors and proteases such as TNF-α, IL-6, Hedgehog, and TGF-β. These cytokines activate an oncogenic network of transcription factors in pancreatic cancer cells leading to tumor growth, survival and invasion4–6 through the regulation of an specific set of target genes (Figure 1). Below we will discuss some of the factors regulated by inflammatory cascades, specially the ones activated in tumoral cells.
Figure 1. Putative crosstalk between oncogenic transcription factors contributing to pancreatic carcinogenesis.
Inflammation induces binding of cytokines and inflammatory ligands to their receptors, which in turn activate the corresponding transcription factor signaling pathways. Binding of HH to the transmembrane protein PTC reverses the inhibitory effect on SMO, which again releases GLI transcription factors from their inhibitory complex and finally initiates nuclear translocation. The activation of the pathway can be blocked by inhibitors of SMO such as cyclopamine and its derivatives currently in clinical studies, GDC-0449 and IPI-926. Interactions with the NF-κB signaling pathway are illustrated by the dashed arrowhead which indicates activation of the HH pathway mainly through the regulation of the expression of its ligand SHH. Binding of EGF or IL-6 activates IKKβ kinase which stimulates degradation of IκBα. Subsequently, NF-κB proteins are released to the nucleus, where they mediate gene transcription alone or in cooperation with STAT proteins. Curcumin, Quercetin, Isoflavone and proteasome or GSK-3 inhibitors interfere with NF-κB signaling mainly through inhibition of NF-κB activation. Thereby, p-STAT3 binding to p-p65 induces histone acetyl transcerase p300 and retains active p65 in the nucleus. Synthetic terpenoids can either block isolated NF-κB and STAT3 activation, or inhibit the interaction of both pathways. A putative binding of p50 to STAT3 is shown by the dashed circle. The STAT3 signaling pathway can as well be activated by the aforementioned inflammatory stimuli. Upon binding of the ligands to the transmembrane receptors (receptor-bound) kinases, such as JAKs, SRC or ABL, are activated and in turn phosphorylate STAT3 proteins. EGFR inhibitor erlotinib as well as tyrosine kinase inhibitors AZD0530 and dasatinib have shown to efficiently block STAT3 activation. The subsequent dimerization leads to nuclear translocation and target gene transcription, partially by interaction with NFAT transcription factors. The interaction on target promoters is not dependent on STAT and NFAT phosphorylation, illustrated by dashed circles. The canonical NFAT signaling pathway is activated by intracellular Ca2+ rises leading to activation of the phosphatase Calcineurin and dephosphorylation of NFAT proteins which shuttle to the nucleus and bind to their target promoters. Calcineurin inhibitors CsA and FK506 block NFAT dephosphorylation and nuclear translocation. NFAT phosphorylation does not exclude nuclear localization and binding to partner proteins, shown by dashed white circles. The dashed arrow indicates a possible interaction with p65, which has been demonstrated for other non-pancreatic tissues. GSK-3 inhibitors successfully disrupt NFAT binding to partner proteins such as STAT3 as well as NFAT transcriptional activity in the nucleus. Finally, the network of inflammatory transcription factors regulate numerous target genes mediating inflammation, growth, invasion, angiogenesis and metastasis, thereby contributing to pancreatic carcinogenesis. PTC, indicates Patched; SMO, Smoothened; HH, Hedgehog; Sufu, SHH, Sonic HH; Suppressor of Fused; EGF(R), epidermal growth factor receptor; IL-6(R), interleukin-6 (receptor); IKKβ, IκB kinase β; IκBα, inhibitor of NF-κB α; GSK-3, Glycogen synthase kinase-3; STAT3, signal transducer and activator of transcription 3; JAK, janus kinase; TNF(R), tumor necrosis factor (receptor); TGF(R), transforming growth factor (receptor); NFAT, nuclear factor of activated T-cells; CsA, Cyclosporine A; COX-2, cyclooxygenase-2; 5-LOX, 5-lipooxygenase; Bcl-2/xL, B cell lymphoma 2/xL; Mcl-1, myeloid cell leukemia sequence 1; cIAP 1/2, cellular inhibitor of apoptosis; XIAP, X-linked inhibitor of apoptosis protein; cFLIP, Cellular FLICE (FADD-like IL-1beta-converting enzyme)-inhibitory protein; CDK 4/6, cyclin-dependent kinase 4/6; MMP-9, matrix-metaloproteinase-9; ICAM-1, inter-Cellular Adhesion Molecule 1;VCAM-1, vascular cell adhesion molecule 1; VEGF, vascular endothelial growth factor; bFGF, basic fibroblast growth factor; HGF, hepatocyte growth factor; VMP1, Vacuole Membrane Protein 1; PDGF, platelet-derived growth factor; CXCR4, CXC-motif chemokine receptor 4.
The transcription factor NF-κB, a master regulator of innate immunity and inflammation, represents a molecular bridge between chronic inflammation and cancer development7–9. In fact, there has been increasing interest in NF-κB’s role in both inflammation-induced carcinogenesis and maintenance of established cancer, where it is constitutively activated10. The NF-κB family is composed of five members that form homodimeric and heterodimeric complexes: RelA (p65), RelB, Rel (cRel), NF-κB1 (p50 and its precursor p105) and NF-κB2 (p52 and its precursor p100). Activation of these proteins is regulated by the canonical NF-κB activation pathway that applies to RelA/p50 dimers, which are sequestered in the cytosol through interactions with inhibitory proteins of the IkB family. Following stimulation by inflammatory cytokines such as TNF-α, IkB is phosphorylated by the IKK complex, leading to their ubiquitylation and subsequent degradation by the proteasome pathway. RelA/p50 dimers are then translocated into the nucleus where they activate the transcription of growth-promoting genes (e.g. cyclin D1, cyclin E, CDK2, and c-Myc) as well as cytokines (e.g. IL-6) (Figure 1). NF-κB has been shown to be constitutively activated in pancreatic cancer where its inhibition enhances the sensitivity of cancer cells to chemotherapeutic agents and death receptor–mediated apoptosis. In addition, NF-κB contributes to control proliferation, cell survival, and invasion of pancreatic cancer cells11,12 induced by inflammatory stimuli originated from the microenvironment.
The functions mediated by NF-κB are at least partially done in cooperation with other factors such as the signal transducer and activator of transcription 3 (STAT3)13–15. This interaction occurs at several subcellular levels and in a highly context-dependent manner. NF-κB controls the expression levels of STAT3 induced by tumor micronenvironment cytokines and growth factors, most notably by the proinflammatory cytokine IL-616,17. Interestingly, active STAT3, in turn can feed back on NF-κB signalling leading to the accumulation of RelA in the nucleus of cancer cells18. The increased activation levels of STAT3 in cancers are almost exclusively secondary to the activation of upstream signaling and, similar to NF-κB do not result from genetic alterations. STAT3 is turned on by IL-6 binding to the extracellular domain of transmembrane cytokine receptors and subsequent activation of receptor-associated tyrosine kinases, such as Janus kinases (JAKs). Phosphorylation of key tyrosine (Tyr705) and serine (Ser727) residues on STAT3 induces formation of transcriptional active STAT3 homodimers associated through reciprocal SH2 domain–phosphotyrosine-705 (pTyr705) interactions. Dimeric STAT3 complexes translocate to the nucleus where they bind to DNA and promote specific gene transcription. In the nucleus of tumor cells and infiltrating immune cells present within the tumor microenvironment, both STAT3 and NF-κB co-regulate numerous genes controlling tumor cell proliferation and survival including induction of c-Myc, cyclin D1 and Bcl-2, as well as epithelial to mesenchymal transdifferentiation (EMT) and migration genes such as E-cadherin, Twist and Snail14,18–20 (Figure 1). Accumulating evidence suggests a crucial role of IL-6 in constitutive STAT3 activation in gastrointestinal cancers including pancreatic cancer, which ultimately determines tumor burden and prognosis. Notably, active STAT3 is often found at the invasive edge of tumors, adjacent to inflammatory cells linking the inflammatory processes to cancer development16,21. Recent reports by Fukuda et al and Lesina et al established that the aberrant activation of IL-6/STAT3 axis promotes PanIN progression and pancreatic cancer development. Moreover, STAT3 represents a critical component of pancreatitis-accelerated PanIN formation and supports cell growth, and metaplasia-associated inflammation22,23.
Recently, it has been reported that tyrosine phosphorylation and activation of the STAT3 transcription factor is stimulated by the transcription factor NFATc1, another oncogenic factor with key roles in gene regulation during pancreatic carcinogenesis24. NFAT factors together with NF-κB belong to the extended NFκB/Rel family of transcription factors and thus share structural similarities of their DNA-binding domains, also known as Rel homology regions (RHR). The canonical mode of DNA binding by the RHR has been well characterized in the structures of several NFκB-DNA complexes and it becomes clear that NFAT can bind to several promoters with kB sites25. NFAT proteins modulate inflammatory processes and participate in the regulation of genes influencing cell growth and differentiation. In resting cells, NFAT resides in the cytosol and in a hyperphosphorylated form. A rise in intracellular calmodulin (CaM)-bound Ca2+ levels activates the heterodimeric phosphatase calcineurin, which dephosphorylates multiple serine residues in NFAT amino terminal region, resulting in the rapid translocation of the protein from the cytoplasm to the nucleus, where it enhances local chromatin acetylation and promotes de novo gene transcription26. Similar to NF-κB and STAT3, ectopic activation of individual NFAT members is now recognized as an important aspect for oncogenic transformation in many different human malignancies including pancreatic cancer. In these tumors, sustained activation of the Ca2+/calcineurin/NFAT signalling and transcription pathway has emerged as a multifunctional and powerful regulatory principle governing growth and survival of transformed cells26–28. Expression and constitutive activation of two members of the family, NFATc1 and NFATc2, is found in PanIN-2 lesions and in the nuclei of most of the invasive tumors. NFAT-dependent cell growth and transformation through c-Myc promoter activation in pancreatic cancer requires the interaction with the ETS-like transcription factor ELK-126,29,30. Similarly, NFAT forms promoter-bound transcription complexes with NF-κB and STAT3 in pancreatic cancer cells in a signalling dependent manner promoting cellular functions associated with pancreatic cancer development and progression (Baumgart and Ellenrieder, personal communication).
Other key oncogenic factors implicated in pancreatic cancer and activated by inflammatory cytokines are the members of the GLI family (GLI1, 2 and 3)31–43, which regulate the expression of genes important for tumor microenvironment (IL-6)33, cell proliferation (CDK2)34, apoptosis (bcl-2) and autophagy (VMP-1) (Martin E. Fernandez-Zapico, personal communication). These zing finger proteins were originally identified as downstream effectors of the HH signaling pathway31–37 (Figure 1), a cascade that is activated during pancreatic inflammatory process and carcinogenesis. As downstream effectors of HH, the GLI factors are activated via two multitransmembrane proteins: patched (PTC) and Smoothened (SMO). In this receptor complex, PTC is the ligand-binding subunit, while SMO represents the signaling component. Upon binding of HH to its receptor (PTC), the inhibitory effect of PTC on SMO is released and signal is triggered leading to the activation of GLI proteins31–32. HH-driven pancreatic tumors are mainly characterized by inappropriate ligand expression, in particular Sonic HH and Indian HH, which is overexpressed in pancreatic tumors, both in mice models and human tumors34–37. Interestingly, GLI activation by the HH ligands seems to be exclusively increased in stromal cells, suggesting an important contribution of the tumor microenvironment to paracrine GLI signaling in pancreatic carcinogenesis31,36,37. These observations may be of therapeutic relevance since pharmacologic inhibition of HH-GLI signaling in a mouse model of pancreatic cancer enhances chemotherapy delivery38. There is an emerging body of experimental work highlighting the role of GLI in pancreatic carcinogenesis independently of HH38–39. Recent reports demonstrate that KRAS regulates GLI activity by increasing GLI1 expression and protein stability and cooperates with GLI2 during pancreatic carcinogenesis39–41. Other studies found that GLI factors are regulated in pancreatic cancer cells through a SMO-independent mechanism by TGF-β39. Activation of this signaling pathway leads to an increase in expression of both GLI1 and GLI2 in pancreatic cancer cells42. Hence, numerous lines of evidence support a role for GLI proteins in pancreatic cancer and suggest this family of transcription factors is a suitable therapeutic target for this dismal disease.
Current strategies to target oncogenic transcription factor in pancreatic cancer
Surgical resection to date represents the best therapy for long-term survival in pancreatic cancer. At diagnosis however, only 15% of the patients are amenable to surgical resection43 and alternative treatment options mostly fail due to a resistance to chemotherapy or radiation44. Currently, the only approved treatments are gemcitabine and erlotinib, both of which are not effective in the majority of patients and most notably, the benefit on survival is measured in weeks45. Based on the central role and intensive crosstalk of inflammatory transcription factors in signaling networks aimed at maintenance of the transformed phenotype in pancreatic cancer, these nuclear molecules represent suitable nodes for therapeutic intervention. In the following paragraph, we discuss few examples of current studies aimed at inhibiting the activity of these oncogenic transcription factors and their target genes in pancreatic carcinogenesis (Figure 1 and Table 1). To date, there are no effective strategies to directly target a transcription factor46, rather, the discussed compounds rather target upstream signaling pathways to block the activation of the respective transcription factor and its signaling.
Table 1.
Clinical trials targeting inflammatory signaling pathways in pancreatic cancer
| Intervention | Mechanism | Clinical trials/status |
|---|---|---|
| Celecoxib and irinotecan/radiotherapy | COX-2 inhibitor, blocks IKK activity | Phase II; terminated |
| Bortezomib (PS-341) and gemcitabine/paclitaxel | 26s proteasome inhibitor; blocks IkB degradation | Phase I/II; completed |
| Curcumin (with gemcitabine) | Inhibits IKK, NF-κB activity and expression | Phase II; completed |
| Curcumin with celecoxib | Phase III; recruiting participants | |
| Genistein with gemcitabine/erlotinib | Blocks IKK activation and activity | Phase II; completed |
| OPB-31121 | STAT3 inhibitor | Phase I; completed |
| Saracatinib (AZD0530) (with gemcitabine) | Src inhibitor; blocks STAT3 phosphorylation | Phase I/II; completed and ongoing/recruiting participants |
| IPI-926-03 with gemcitabine | Hedgehog inhibitor | Phase I/II; recruiting participants |
| GDC-0449 with erlotinib/gemcitabine/paclitaxel | Hedgehog inhibitor | Phase I/II; recruiting participants |
Information about closed and ongoing trials found on the NIH web sites:
www.clinicaltrials.gov. Accessed April 7, 2011
www.cancer.gov/clinicaltrials. Accessed March 11, 2011
Dietary ingredients such as curcumin, quercetin and isoflavone genistein have shown to efficiently suppress NF-κB machinery and in turn exert antitumor and proapoptotic effects in numerous murine tumor models and in Phase II trials where they potentiated antitumor activity of gemcitabine47–51. Synthetic terpenoids have proven to efficiently block NF-κB as well as STAT3 activation and subsequently prolonged survival in a transgenic mouse model of pancreatic cancer. Interestingly, one proposed mechanism mediating its antitumor properties is the blockade of the NF-κB-STAT3 crosstalk52. Anti-inflammatory compounds like aspirin and sulindac further inhibited NF-κB in a genetically engineered mouse model and thus delayed the progression of PanINs and cancer formation, highlighting the significance of inflammation-induced transcription factors and their networks in pancreatic cancer progression53,54. The effects of aspirin on tumorigenesis remain to be further elucidated, as other studies could not demonstrate inhibitory effects on cancer growth55, rather suggesting a tumor preventive function. Finally, several proteasome inhibitors (e.g. bortezomib), which prevent the degradation of IkB and in turn NF-κB release could efficiently block tumor growth in preclinical models with and without chemotherapy56.
Genetic silencing of STAT3 suppresses tumor growth, invasion and metastasis in a pancreatic cancer xenograft model and therefore underscores the requirement of STAT3 for pancreatic cancer growth57. Several compounds inhibiting the activation of STAT3 in pancreatic cancer cells have been evaluated in preclinical models. As the mechanism of persistent STAT3 activation in tumors has been attributed to phosphorylation by tyrosine kinases like JAK, SRC family kinases, or EGFR, most studies aim to assess inhibitors of upstream activating pathways. Treatment with AZD0530, an SRC inhibitor, thus resulted in downregulation of p-STAT3 and significantly inhibited tumor growth in pancreatic cancer mouse models58. These findings were confirmed by Nagaraj et al who used dasatinib to block SRC kinase in vivo. In addition, combined inhibition of SRC and EGFR with dasatinib, erlotinib and gemcitabine synergistically blocked constitutively activated STAT3 and consequently growth in a mouse model of pancreatic cancer59.
The genetic knockdown of NFAT remarkably reduced in vivo tumor growth of pancreatic cancer cells29,30. Two widely used immunosuppressive agents, Cyclosporine A (CsA) and FK506, act as potent calcineurin-NFAT inhibitors. Their use in cancer therapy is limited, however, due to high side effects and to the fact that long-term application is accompanied by increased incidence of cancer. Consequently, a tumor-targeted therapy is required. Several inhibitors structurally and functionally related to CsA and FK506 that exhibit fewer side effects have been developed but remain to be tested in terms of efficacy, stability, and bioavailability in preclinical tumor models27. Nonetheless, small-molecule inhibitors of NFAT are preferred for clinical use. Further strategies to block NFAT activity in pancreatic cancer include targeting of endogenous activators of NFAT, such as GSK-3 (V. Ellenrieder, personal communication). Recent studies revealed that the inhibition of GSK-3 kinase activity disrupts STAT3-NFAT interaction and consequently reduces NFAT transcriptional activity and cell growth in pancreatic xenograft mouse models. Moreover it acts to stabilize NFAT protein and accordingly enhances oncogenic NFAT functions (Baumgart and Ellenrieder, personal communication). GSK-3 is also a potent modulator of NF-κB activity in gastrointestinal cancers. Several authors confirmed the abrogation of NF-κB activity and target gene transcription upon pharmacologic GSK-3 depletion in mouse models of pancreatic cancer60,61. Since chronic inflammation represents a risk factor for the development of pancreatic cancer, inflammatory pathways should be considered as critical therapeutic targets. Notably, GSK-3 regulates the transcription factors NF-κB, STAT, NFAT and possibly GLI, in different cells present within the tumor microenvironment62. As these transcription factors are also correlated to inflammatory responses they emerged as potential drug targets in inflammatory diseases. Inflammation is frequently linked to carcinogenesis and as such these proteins primarily represent key modulators of inflammatory processes promoting cancer development. This renders GSK-3 an interesting tool to repress (inflammation-mediated) carcinogenesis.
The use of chemical HH-GLI pathway inhibitors, such as cyclopamine and its derivatives in clinical trials (GDC-0449 and IPI-926) demonstrates a reduced tumor burden and metastasis in mouse models36–39,63. Furthermore, the effect of a SMO antagonist on the efficacy of chemotherapy was recently analyzed in a genetic mouse model of pancreatic cancer. Interestingly, delivery and efficacy of gemcitabine could be improved, which was mainly mediated through depletion of tumor-associated stromal tissue. These findings most notably indicate a contribution of the HH-GLI pathway to inflammation-linked chemoresistance36. It has also been shown that NF-κB directly regulates HH expression in a model of inducible NF-κB activation in pancreatic acinar cells. Therein, genetic silencing of HH significantly reduced the IKK2-mediated increase of in vivo tumor growth, which suggests an intense interaction between NF-κB and HH signaling pathways in oncogenesis64 and supports the rationale of a combination therapy targeting these two oncogenic pathways in pancreatic cancer.
In summary, oncogenic transcription factor proteins are powerful molecules involved in the regulation of cell proliferation, differentiation and apoptosis. Furthermore, these factors have been demonstrated to play an important role in the pathogenesis of pancreatic cancer. Although direct interference with the activity of transcription factors would represent the most specific strategy, it remains a challenging and moreover, a unidirectional task46 due to intensive crosstalk of inflammatory transcription factors. As numerous oncogenic and inflammatory pathways converge on these factors, targeting central upstream regulators and subsequently the complete network appears feasible. This, in contrast to isolated chemotherapy, could block the effects of multiple pathways that contribute to malignancy. Targeting transcription factors such as NF-κB or STAT3 that moreover mediate resistance to apoptosis13 and chemoresistance65, a common feature of pancreatic cancer, would offer a favorable option. Nonetheless, there are still limitations for the application of the aforementioned drugs. At this, some display low bioavailability and as a consequence, low tumor tissue levels in vivo50. As STAT3, NF-κB, or NFAT factors exert global effects on functions and development of immune cells, efforts to suppress inflammatory signaling could nevertheless affect immunsurveillance and in turn tumor-suppressive responses on long-term application. Thus, understanding the role of transcription factors in modulating gene expression and pancreatic carcinogenesis mainly in terms of inflammation will lead to more effective clinical tools for diagnosis, prognosis, treatment and prevention strategies for this dismal disease. Table 1 summarizes current and closed clinical trials evaluating inhibitors of NF-κB, STAT3 and GLI transcription factors in the treatment of pancreatic cancer.
Acknowledgments
We thank Angela McCleary-Wheeler and Gaurav Aggarwal for critical reading of the manuscript and helpful advice. This work was generously supported by the Deutsche Forschungsgemeinschaft (V.E.; KFO210, SFB-TR17), the LOEWE-Schwerpunkt “Tumor and Inflammation” (V.E.) and the Max-Eder program of the German Cancer Research Foundation (V.E., 70-3022-El I), Novartis Foundation (V.E.), Schulze Center for Novel Therapeutics (M.E.F-Z), Mayo Clinic Cancer Center (M.E.F-Z), the National Institutes of Health CA136526 (M.E.F-Z), Mayo Clinic Pancreatic SPORE P50 CA102701 (M.E.F-Z), and Mayo Clinic Center for Cell Signaling in Gastroenterology P30 DK84567 (M.E.F.-Z.).
References
- 1.Löhr M, Klöppel G, Maisonneuve P, et al. Frequency of K-ras mutations in pancreatic intraductal neoplasias associated with pancreatic ductal adenocarcinoma and chronic pancreatitis: a meta-analysis. Neoplasia. 2005;7:17–23. doi: 10.1593/neo.04445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hingorani SR, Petricoin EF, Maitra A, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 3.Morris JP, 4th, Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10:683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerra C, Schuhmacher AJ, Cañamero M, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 5.Erkan M, Reiser-Erkan C, Michalski CW, et al. Tumor microenvironment and progression of pancreatic cancer. Exp Oncol. 2010;32:128–131. [PubMed] [Google Scholar]
- 6.Greer JB, Whitcomb DC. Inflammation and pancreatic cancer: an evidence-based review. Curr Opin Pharmacol. 2009;9:411–418. doi: 10.1016/j.coph.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 7.Holcomb B, Yip-Schneider M, Schmidt CM. The role of nuclear factor kappaB in pancreatic cancer and the clinical applications of targeted therapy. Pancreas. 2008;36:225–235. doi: 10.1097/MPA.0b013e31815b3207. [DOI] [PubMed] [Google Scholar]
- 8.Billadeau DD. Primers on molecular pathways. The glycogen synthase kinase-3beta. Pancreatology. 2007;7:398–402. doi: 10.1159/000108955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Algül H, Adler G, Schmid RM. NF-kappaB/Rel transcriptional pathway: implications in pancreatic cancer. Int J Gastrointest Cancer. 2002;31:71–78. doi: 10.1385/IJGC:31:1-3:71. [DOI] [PubMed] [Google Scholar]
- 10.Karin M. NF-kappaB as a critical link between inflammation and cancer. Cold Spring Harb Perspect Biol. 2009;1:a000141. doi: 10.1101/cshperspect.a000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maier HJ, Schmidt-Strassburger U, Huber MA, et al. NF-kappaB promotes epithelial-mesenchymal transition, migration and invasion of pancreatic carcinoma cells. Cancer Lett. 2010;295:214–228. doi: 10.1016/j.canlet.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Kong R, Sun B, Jiang H, et al. Downregulation of nuclear factor-kappaB p65 subunit by small interfering RNA synergizes with gemcitabine to inhibit the growth of pancreatic cancer. Cancer Lett. 2010;291:90–98. doi: 10.1016/j.canlet.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 13.Greten FR, Weber CK, Greten TF, et al. Stat3 and NF-kappaB activation prevents apoptosis in pancreatic carcinogenesis. Gastroenterology. 2002;123:2052–2063. doi: 10.1053/gast.2002.37075. [DOI] [PubMed] [Google Scholar]
- 14.Bollrath J, Greten FR. IKK/NF-kappaB and STAT3 pathways: central signalling hubs in inflammation-mediated tumour promotion and metastasis. EMBO Rep. 2009;10:1314–9. doi: 10.1038/embor.2009.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He G, Karin M. NF-κB and STAT3 - key players in liver inflammation and cancer. Cell Res. 2011;21:159–168. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee H, Herrmann A, Deng JH, et al. Persistently activated Stat3 maintains constitutive NF-kappaB activity in tumors. Cancer Cell. 2009;15:283–293. doi: 10.1016/j.ccr.2009.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grivennikov SI, Karin M. Inflammation and oncogenesis: a vicious connection. Curr Opin Genet Dev. 2010;20:65–71. doi: 10.1016/j.gde.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu H, Jove R. The STATs of cancer--new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 21.Bromberg J, Wang TC. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell. 2009;15:79–80. doi: 10.1016/j.ccr.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lesina M, Kurkowski MU, Ludes K, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–469. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Fukuda A, Wang SC, Morris JP, 4th, et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19:441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagnuas L, Clipstone NA. Deregulated NFATc1 activity transforms murine fibroblasts via an autocrine growth factor-mediated Stat3-dependent pathway. J Cell Biochem. 2009;108:237–248. doi: 10.1002/jcb.22245. [DOI] [PubMed] [Google Scholar]
- 25.Serfling E, Berberich-Siebelt F, Avots A, et al. NFAT and NF-kappaB factors-the distant relatives. Int J Biochem Cell Biol. 2004;36:1166–1170. doi: 10.1016/j.biocel.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 26.König A, Fernandez-Zapico ME, Ellenrieder V. Primers on Molecular Pathways - The NFAT Transcription Pathway in Pancreatic Cancer. Pancreatology. 2010;10:416–422. doi: 10.1159/000315035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mancini M, Toker A. NFAT proteins: emerging roles in cancer progression. Nat Rev Cancer. 2010;9:810–820. doi: 10.1038/nrc2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller MR, Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010;10:645–656. doi: 10.1038/nri2818. [DOI] [PubMed] [Google Scholar]
- 29.Buchholz M, Schatz A, Wagner M, et al. Overexpression of c-myc in pancreatic cancer caused by ectopic activation of NFATc1 and the Ca2+/calcineurin signaling pathway. EMBO J. 2006;25:3714–3724. doi: 10.1038/sj.emboj.7601246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.König A, Linhart T, Schlengemann K, et al. NFAT-induced histone acetylation relay switch promotes c-Myc-dependent growth in pancreatic cancer cells. Gastronenterology. 2010;138:1189–1199. doi: 10.1053/j.gastro.2009.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saqui-Salces M, Merchant JL. Hedgehog signaling and gastrointestinal cancer. Biochim Biophys Acta. 2010;1803:786–795. doi: 10.1016/j.bbamcr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stecca B, Ruiz I, Altaba A. Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J Mol Cell Biol. 2010;2(2):84–95. doi: 10.1093/jmcb/mjp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elsawa SF, Almada LL, Ziesmer SC, et al. GLI2 mediates cytokine crosstalk in the tumor microenvironment. J Biol Chem. 2011;286:21524–21534. doi: 10.1074/jbc.M111.234146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelleher FC. Hedgehog signaling and therapeutics in pancreatic cancer. Carcinogenesis. 2011;32:445–451. doi: 10.1093/carcin/bgq280. [DOI] [PubMed] [Google Scholar]
- 35.Thayer SP, di Magliano MP, Heiser PW, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425:851–856. doi: 10.1038/nature02009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theunissen JW, de Sauvage FJ. Paracrine Hedgehog signaling in cancer. Cancer Res. 2009;69:6007–6010. doi: 10.1158/0008-5472.CAN-09-0756. [DOI] [PubMed] [Google Scholar]
- 37.Tian H, Callahan CA, DuPree KJ, et al. Hedgehog signaling is restricted to the stromal compartment during pancreatic carcinogenesis. Proc Natl Acad Sci U S A. 2009;106:4254–4259. doi: 10.1073/pnas.0813203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Olive KP, Jacobetz MA, Davidson CJ, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science. 2009;324:1457–1461. doi: 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nolan-Stevaux O, Lau J, Truitt ML, et al. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009;23(1):24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji Z, Mei FC, Xie J, et al. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J Biol Chem. 2007;282:14048–14055. doi: 10.1074/jbc.M611089200. [DOI] [PubMed] [Google Scholar]
- 41.Pasca di Magliano M, Sekine S, Ermilov A, et al. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes Dev. 2006;20:3161–3173. doi: 10.1101/gad.1470806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dennler S, André J, Alexaki I, et al. Induction of sonic hedgehog mediators by transforming growth factor-beta: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007;67:6981–6986. doi: 10.1158/0008-5472.CAN-07-0491. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt CM, Powell ES, Yiannoutsos CT, et al. Pancreaticoduodenectomy: a 20-year experience in 516 patients. Arch Surg. 2004;139:725–7. doi: 10.1001/archsurg.139.7.718. [DOI] [PubMed] [Google Scholar]
- 44.Lionetto R, Pugliese V, Bruzzi P, et al. No standard treatment is available for advanced pancreatic cancer. Eur J Cancer. 1995;31A:882–887. doi: 10.1016/0959-8049(94)00445-5. [DOI] [PubMed] [Google Scholar]
- 45.Burris HA, 3rd, Moore MJ, Andersen J, et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: a randomized trial. J Clin Oncol. 1997;15:2403–2413. doi: 10.1200/JCO.1997.15.6.2403. [DOI] [PubMed] [Google Scholar]
- 46.Darnell JE. Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 47.Li L, Braiteh FS, Kurzrock R. Liposome-encapsulated curcumin: in vitro and in vivo effects on proliferation, apoptosis, signaling, and angiogenesis. Cancer. 2005;104:1322–1331. doi: 10.1002/cncr.21300. [DOI] [PubMed] [Google Scholar]
- 48.Jutooru I, Chadalapaka G, Lei P, et al. Inhibition of NFkappaB and pancreatic cancer cell and tumor growth by curcumin is dependent on specificity protein down-regulation. J Biol Chem. 2010;285:25332–25344. doi: 10.1074/jbc.M109.095240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mouria M, Gukovskaya AS, Jung Y, et al. Food-derived polyphenols inhibit pancreatic cancer growth through mitochondrial cytochrome C release and apoptosis. Int J Cancer. 2010;98:761–769. doi: 10.1002/ijc.10202. [DOI] [PubMed] [Google Scholar]
- 50.Dhillon N, Aggarwal BB, Newman RA, et al. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin Cancer Res. 2008;14:4491–4498. doi: 10.1158/1078-0432.CCR-08-0024. [DOI] [PubMed] [Google Scholar]
- 51.Banerjee S, Zhang Y, Ali S, et al. Molecular evidence for increased antitumor activity of gemcitabine by genistein in vitro and in vivo using an orthotopic model of pancreatic cancer. Cancer Res. 2005;65:9064–9072. doi: 10.1158/0008-5472.CAN-05-1330. [DOI] [PubMed] [Google Scholar]
- 52.Liby KT, Royce DB, Risingsong R, et al. Synthetic triterpenoids prolong survival in a transgenic mouse model of pancreatic cancer. Cancer Prev Res (Phila) 2010;3:1427–1434. doi: 10.1158/1940-6207.CAPR-10-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto Y, Yin MJ, Lin KM, et al. Sulindac inhibits activation of the NF-kappaB pathway. J Biol Chem. 1999;274:27307–27314. doi: 10.1074/jbc.274.38.27307. [DOI] [PubMed] [Google Scholar]
- 54.Fendrich V, Chen NM, Neef M, et al. The angiotensin-I-converting enzyme inhibitor enalapril and aspirin delay progression of pancreatic intraepithelial neoplasia and cancer formation in a genetically engineered mouse model of pancreatic cancer. Gut. 2010;59:630–637. doi: 10.1136/gut.2009.188961. [DOI] [PubMed] [Google Scholar]
- 55.Sclabas GM, Uwagawa T, Schmidt C, et al. Nuclear factor kappa B activation is a potential target for preventing pancreatic carcinoma by aspirin. Cancer. 2005;103:2485–2490. doi: 10.1002/cncr.21075. [DOI] [PubMed] [Google Scholar]
- 56.Lenz HJ. Clinical update: proteasome inhibitors in solid tumors. Cancer Treat Rev. 2003;29(Suppl 1):41–48. doi: 10.1016/s0305-7372(03)00082-3. [DOI] [PubMed] [Google Scholar]
- 57.Qiu Z, Huang C, Sun J, et al. RNA interference-mediated signal transducers and activators of transcription 3 gene silencing inhibits invasion and metastasis of human pancreatic cancer cells. Cancer Sci. 2007;98:1099–1106. doi: 10.1111/j.1349-7006.2007.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rajeshkumar NV, Tan AC, De Oliveira E, et al. Antitumor effects and biomarkers of activity of AZD0530, a Src inhibitor, in pancreatic cancer. Clin Cancer Res. 2009;15:4138–4146. doi: 10.1158/1078-0432.CCR-08-3021. [DOI] [PubMed] [Google Scholar]
- 59.Nagaraj NS, Washington MK, Merchant NB. Combined blockade of Src kinase and epidermal growth factor receptor with gemcitabine overcomes STAT3-mediated resistance of inhibition of pancreatic tumor growth. Clin Cancer Res. 2011;17:483–493. doi: 10.1158/1078-0432.CCR-10-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ougolkov AV, Fernandez-Zapico ME, Savoy DN, et al. Glycogen synthase kinase-3beta participates in nuclear factor kappaB-mediated gene transcription and cell survival in pancreatic cancer cells. Cancer Res. 2005;65:2076–2081. doi: 10.1158/0008-5472.CAN-04-3642. [DOI] [PubMed] [Google Scholar]
- 61.Wilson W, 3rd, Baldwin AS. Maintenance of constitutive IkappaB kinase activity by glycogen synthase kinase-3alpha/beta in pancreatic cancer. Cancer Res. 2008;68:8156–63. doi: 10.1158/0008-5472.CAN-08-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beurel E, Michalek SM, Jope RS. Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3) Trends Immunol. 2010;31:24–31. doi: 10.1016/j.it.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feldmann G, Habbe N, Dhara S, et al. Hedgehog inhibition prolongs survival in a genetically engineered mouse model of pancreatic cancer. Gut. 2008;57:1420–1430. doi: 10.1136/gut.2007.148189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kasperczyk H, Baumann B, Debatin KM, et al. Characterization of sonic hedgehog as a novel NF-kB target gene that promotes NF-kB mediated apoptosis resistance and tumor growth in vivo. FASEB J. 2009;23:21–33. doi: 10.1096/fj.08-111096. [DOI] [PubMed] [Google Scholar]
- 65.Arlt A, Gehrz A, Müerköster S, et al. Role of NF-kappaB and Akt/PI3K in the resistance of pancreatic carcinoma cell lines against gemcitabine-induced cell death. Oncogene. 2003;22:3243–3251. doi: 10.1038/sj.onc.1206390. [DOI] [PubMed] [Google Scholar]