Abstract
Cancer cells possess fundamentally altered metabolism that provides a foundation to support tumorigenicity and malignancy. Our understanding of the biochemical underpinnings of cancer has benefited from the integrated utilization of large-scale profiling platforms (e.g. genomics, proteomics, and metabolomics), which, together, can provide a global assessment of how enzymes and their parent metabolic networks become altered in cancer to fuel tumor growth. This review presents several examples of how these integrated platforms have yielded fundamental insights into dysregulated metabolism in cancer. We will also discuss questions and challenges that must be addressed to more completely describe, and eventually control, the diverse metabolic pathways that support tumorigenesis.
INTRODUCTION
Cancer cells have fundamentally altered cellular metabolism which directly contributes to tumorigenicity and malignancy. Deciphering the full scope of dysregulated metabolism in cancer and its relevance to disease pathogenesis and potential therapeutic relevance requires the advancement of technologies to identify altered enzymes and metabolites in cancer. This review will discuss how large-scale profiling methods, such as genomics, proteomics, and metabolomics have been innovatively used to elucidate metabolic pathways that drive tumorigenesis and metastasis. Not only have such large-scale endeavors been useful in providing fundamental insights into the basic biochemistry that defines cancer cells, but they have also led to the discovery of potential targets for cancer therapy. We will also discuss challenges facing the field of cancer metabolism.
Fundamental to the proliferation of a transformed cell is first and foremost the ability to rapidly and robustly biosynthesize essential biomolecules required for cell division. The study of cancer metabolism has hence primarily focused on pathways that, when altered, can lead to the aberrant production or consumption of essential biomolecules such as glucose, amino acids, nucleotides, and lipids (DeBerardinis et al., 2008a; Deberardinis et al., 2008b). Beyond the synthesis of biomolecules, studies have also shown that cancer cells rewire, mutationally activate, and/or transcriptionally upregulate metabolic pathways that produce oncogenic signaling molecules that in turn fuel tumor growth and malignancy (Cairns et al., 2011; Dang et al., 2009b; Nomura et al., 2010a). For many of these pathways, large-scale profiling platforms and innovative discovery-based approaches played critical roles in uncovering connections to cancer pathogenicity.
The Regulation of Pyruvate Kinase and its Role in Glucose Metabolism in Cancer
In 1929, Otto Warburg noted that transformed cells consume glucose at an abnormally high rate (Warburg, 1956). However, rather than leading to an increase in cellular energy via the citric acid cycle, Warburg showed that this increased glycolytic flux instead leads to the production of lactate, even under non-hypoxic conditions (Warburg, 1956). While this “Warburg effect” appeared to be an irrefutable and universal property of most cancer cells, what had remained enigmatic for some time was the reason for and mechanism by which cancer cells adopt this switch to aerobic glycolysis. Nearly 80 years later, critical insights have been made demonstrating how cancer cells exhibit multiple additional levels of regulation on glycolysis, which collectively divert carbon from glucose towards the synthesis of molecular building blocks such as amino acids, nucleic acids, and lipids, for the purpose of generating ample protein, DNA, and cellular membranes for proliferation. Many of these discoveries have been made with the help of innovative large-scale genomic, proteomic, and metabolomic profiling platforms that have allowed scientists to delve deeper into aspects of cancer metabolism. Christofk et al. in 2008 demonstrated that a single switch of pyruvate kinase from the M1 (PKM1) to M2 (PKM2) splice isoform is sufficient to shift cellular metabolism to favor aerobic glycolysis (Christofk et al., 2008a). They then further showed that PKM2-expressing cells consume less oxygen and produce more lactate than PKM1-expressing cells and that replacement of PKM2 with PKM1 in cancer cells quite provocatively reverses this metabolic phenotype that embodies the Warburg effect (Christofk et al., 2008a). Christofk et al went further to develop cells that stably express mouse PKM1 or PKM2 in the human lung cancer cell line H1299 in the background of knocking down endogenous PKM2. Quite provocatively, mice injected with the PKM1 cells showed a significant delay in tumor development as compared with those injected with PKM2-expressing cells, which developed much larger tumors. These studies showed that PKM2 expression provides a selective growth advantage for tumor cells in vivo prompting investigations into the metabolic and regulatory mechanisms behind the action of PKM2 in cancer.
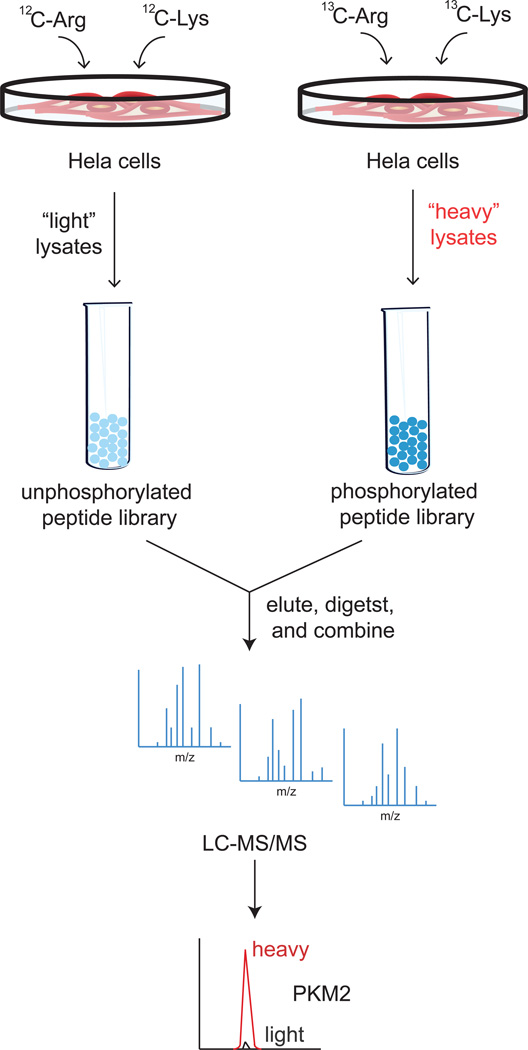
Subsequent proteomic studies have uncovered that PKM2, unlike PKM1, cannot constitutively maintain its active tetrameric structure due to multiple additional levels of post-translational regulation found specifically on PKM2 that leads to overall decreased pyruvate kinase activity (Anastasiou et al., 2011; Christofk et al., 2008b; Hitosugi et al., 2009; Lv et al., 2011) (Figure 1). When searching for phosphotyrosine (pTyr)-binding proteins from cell lysates using a SILAC (stable isotope labeling of amino acids in cell culture)-based quantitative proteomic enrichment strategy with a phosphotyrosine peptide library affinity matrix, Christofk et al found that PKM2 selectively and directly binds to phosphotyrosine peptides, resulting in the displacement of the activating cofactor fructose-1,6-bisphosphate, thereby inhibiting PKM2 activity (Christofk et al., 2008b). Christofk et al labeled HeLa cells with heavy isotopic 13C-lysine and 13C-arginine or normal isotopic 12C-lysine and 12C-arginine, followed by enrichment of phosphotyrosine binding proteins by flowing heavy cell lysates over a phosphotyrosine peptide library versus light cell lysates over a corresponding unphosphorylated peptide library. Upon proteomic analysis, pyruvate kinase was among proteins that exhibited a significantly higher SILAC heavy to light ratio (Christofk et al., 2008b) (Figure 2). The authors then showed that a mutant form of PKM2 that can no longer bind phosphotyrosine peptides and thereby exhibits greater PKM2 activity, leads to enhanced oxygen consumption and decreased lactate production, and that PKM2-expressing cells divert upstream glycolytic intermediates to an anabolic, rather than catabolic fate. In 2009, using a phosphoproteomic strategy of enriching tyrosine-phosphorylated peptides, Hitosugi et al found that PKM2 is also phosphorylated by signaling conferred by the oncogenic fibroblast growth factor receptor type 1, which in turn also inhibits the formation of the active tetrameric form of PKM2 by disrupting its interaction with fructose-1,6-bisphosphate. The authors then demonstrated that a mutant form of PKM2 that is incapable of becoming phosphorylated leads to lactate reduction and increased oxygen consumption—a metabolic feature also observed when PKM2 is replaced with the constitutively active PKM1 (Hitosugi et al., 2009). A recent study has provided yet another mode of regulation upon PKM2 that leads to a diversion of glycolytic flux. Anastasiou et al., 2011 showed that Cys358 on PKM2 is an oxidative sensor that becomes oxidized to inhibit PKM2 activity (Anastasiou et al., 2011). This inhibition of PKM2 activity then leads to the build-up of glucose-6-phosphate, which can subsequently get diverted to the pentose phosphate pathway to generate sufficient antioxidant response, via the reduction of glutathione, for the cancer cell (Figure 1). The authors showed that a C358S mutant of PKM2 leads to decreased glutathione (GSH) and increased oxidative stress, leading to impairments in proliferation and tumor xenograft growth (Anastasiou et al., 2011). Lv et al recently also identified an acetylated lysine on PKM2, using a proteomic strategy in which the authors immunoprecipitated acetyl-lysine peptides from tryptically-digested cytosolic proteomes of LNCaP prostate cancer cells and primary prostate tumors (Lv et al., 2011), and analyzed eluting peptides by LC-MS/MS. The authors showed that K305 acetylation on PKM2 is stimulated by glucose and inhibits PKM2 activity through subsequent autophagic degradation, leading to enhanced tumorigenicity. They further showed that mutating the lysine targeted for acetylation to an acetylation-mimic glutamine leads to enhanced proliferation and tumor growth (Lv et al., 2011). Collectively, a variety of proteomic approaches have been used to provide clear evidence that PKM2 is subject to multiple modes of post-translational regulation that function to retard the last step of glycolysis, thereby leading to an accumulation of upstream glycolytic intermediates. Moreover, cancer cells can divert these upstream glycolytic intermediates towards various anabolic processes in order to maintain their proliferative and tumorigenic capacity.
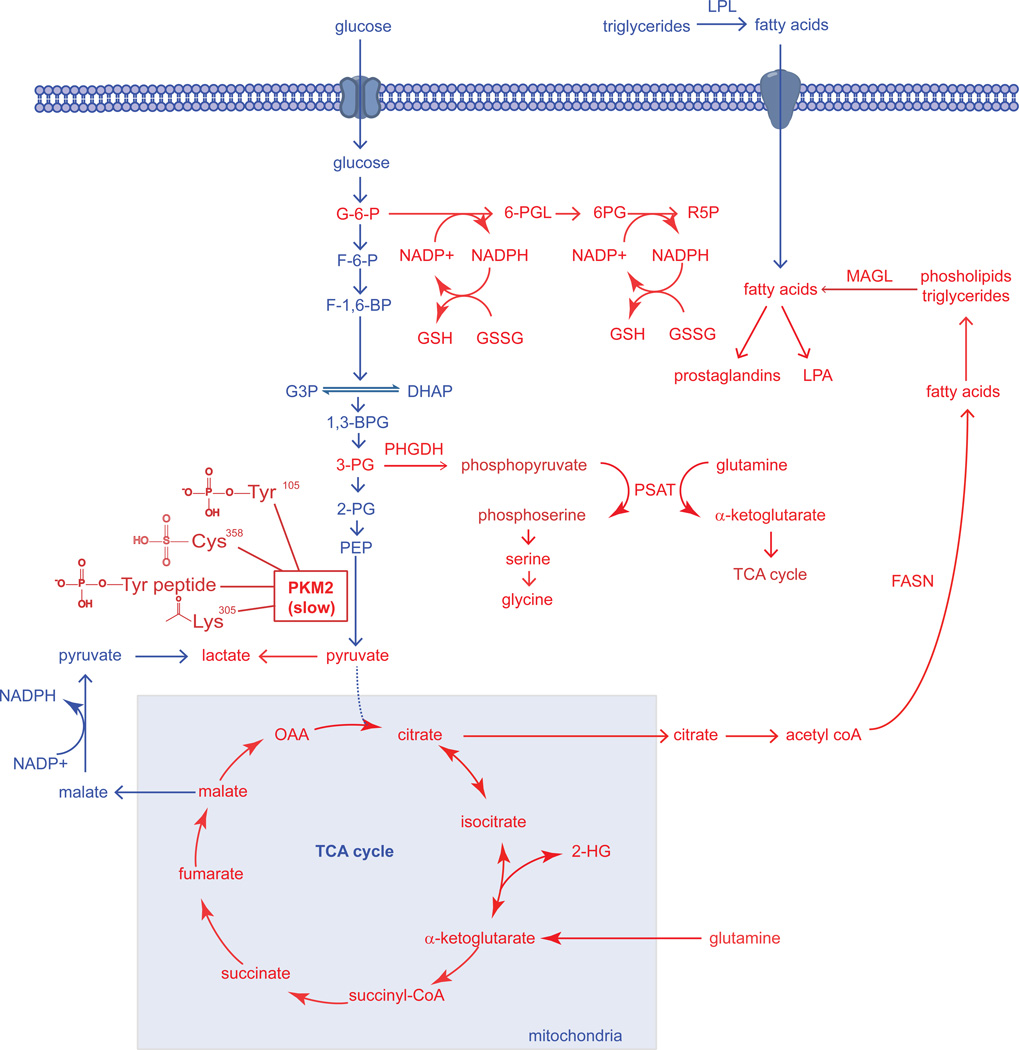
Figure 1. Dysregulated Metabolic Pathways in Cancer Cells Identified Thus Far.
The metabolic pathways that sustain the proliferative nature of a cancer cells are very much the same pathways that constitute the metabolism of normal cells. However, cancer cells are able to aberrantly rewire many of these normal pathways to meet their excessive needs for growth and proliferation. In the figure above, we see that pathways that have been revealed to be essential in cancer cells (shown in Red) are pathways that are fundamentally important for the synthesis of biological macromolecules, antioxidants, and signaling molecules that facilitate cellular growth, survival, and progression. The “Warburg effect,” the preferential shunting of pyruvate to lactate under conditions of normoxia, is facilitated by the switch from the M1 splice isoform of pyruvate kinase to the M2 splice isoform, which is in-general less active due to several modes of regulation on PKM2 activity in the form of protein-protein interactions or posttranslational modifications which all collectively inhibit PKM2 activity (Anastasiou et al., 2011; Christofk et al., 2008b; Hitosugi et al., 2009; Lv et al., 2011). The slow nature of this M2 splice isoform allows for the buildup of upstream glycolytic intermediates rather than favoring the flux of glycolytic carbon through the TCA cycle. Two of the important upstream glycolytic intermediates that are essential for synthesizing necessary biological macromolecules are glucose-6-phosphate (G6P) and 3-phosphoglycerate (3PG). The 3-step formation of Ribose-5-phosphate (R5P) via 6-phosphogluconolactone (6-PGL) and 6-phosphogluconate (6-PG), involves the production of reducing equivalent in the form of NADPH, which in-turn can be used as the primary reducing power in the production of glutathione (GSH) from its oxidized precursor glutathione disulfide (GSSG) (Anastasiou et al., 2011). It was also discovered that a significant portion of glycolytic carbon is diverted towards serine and glycine biosynthesis by phoshphoglycerate dehydrogenase (PHGDH), which catalyzes the conversion of 3-PG to phosphopyruvate. Phosphopyruvate is then converted to phosphoserine, concurrently with the anaplerotic generation of α-ketoglutarate by glutamine to supply the TCA cycle (Locasale et al., 2011a; Possemato et al., 2011). Glutaminolysis-derived α-ketoglutarate provides reducing power in the form of NAPDH and regenerates oxaloacetate (OAA) to form citrate which are both necessary for the production of fatty acids by fatty acid synthase (FASN) (DeBerardinis et al., 2007; Metallo et al., 2012). Under hypoxia or mitochondrial dysfunction, α-ketoglutarate can also undergo reductive carboxylation to isocitrate and then to citrate to support lipogenesis (Mullen et al., 2012). Upon hypoxia or if IDH1 or IDH2 is mutated, these enzymes can also form the oncometabolite 2-hydroxyglutarate (2-HG) (Dang et al., 2010; Wise et al., 2011). Upon synthesis of fatty acids and esterification onto phospholipids or triglycerides, these esterified lipids are mobilized through lipolytic processes involving enzymes such as monoacylglycerol lipase (MAGL), which can lead to the formation of oncogenic lipid signaling molecules including lysophosphatidic acid (LPA) and prostaglandins (Nomura et al., 2010b). Fatty acids can also arise from extracellular sources by hydrolysis of triglycerides by lipoprotein lipase (LPL).
Figure 2. Discovering PKM2 as a Phosphopeptide Binding Protein using Quantiative Proteomics.
Christofk et al labeled HeLa cells with heavy isotopic 13C-lysine and 13C-arginine or normal isotopic 12C-lysine and 12C-arginine. The heavy labeled cells lysates were collected and flowed over a phosphorylated peptide library while the light labeled lysates were flowed over an unphosphorylated peptide library. The eluents were then digested and subsequently analyzed by LC/MS/MS. By identifying proteins that exhibited a high ratio of heavy to light label, phosphoproteins could be identified. One such protein that exhibited a particularly high SILAC heavy to light ratio was Pyruvate Kinase M2 (PKM2) (Christofk et al., 2008b).
While the effects of posttranslational regulation on the metabolic functionality of PKM2 are now well-understood, the mechanism whereby PKM2 drives tumurogencity and the extent to which PKM2 alters glucose metabolism in cancer cells has remained enigmatic. However, recent studies showing non-catalytic functions of this enzyme have provided valuable insights towards answering these unsettled questions. Yang et al found that epidermal growth factor receptor (EGFR) activation induces nuclear translocation of PKM2, but not PKM1, where a lysine residue 433 of PKM2 binds to c-Src-phosphorylated Y333 of β-catenin (Yang et al., 2011). The authors showed that this is a requisite step for promoter recruitment of these two proteins to initiate cyclin D1 expression, which is fundamental to cell proliferation and brain tumor development. Luo et al found that PKM2, but not PKM1, interacts directly with the HIF-1α subunit and promotes transactivation of HIF-1 target genes by enhancing HIF-1 binding and p300 recruitment to hypoxia response elements (Luo et al., 2011). These authors further showed that PKM2 interacts with prolyl hydroxylase 3 (PHD3) to enhance PKM2 binding to HIF-1α as well as its co-activator functions. In addition, they showed that PHD3 knockdown not only reduces glucose uptake and lactate production but also increases oxygen consumption. Collectively, these studies show that the non-catalytic roles of PKM2 are also critical in reprogramming the metabolism of cancer cells and driving cell proliferation and tumor growth.
Based on recent studies showing that activating PKM2 may reverse the Warburg effect, valiant efforts have been made in recent years to develop activators of PKM2 that enhance the enzymatic activity of PKM2, towards potential cancer therapy. These activators were identified through quantitative high-throughput screening efforts of nearly 300,000 small-molecules of the NIH Molecular Libraries Small Molecule Repository using an ATP-generation assay coupled to luminescence (Boxer et al., 2010; Jiang et al., 2010; Walsh et al., 2011). These small-molecules primarily consist of N,N’-diarylsulfonamides, thieno[3,2-b]pyrrole[3,2-d]pyridazinones, and 2-oxo-N-aryl-1,2,3,4-tetrahydroquinoline-6-sulfonamides. One of these small-molecules, DASA-10, was recently shown to enhance cancer cell death under conditions of oxidative stress (Anastasiou et al., 2011). While activators of PKM2 activity have the potential to become promising therapeutic strategies for cancer, pharmacological or genetic therapies to downregulate PKM2 expression, thereby impairing the non-catalytic protein-protein interactions of PKM2, may also provide unique avenues for cancer therapy.
Aberrant Amino Acid and TCA Cycle Metabolism Underlying Cancer
In addition to altered glucose metabolism, cancer cells also exhibit fundamental alterations in amino acid metabolism that contribute to tumorigenicity. Metabolic flux analysis using either NMR or mass spectrometry is a powerful approach towards mapping altered metabolic flux in cancer cells by tracing the incorporation of 13C-labeled metabolites arising from 13C-glucose or 13C-glutamine treatment of cells (Figure 3). The metabolism of glutamine to lactate through “glutaminolysis”, has been shown to be a particularly important anaplerotic driving-force for cancer cell proliferation (DeBerardinis et al., 2008a). Anaplerosis refers to the replenishment of mitochondrial citric acid carbon pool. Glutamine can drive anaplerosis by providing the mitochondria with precursors for the synthesis of nucleotides, amino acids, and lipids. DeBerardinis et al utilized 13C-labeled nutrients (e.g. 13C-glucose or 13C-glutamine) coupled with 13C NMR spectroscopy as a method to selectively enrich metabolites in real time, allowing for the measurement of metabolic fluxes, towards understanding how biochemical pathways become rewired in cancer. Through these studies, DeBerardinis et al found that cancer cells utilize the tricarboxylic acid (TCA) cycle primarily to generate building blocks, in particular, citrate which is exported out of the mitochrondria to provide acetyl CoA for fatty acid and phospholipid biosynthesis (DeBerardinis et al., 2007) (Figure 1, 3). They also found that glutamine dependent anaplerosis, the process of glutamine replenishing TCA cycle intermediates, satisfied the large cellular demand for reducing power (NAPDH generation) and oxaloacetate renewal (via conversion of glutamine to α-ketoglutarate) that is necessary for continued (DeBerardinis et al., 2007) fatty acid synthesis and TCA flux.
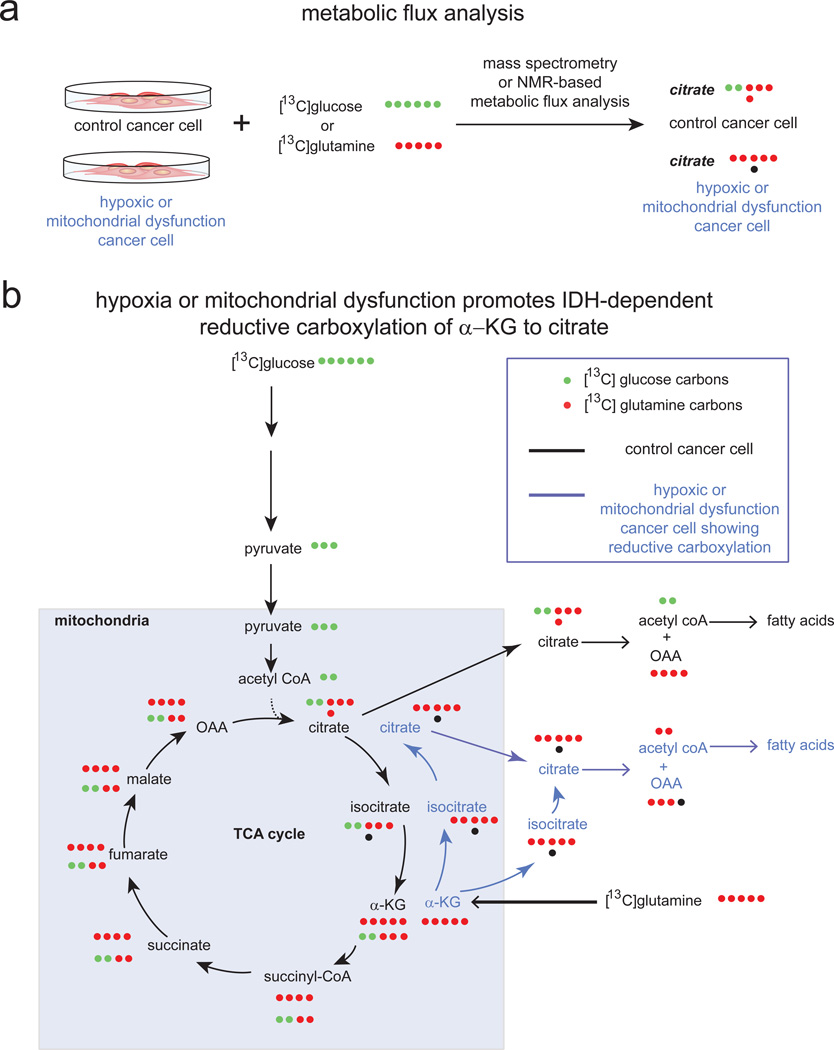
Figure 3. Metabolic Flux Analysis Reveals Dysregulated Cancer Cell Metabolism under Hypoxia or Mitochondrial Dysfunction.
a. Recent studies have performed metabolic flux analysis on cancer cells labeled with [13C]glucose or [13C]glutamine to dissect dysregulated metabolism of cancer cells exposed to hypoxia or mitochondrial dysfunction. b. These studies revealed that cancer cells under normoxia can form citrate through both glycolysis (bottom row of labeled carbons in TCA cycle) and glutaminolysis (top row of labeled carbons in TCA cycle). Upon hypoxia or mitochondrial dysfunction, cancer cells undergo a metabolic switch in citrate is produced through glutamine-dependent reductive carboxylation of α–ketoglutarate (α–KG) to form isocitrate via IDH1 or IDH2 and then to citrate to support de novo lipogenesis (blue arrows). Shown in the figure are [13C]-labeled carbons arising either from glucose (in green) or glutamine (in red) after one pass through the TCA cycle. Carbon atoms arising from CO2 are labeled as black dots.
Another route for the production of α-ketoglutarate within the cells is via a reaction catalyzed by the TCA cycle enzyme isocitrate dehydrogenase1 (IDH1). IDH1 catalyzes the oxidative decarboxylation of isocitrate to α-ketoglutarate with concomitant reduction of NADP+ to NADPH. A genome wide analysis consisting of sequencing protein-coding genes and RNA sequencing of glioma and acute myeloid leukemia patients identified mutations in the active site of the enzyme IDH1 as a common feature of a major subset of low-grade gliomas and secondary glioblastomas and leukemic cancers (Mardis et al., 2009; Parsons et al., 2008). These mutations were found to occur at a single amino acid residue of IDH1, arginine 132, which is most commonly mutated to histidine. In an effort to understand the effects of such a mutation on the cellular metabolome, Dang et al stably transfected U87MG glioblastoma cells with both myc-tagged wild-type IDH1 as well as R132 mutant IDH1 and compared the metabolomes of these cells using an untargeted liquid chromatography/mass-spectrometry (LC/MS) based untargeted metabolomic profiling approach, which allows for an unbiased comparison of metabolites altered between two groups. The authors identified a novel oncometabolite 2-hydroxyglurarate (2-HG) that was dramatically increased in cells that contained this R132 mutation in IDH1 (Dang et al., 2009a). Surprisingly, while the wild type IDH1 produced α-ketoglutarate and NADPH, in vitro assays revealed that the R132 mutant IDH1 consumed NADPH and reduced α-ketoglutarate to 2-HG (Dang et al., 2009a) (Figure 1). These studies provided the first evidence for a mutated enzyme in cancer conferring, not only a loss of endogenous function, but also a neomorphic function to yield an unforeseen metabolite, and underscored the utility of using unbiased and untargeted metabolomic approaches to study cancer metabolism.
Further metabolomic profiling by Reitman et al revealed that both mutation in IDH1 and separate treatment with 2-HG resulted in a host of downstream metabolic changes including changes in amino acids, glutathione metabolites, choline derivatives, and TCA cycle intermediates (Reitman et al., 2011). Specifically, using targeted LC-MS/MS they also found that the N-acetylated amino acids NAA and NAAG (most commonly found as dipeptides in the brain) were significantly lowered in cells containing a mutated IDH1 (Reitman et al., 2011).
In addition to its role in eliciting direct metabolic changes, IDH1 derived 2-HG has also recently been shown to play an epigenetic role in cancer, primarily through an increase in CpG island methylation. Using a large scale genome-wide analysis of DNA methylation, Noushmehr et al profiled promoter DNA methylation alterations in 272 glioblastoma tumors and found that a distinct subset of these glioblastomas were positive for this CpG island methylator phenotype (G-CIMP) (Noushmehr et al., 2010). Strikingly, these G-CIMP-positive samples were tightly correlated with mutations in IDH1. Figueroa et al further extended this correlation between IDH1 mutation and DNA methylation to other cancers types as they showed that IDH1/IDH2 mutant AML is associated with more extensive promoter hypermethylation compared to other AML subtypes (Figueroa et al., 2010). The mechanism underlying the role of 2-HG in driving DNA methylation is still unclear, however recent evidence from Xu et al has shown that 2-HG can act as a competitive inhibitor of α-ketoglutarate dependent demethylases, including histone demethylases and the TET family of 5-methylcytosine hydroxylases, thereby explaining this correlation between IDH1 derived 2-HG and CpG island hypermethylation (Xu et al., 2011). Recent studies have also shown that mutations in IDH1 are sufficient to establish the glioma hypermethylator phenotype, showing that introduction of mutant IDH1 into primary human astrocytes alters specific histone marks, induces extensive DNA hypermethylation, and reshapes the methylome in a similar manner to the changes observed in glioma CpG island methylator phenotype-positive lower-grade gliomas (Turcan et al., 2012). Intriguingly, Lu et al also showed that introduction of mutant IDH into immortalized astroyctes resulted in progressive accumulation of histone methylation. They also showed that 2-HG was associated with repression of the inducible expression of lineage-specific differentiation genes in adipocytes and blocked differentiation through inhibiting H3K9 demethylase KDM4C, and that genetic knockdown of this enzyme was sufficient to block differentiation (Lu et al., 2012).
Interestingly, recent studies have shown that under hypoxia or mitochondrial dysfunction, cancer cells undergo a switch in which citrate, an important lipogenic precursor, is produced not from glycolytic carbon, but primarily from glutamine via reductive carboxylation of α–ketoglutarate to isocitrate via IDH1 or IDH2 (Metallo et al., 2012; Mullen et al., 2012; Scott et al., 2011; Wise et al., 2011). These studies mapped this reductive carboxylation pathway using metabolic flux analysis measuring incorporation of 13C-labeled carbons arising from 13C-glucose or 13C-glutamine labeling in cells by mass spectrometry or NMR. These studies also showed that knocking down IDH2 impairs cell proliferation (Wise et al., 2011). Furthermore, Metallo et al showed that cancer cells deficient in the Von Hippel-Lindau (VHL) tumor suppressor, a protein frequently lost in renal cell carcinoma, preferentially utilize reductive glutamine metabolism for lipogenesis even under normoxic conditions. In a separate study, Wise et al intriguingly showed that 2-HG, originally thought to arise only from mutated neomorphic IDH, can in fact be formed in cells possessing wild-type IDH1 and IDH2 also through reductive metabolism by IDH2 in the mitochondria (Wise et al., 2011).
Collectively, mass spectrometry and NMR-based metabolomic studies have provided valuable insights into how cancer cells undergo a metabolic switch that consists of glutamine-dependent anaplerotic pathways that can convert α-ketoglutarate through either wild-type or mutated IDH1 and IDH2, to isocitrate through reductive mechanisms, which in-turn can support lipid biosynthesis to fuel cancer cell proliferation. Untargeted and unbiased metabolomic approaches have also uncovered neomorphic roles associated with mutant IDH which generates the oncometabolite 2-HG. Subsequent studies have shown this oncometabolite to be demethylase inhibitor, leading to epigentic changes that may drive cancer.
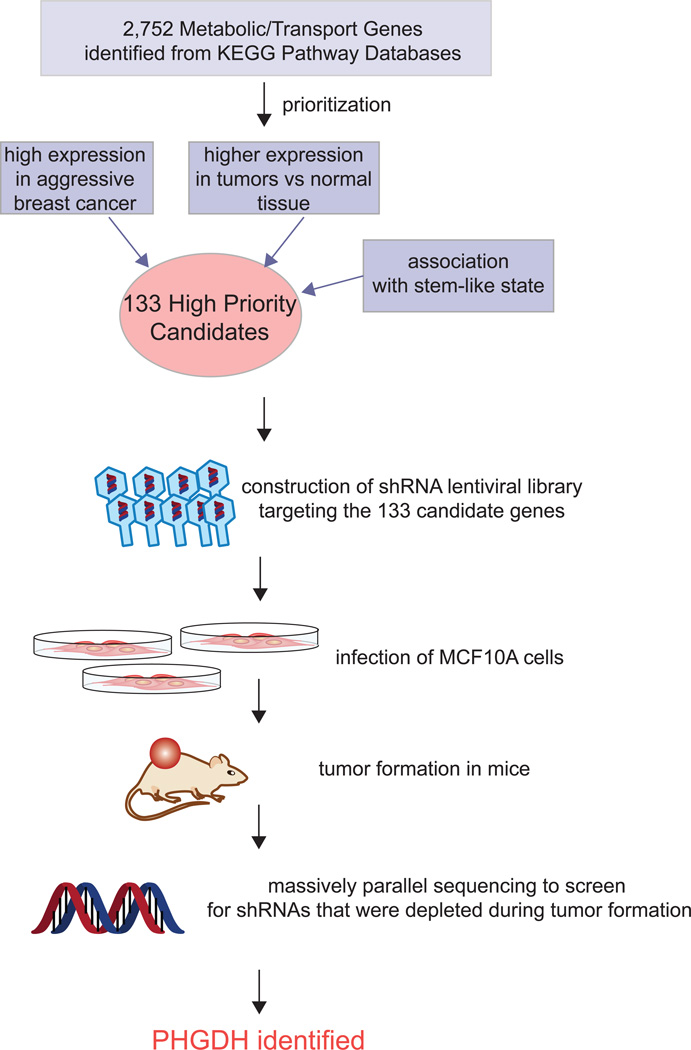
Recent experimental approaches have revealed that many other amino acid metabolic pathways are also dysregulated in cancer and contribute to cancer aggressiveness. Locasale et al found that in some cancer cells a relatively large amount of glycolytic carbon is diverted into serine and glycine metabolism (Locasale et al., 2011a) (Figure 1). They showed this using two independent metabolomic approaches. They first used sensitivity-enhanced nuclear magnetic resonance (NMR)-based two-dimensional heteronuclear single quantum correlation spectroscopy (HSQC) to quantify steady-state levels of glucose-derived metabolites in HEK293T cells following 24 h of labeling with 13C-glucose. While, as expected, lactate levels were extremely high, surprisingly, the glycolytic flux towards glycine was also equally high. In the second experimental approach, using targeted LC-MS/MS-based measurements of metabolites, they found that a substantial portion of 13C-glucose was diverted from 3-phosphoglycerate to the serine and glycine biosynthetic pathways, on par with incorporation of glucose carbons into nucleotides. The first committed step in this pathway is the oxidation of the glycolytic intermediate 3-phosphoglycerate to 3-phosphohydroxypyruvate by the enzyme phosphoglycerate dehydrogenase (PHGDH). Using a functional genomics approach, Locasale identified PHGDH as a frequently amplified gene in a pooled analysis of somatic copy number alterations across 3,131 cancer samples, most notably in melanoma cells(Locasale et al., 2011a). Concurrent with the study from Locasele et al, Possemato et al. devised an elegant negative selection in vivo RNAi genomics screen which also identified PHGDH as an important metabolic pathway in tumorigenesis and cell proliferation (Possemato et al., 2011) (Figure 4). They first cross-referenced maps of metabolic pathways with the KEGG database to compile a comprehensive list of 2,752 genes encoding all known human metabolic enzymes and transporters. From this list, genes were prioritized based on their association with cancer and stem cell-like properties to a set of 133 metabolic enzyme and transporter genes. They then screened for RNA interference oligonucleotides for each of these 133 genes that become depleted during breast tumor formation in mice. Sixteen hits were identified, including PHDGH. They further showed that PHGDH is in a genomic region of recurrent copy number gain in breast cancer and PHGDH protein levels are elevated in 70% of estrogen receptor (ER)-negative breast cancers. They further demonstrated that knocking down PHGDH reduces cell proliferation in breast cancer cells. Through measurement of steady-state levels and incorporation of isotopic 13C-glutamine in cells followed by targeted LC/MS analysis, the authors also found that PHGDH knockdown results in deficiencies in the levels of multiple TCA cycle intermediates and anaplerosis of glutamine to alpha-ketoglutarate (Possemato et al., 2011). They found that the serine synthesis pathway is responsible for approximately 50% of the net conversion of glutamine to alpha-ketoglutarate. Collectively, these two parallel studies using innovative genomic and metabolomic tools show how PHGDH promotes cancer pathogenicity by diverting glycolytic flux to serine and glycine biosynthetic pathways.
Figure 4. Functional Genomic approach to Discover PHGDH as an Important Metabolic Enzyme in Cancer.
Possemato et al used an elegant negative selection in vivo RNAi screen to identify metabolic enzymes necessary to support breast tumorigenesis. A prioritized list of 133 metabolic enzymes and metabolite-transport genes were individually knocked down by shRNA lentiviral constructs in MCF10a cells, combined together and collectively injected into mice. Upon tumor formation, the resultant cancers were screened by sequencing efforts to identify genes, for which the knockdown was selected against. From this functional genomic screen, a total of 16 genes were identified, one of which was PHGDH, involved in serine and glycine biosynthesis (Possemato et al., 2011).
Dysregulated Lipid Metabolism that Supports Cellular Membrane Biosynthesis and Oncogenic Lipid Signaling Pathways
Dysregulated lipid metabolism and heightened de novo lipogenesis are established hallmarks of cancer (Menendez and Lupu, 2007a; Wymann and Schneiter, 2008). Tumor cells synthesize fatty acids for the purpose of both membrane synthesis as well as for the generation of lipid signaling molecules to fuel cell proliferation and cancer malignancy (Menendez and Lupu, 2007b). These lipid signaling pathways that are critically important in driving almost every aspect of cancer progression, include phosphatidyl inositol, lysophosphatidic acid (LPA), sphingosine-1-phosphate, and prostaglandin signaling pathways (Engelman et al., 2006; Mills and Moolenaar, 2003; Pyne and Pyne, 2010; Wang and Dubois, 2010).
In 1994, Kuhajda et al discovered fatty acid synthase (FASN) as an oncogenic protein that was found in tumor cells from breast cancer patients with markedly poorer prognosis (Kuhajda et al., 1994). Since this time, understanding dysregulated lipid metabolism in cancer cells has gained considerable interest. Several studies have shown that genetic and pharmacological ablation of FASN leads to impairments in cancer pathogenicity, specifically cell cycle arrest and apoptosis (Menendez and Lupu, 2007b). Several mechanisms for FASN inhibition-based cytotoxicity have been proposed including phospholipid depletion, alterations in lipid rafts, inhibition in DNA replication, malonyl coA buildup and associated toxicity, and inhibition of anti-apoptotic proteins such as Akt. These studies have been reviewed previously and will not be discussed further in this perspective (Menendez and Lupu, 2007b). Common FASN inhibitors that have been used to study the role of FASN in cancer include cerulenin and its derivative C75, epigallocatechin-3-gallate, triclosan, and orlistat which target different regions of the FASN complex and elicit apoptotic cell death in cancer cells (Menendez and Lupu, 2007a). However, none of these inhibitors are likely very selective for FASN and are not suitable for clinical development (Vazquez et al., 2008). Newer pharmacological inhibitors of FASN that are significantly more potent and selective have arisen from the pharmaceutical industry including GSK837149A from GlaxoSmithKline, identified from an impurity found in an active hit from a high-throughput screen utilizing an NAPDH consumption assay (Vazquez et al., 2008). Merck has also recently identified the broad-spectrum antibiotic platensimycin as a selective mammalian fatty acid synthase inhibitor (Wu et al., 2011).
While the de-novo synthesis of lipids has clearly been shown to be essential to conferring cancer malignancy, the mobilization of esterified lipids is also necessary to remodel cellular lipids into protumorigenic lipid signaling molecules (Nomura et al., 2010b; Wymann and Schneiter, 2008). A recent study utilized a chemoproteomic platform termed activity based protein profiling (ABPP) to identify monoacylglycerol lipase (MAGL) as a highly expressed serine hydrolase in multiple human aggressive cancer cells and primary tumors (Nomura et al., 2011; Nomura et al., 2010b) (Figure 5). ABPP uses active-site directed chemical probes to interrogate the functional state of large numbers of enzymes directly in native complex proteomes (Nomura et al., 2010a). Because activity-based probes label the active sites of their enzyme targets, they can form the basis for a competitive screen for enzyme inhibitors. Long et al used this approach to develop a highly potent and selective inhibitor of MAGL, JZL184, previous to this study (Long et al., 2009). Pharmacological and genetic ablation of MAGL in aggressive cancer cells lines using JZL184 or RNA interference resulted in impairments in cancer cell aggressiveness and tumorigenicity, while the overexpression of MAGL in non-aggressive cancer enhanced cancer cell pathogenicity. Using a mass-spectrometry-based untargeted metabolomics approach termed discovery metabolite profiling (DMP), the authors then showed that MAGL blockade leads to elevations in the anti-tumorigenic endocannabinoid signaling lipid 2-arachidonoylglycerol and reductions in free fatty acid (FFA) levels in aggressive cancer cells which in-turn reduce the downstream levels of protumorigenic signaling lipids such as LPA and prostaglandins (Nomura et al., 2011; Nomura et al., 2010b) (Figure 1, Figure 5). The pathogenicity deficits observed by MAGL knockdown were rescued by addition of fatty acids, LPA, or PGE2 in vitro or a high-fat diet in vivo. These results taken together demonstrate how cancer cells can coopt a lipolytic enzyme to translate their lipogenic state into an array of pro-tumorigenic signals.
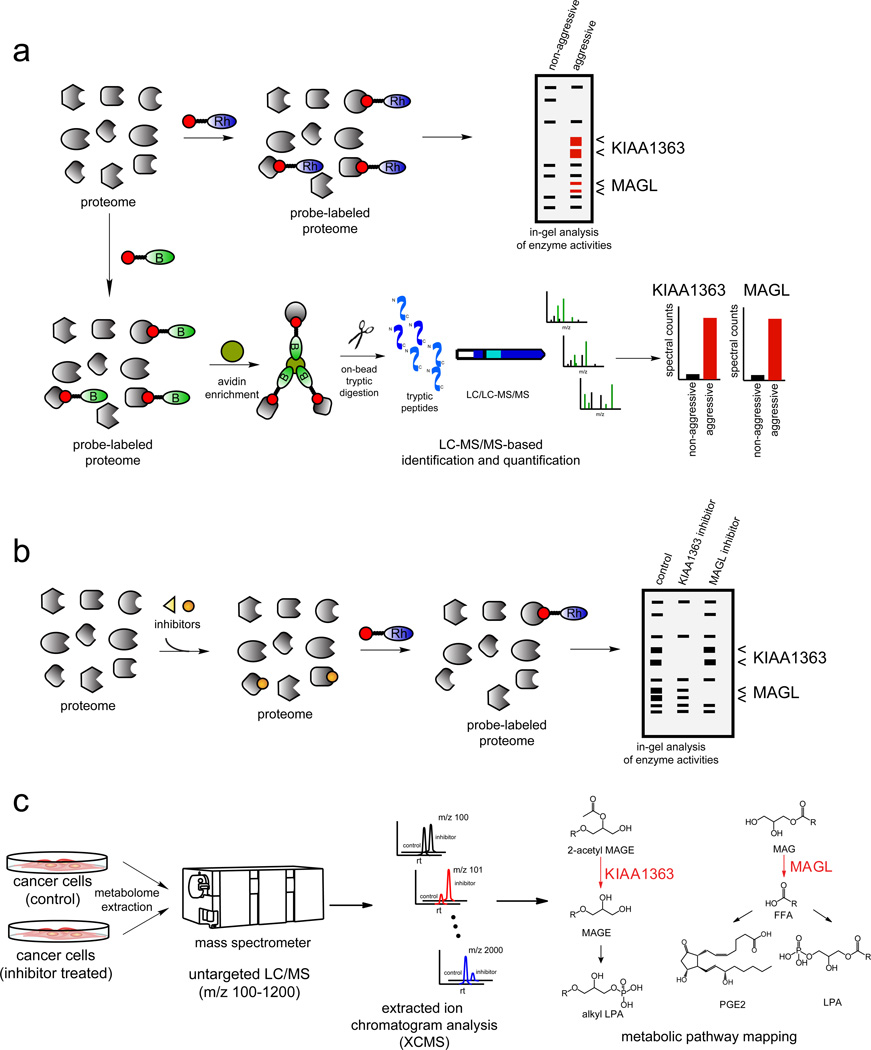
Figure 5. Activity-Based Protein Profiling (ABPP) Coupled with Untargeted Metabolomics in Annotating Dysregulated Enzyme Activities in Aggressive Cancers.
a) ABPP uses active site-directed probes to assess the functional state of large numbers of enzymes directly in complex proteomes (Nomura et al., 2010a). Activity-based probes (ABPs) consist of a reactive group, a spacer arm, and a detection handle (e.g. fluorophore such as a rhodamine (Rh) or biotin (B)). In a typical ABPP experiment, a proteome is reacted with the activity-based probe and readout either by fluorescence on a 1D-SDS-PAGE gel for rhodamine-ABPs (above), or by avidin enrichment, on-bead tryptic digest, and identification and quantification of peptides by Multidimensional Protein Identification Technology (MudPIT) for biotinylated-ABPs (below) (Nomura et al., 2010a). Through ABPP analysis of the serine hydrolase proteome with the serine hydrolase ABP fluorophosphonate (FP)-Rh or FP-biotin, KIAA1363 and MAGL were identified as upregulated in multiple human aggressive cancer cells and primary tumors (Jessani et al., 2002; Nomura et al., 2010b). b) ABPP can also be used in a competitive format to assess potency and selectivity of inhibitors in complex proteomes. Inhibitors can compete with the ABP and enzyme inhibition will be read-out by loss of fluorescence on a SDS-PAGE gel (using a rhodamine-ABP) or loss of spectral counts by mass spectrometry (using a biotintylated-ABP). Competitive ABPP was used to develop selective inhibitors of KIAA1363 and MAGL (Chang et al., 2011b; Chiang et al., 2006; Long et al., 2009). c) With selective inhibitors in hand, the metabolic roles of KIAA1363 and MAGL were elucidated using untargeted LC-MS-based metabolomic approaches in which metabolomes were extracted and analyzed by LCMS, broadly scanning for metabolites across a large mass range. To complement the large amount of data that arises from untargeted metabolomic analysis, powerful software tool XCMS was used for quantitation and identification of ions within LC/MS datasets (Smith et al., 2006) which aligns, quantifies, and statistically rank ions that are altered between two sets of metabolomic data. This methodology was used to annotate KIAA1363 and MAGL as enzymes that regulates ether lipid and fatty acid networks, respectively (Chiang et al., 2006; Nomura et al., 2010b).
Consistent with Nomura et al’s findings that an increase in dietary fat intake could potentially drive cancer cell pathogenesis, Kuemmerl et al were able to demonstrate that some cancer cells also rely on the presence of another lipolytic enzyme, lipoprotein lipase (LPL), to acquire fatty acids from circulating dietary triglycerides, thereby fueling their growth and proliferation. Using gene expression profiling methods, Kuemmerl et al revealed that select breast and prostate cancer cells express high levels of both LPL and the fatty acid transporter, CD36 (Kuemmerle et al., 2011). In the cells in which there was no substantial increase in LPL gene expression, they showed, through further transcriptomic analysis, that these cells maintained extremely high expression of genes that promote endogenous fatty acid synthesis, most notably FASN. Furthermore, upon pharmacological blockade of endogenous fatty acid synthesis, exogenous delivery of LPL along with triglyceride rich lipoprotein particles served to rescue the malignant phenotype that was previously lost upon inhibition of endogenous fatty acid production (Kuemmerle et al., 2011) (Figure 1). Nieman et al recently discovered that primary human omental adipocytes promote homing, migration, and invasion of ovarian cancer cells, through direct transfer of lipids from adipocytes to ovarian cancer cells to promote tumor growth. Through protein array profiling, the authors found an upregulation in fatty acid binding protein 4 (FABP4) in omental metastases as compared to primary ovarian tumors, and that FABP4 deficiency substantially impaired ovarian tumor metastasis in mice (Nieman et al., 2011). Taken together, these results indicate that cancer cells must increase their production of global fatty acids in one of 3 ways: endogenous production (via FASN), release of esterified fatty acids (via MAGL), or release and absorption of fatty acids from dietary sources (via LPL and FABP4).
In addition to fatty acid-derived glycerolipids, ether lipid metabolism has also been known to be dysregulated in cancers since the pioneering work performed in the 1960s by Snyder and Wood (Snyder and Wood, 1969; Wood and Snyder, 1967), but the metabolic pathways that drive this metabolic transition were not well understood. Studies utilizing ABPP platforms identified the serine hydrolase KIAA1363 as one such driver of ether lipid metabolism that is upregulated across multiple human aggressive cancer cells and primary tumors (Chiang et al., 2006; Jessani et al., 2005). Several potent and selective pharmacological carbamate inhibitors of KIAA1363 have been developed through competitive ABPP approaches, such as AS115 and JW480 as well as an activity-based imaging probe JW576, which are all based off of the carbamate scaffold (Chang et al., 2011a; Chang et al., 2011b; Chiang et al., 2006). Both pharmacological and genetic blockade of KIAA1363 has been shown to impair cancer cell migration, invasion, and in vivo tumorigenicity through lowering ether lipids and oncogenic signaling lipids such as alkyl lysophosphatidic acid (Chang et al., 2011b; Chiang et al., 2006) (Figure 5).
Challenges in the Field of (Cancer) Metabolism
The field of cancer metabolism has thus far focused on major commonly known biochemical pathways, such as glycolysis, TCA cycle, fatty acid synthesis, and amino acid metabolism (Lunt and Vander Heiden, 2011). While this has been critical at delineating the foundational structure of altered metabolism in cancer, looking towards the future, we need to expand our investigations and technologies to more comprehensively encompass different aspects of metabolism. This endeavor, however, is hindered by our incomplete understanding of the metabolic pathways that operate in normal human cells, let alone in cancerous ones. We also lack pharmacological tools to interrogate even very well-understood biochemical pathways, as evidenced by a dearth of specific inhibitors for enzymes in glycolysis or TCA cycle. Metabolic pathways are likely quite plastic and adaptable in cancer cells, so we view the development of selective chemical probes to perturb these networks in a rapid and temporally controlled manner as an important complementary approach to gene knockouts or knockdowns, which typically create models of prolonged target disruption.
Let’s consider a hypothetical situation that underscores the technical challenges that remain for researchers interested in understanding and controlling cancer metabolism. Presume that one finds through a genome or proteome-wide search a poorly characterized enzyme that highly correlated with cancer; how does one gain understanding of the biochemical function that this enzyme performs in tumor cells and integrate its activity into the larger biochemical and signaling networks that drive pathogenesis? In this review, we have presented instances where advanced proteomic and metabolomic approaches have achieved this goal. However, the metabolome rivals, if not surpasses, the genome and proteome in terms of its physicochemical content and complexity. As such, current analytical technologies, even when used in combination, likely do not approach providing a complete picture of the metabolome. Here, improvements in methods for metabolite enrichment, separation, and identification, as well as continued advances in MS and NMR platforms should enable researchers to delve deeper and deeper into the highly diversified metabolomic landscape of cells and tissues.
If a comprehensive and global understanding of metabolic pathways in cancer is to be achieved, the insights gained from metabolomic mapping of enzyme functions in cancer must be coupled to the assembly of biochemical reactions into pathways and larger metabolic networks. Essential to this process is mapping the metabolic flux through these pathways and networks, not only in situ in cells, but also in living systems or in live tumors in human patients and modeling these fluxes to identify critical nodes for therapeutic intervention (Wahrheit et al., 2011). There are indeed powerful strategies for measuring both in situ and in vivo metabolic flux using isotopic tracers including monitoring 13C-labeled substrates and their metabolic fates (Zamboni, 2011) or tracing D2O heavy water (Hellerstein and Murphy, 2004) in lipid, nucleic acid, and amino acid metabolism. While single isotopic tracers are an extremely valuable tool that can be used to probe metabolic networks, the application of multiple tracers simultaneously has provided an even more thorough understanding of metabolic flux. To this end, a recent paper by Walther et al has elegantly elucidated a method that allows researchers to determine the optimal tracer combination for a particular metabolic pathway (Walther et al., 2012). More recently, Hiller et al have developed a new methodological approach to trace stable isotopes in non-targeted metabolomics data (Hiller et al., 2011). These endeavors will have to be improved and further implemented in the study of cancer metabolism with better isotopic or chemical labeling strategies coupled with improvements in imaging, magnetic resonance, mass-spectrometry-based platforms as well as modeling tools, to better dissect cancer metabolism and use these approaches to diagnose and treat cancer patients.
Beyond technical challenges, there are also conceptual advances that must be made. Much of the field of cancer metabolism, for instance, has focused to date on pathways that are altered during the transformation process that initiates cancer. Less is understood about the metabolism that drives specific stages of tumor progression, malignancy, and eventually metastasis and drug-resistance. It is possible that even the same general metabolic pathway may contribute in different ways to individual stages of cancer. Take as an example lipogenesis. FASN is highly upregulated during the transformation process and contributes substantially to proliferation and survival, presumably by generating cellular membranes required for cell division, as well as possibly substrate for energy production by fatty acid oxidation. However, on its own, FASN does not appear to produce lipid changes that promote high malignancy (Migita et al., 2009). Upregulation of MAGL, on top of a high FASN background, on the other hand, may promote this conversion to an aggressive state by liberating fatty acids for the biosynthesis of pro-migratory signaling lipids (e.g., lysophospholipids, eicosanoids). Only by examining cancer cells (or tumors) at different stages of disease progression, as opposed to more classical comparisons of normal and transformed cells, can such examples of co-adapting metabolic pathways be uncovered.
Recent work also suggests that malignant progression of cancer is accompanied by an increased proportion of cancer stem/precursor cells (CSC) within the tumor (Pece et al., 2010) and that activation of the epithelial-to-mesenchymal transition (EMT) is associated with CSC properties, including expression of the stem cell-associated antigenic profile (e.g. CD44+/CD24−/low for breast cancer), self-renewal capabilities, resistance to conventional therapies, cancer aggressiveness, and poor clinical outcome for multiple tumor types (Chaffer and Weinberg, 2011; Gupta et al., 2009b; Pece et al., 2010; Sabbah et al., 2008). Several studies have linked EMT directly to CSCs and aggressive properties of cancer (Cordenonsi et al., 2011; Gupta et al., 2009a; Mani et al., 2008), and have shown that CSCs or cells that have undergone EMT are highly resistant to traditional cancer therapies(Barr et al., 2008; Buck et al., 2008; Zielske et al., 2010), indicating that current therapeutic strategies preferentially target the non-CSC population. Despite the potential importance of this CSC population, the metabolic alterations that underlie EMT or CSCs remain poorly understood, although emerging data suggest that human cancer cell lines possessing EMT/stem-like features also dislpay a uniuqe set of metabolic enzymes (Nomura et al., 2011). Disrupting these pathways with small molecule inhibitors or RNA-interference probes should inform on their specific contributions to the biochemistry and malignant properties of CSCs.
In addition to identifying dysregulated metabolic pathways that underlie cancer initiation and progression, understanding the mechanism by which these pathways become dysregulated will also be essential for the diagnosis and treatment of cancer patients. Identifying the upstream “master regulator” may provide nodes that can be targeted to shut down entire metabolic programs that drive cancer. For example, a recent study by Gaglio et al showed oncogenic Kras to be essential for the metabolic reprogramming of cancer cells by enhancing glycolytic activity, decreasing TCA flux, and increasing the utilization of glucose for anabolic means (Gaglio et al., 2011). More recently, a study by Ying et al showed that oncogenic Kras maintains pacreatic tumors through regulation of anabolic glucose metabolism, channeling glucose intermediates into both the hexosamine biosynthesis and pentose phosphate pathways (Ying et al., 2012). PHGDH copy number appears to be highly amplified in human cancers, which accounts for its subsequent metabolic and oncogenic effects (Locasale et al., 2011b). However, the mechanism behind what drives other dysregulated metabolic pathways are not as well understood, and some of these pathways are likely turned on by upstream signaling or post-translational mechanisms.
Beyond profiling metabolic enzymes implicated in transformation, stemness, malignancy, and metastasis, it still remains unclear in any of these instances whether the dyregulated enzymes, when pharmacologically targeted, will have a strong enough effect on disease progression to be therapeutic. Drug programs that target a metabolic enzyme would benefit from identifying genetically-defined cancer sub-types (or resistance phenotypes) that are mechanistically tied to the disrupted pathways, so that one can properly select patients for therapeutic testing. Such mechanistic links, in most cases, have yet to be established. Drugs that target cancer metabolism will also likely have to be combined with current chemotherapeutic regimens to exhibit or maximize therapeutic efficacy. The optimal combination of these drugs or whether we can reduce current chemotherapy dosing regimens when combined together with cancer metabolism-based therapy is not well understood. For malignancy-related enzymes, the problem may even more challenging since they may contribute principally to processes like metastasis, which are more difficult to monitor preclinically and clinically compared to simple tumor growth.
Conclusions
Ever since Warburg first proposed that cancer may be a disease defined by aberrant metabolism, scientists have sought to define the specific biochemical pathways that enable previously normal cells to take on a deviant tumorigenic state. This review has highlighted many of the unique discovery platforms and global profiling approaches that are leading the way to identify new biochemical nodes that contribute to cancer development and malignancy. These include multiple inhibitory controls on glycolysis (e.g. PKM2) that divert flux of glycolytic carbons into anaplerotic pathways for generation of biosynthetic precursors, such as amino acids through PHGDH, or redox potential through the pentose phosphate pathway. Also included is a fundamental rewiring of cancer cells to exert heightened utilization of other carbon sources such as glutamine to supply TCA cycle anaplerosis; in certain cases, this occurs through IDH1/2-mediated reductive carboxylation to generate intermediates such as citrate that fuel de novo lipogenesis through FASN. Beyond the synthesis of cellular membrane lipids required for proliferation, studies have also found that these lipids are mobilized by distinct enzymatic pathways in cancer cells (e.g. MAGL, LPL, KIAA1363) to create building blocks for protumorigenic signaling lipids and for use as an energy source. Quite intriguingly, certain enzymes can even acquire neomorphic activities through mutation, such as mutant IDH, which generate novel oncogenic biomolecules that have wide-spread metabolic and epigenetic consequences. These findings have arisen out of the integrated utilization of genomic (deep genome sequencing and RNA sequencing), proteomic (post-translational mapping and functional proteomics), and metabolomic (steady-state, targeted/untargeted, and metabolic flux analysis) approaches. We expect that the continued efforts to probe biochemical pathways in cancer cells using such large-scale profiling methods will strengthen our understanding of dysregulated metabolism in cancer and identify potential avenues for therapeutic intervention.
Acknowledgements
We thank the members of the Nomura and Cravatt laboratories for helpful discussion and critical reading of the manuscript. This work was supported by the National Institutes of Health (R00DA030908 (DKN, DIB)).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, Cantley LC. Inhibition of pyruvate kinase m2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr S, Thomson S, Buck E, Russo S, Petti F, Sujka-Kwok I, Eyzaguirre A, Rosenfeld-Franklin M, Gibson NW, Miglarese M, Epstein D, Iwata KK, Haley JD. Bypassing cellular EGF receptor dependence through epithelial-tomesenchymal-like transitions. Clin Exp Metastas. 2008;25:685–693. doi: 10.1007/s10585-007-9121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer MB, Jiang JK, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, Veith H, Leister W, Austin CP, Park HW, Inglese J, Cantley LC, Auld DS, Thomas CJ. Evaluation of Substituted N,N '-Diarylsulfonamides as Activators of the Tumor Cell Specific M2 Isoform of Pyruvate Kinase. J Med Chem. 2010;53:1048–1055. doi: 10.1021/jm901577g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck E, Eyzaguirre A, Rosenfeld-Franklin M, Thomson S, Mulvihill M, Barr S, Brown E, O'Connor M, Yao Y, Pachter J, Miglarese M, Epstein D, Iwata KK, Haley JD, Gibson NW, Ji QS. Feedback Mechanisms Promote Cooperativity for Small Molecule Inhibitors of Epidermal and Insulin-Like Growth Factor Receptors. Cancer research. 2008;68:8322–8332. doi: 10.1158/0008-5472.CAN-07-6720. [DOI] [PubMed] [Google Scholar]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nature reviews. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- Chaffer CL, Weinberg RA. A Perspective on Cancer Cell Metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- Chang JW, Moellering RE, Cravatt BF. An Activity-Based Imaging Probe for the Integral Membrane Hydrolase KIAA1363. Angew Chem Int Ed Engl. 2011a doi: 10.1002/anie.201107236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang JW, Nomura DK, Cravatt BF. A potent and selective inhibitor of KIAA1363/AADACL1 that impairs prostate cancer pathogenesis. Chemistry & biology. 2011b;18:476–484. doi: 10.1016/j.chembiol.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chemistry & biology. 2006;13:1041–1050. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008a;452 doi: 10.1038/nature06734. 230-U274. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008b;452 doi: 10.1038/nature06667. 181-U127. [DOI] [PubMed] [Google Scholar]
- Cordenonsi M, Zanconato F, Azzolin L, Forcato M, Rosato A, Frasson C, Inui M, Montagner M, Parenti AR, Poletti A, Daidone MG, Dupont S, Basso G, Bicciato S, Piccolo S. The Hippo Transducer TAZ Confers Cancer Stem Cell-Related Traits on Breast Cancer Cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Heiden MGV, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009a;462 doi: 10.1038/nature08617. 739-U752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009b;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2010;465:966. doi: 10.1038/nature09132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell metabolism. 2008a;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. P Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Current opinion in genetics & development. 2008b;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet. 2006;7:606–619. doi: 10.1038/nrg1879. [DOI] [PubMed] [Google Scholar]
- Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li YS, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun ZX, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Lowenberg B, Licht JD, Godley LA, Delwel R, Valk PJM, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 Mutations Result in a Hypermethylation Phenotype, Disrupt TET2 Function, and Impair Hematopoietic Differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglio D, Metallo CM, Gameiro PA, Hiller K, Danna LS, Balestrieri C, Alberghina L, Stephanopoulos G, Chiaradonna F. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol 7. 2011 doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nature Medicine. 2009a;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Onder TT, Jiang GZ, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of Selective Inhibitors of Cancer Stem Cells by High-Throughput Screening. Cell. 2009b;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellerstein MK, Murphy E. Stable isotope-mass spectrometric measurements of molecular fluxes in vivo: emerging applications in drug development. Current opinion in molecular therapeutics. 2004;6:249–264. [PubMed] [Google Scholar]
- Hiller K, Metallo C, Stephanopoulos G. Elucidation of Cellular Metabolism Via Metabolomics and Stable-Isotope Assisted Metabolomics. Current pharmaceutical biotechnology. 2011;12:1075–1086. doi: 10.2174/138920111795909096. [DOI] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, Xie J, Gu TL, Polakiewicz RD, Roesel JL, Boggon TJ, Khuri FR, Gilliland DG, Cantley LC, Kaufman J, Chen J. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Science signaling. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessani N, Liu Y, Humphrey M, Cravatt BF. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. P Natl Acad Sci USA. 2002;99:10335–10340. doi: 10.1073/pnas.162187599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3rd, Jeffrey SS, Cravatt BF. A streamlined platform for high-content functional proteomics of primary human specimens. Nature methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- Jiang JK, Boxer MB, Vander Heiden MG, Shen M, Skoumbourdis AP, Southall N, Veith H, Leister W, Austin CP, Park HW, Inglese J, Cantley LC, Auld DS, Thomas CJ. Evaluation of thieno[3,2-b]pyrrole[3,2-d]pyridazinones as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett. 2010;20:3387–3393. doi: 10.1016/j.bmcl.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuemmerle NB, Rysman E, Lombardo PS, Flanagan AJ, Lipe BC, Wells WA, Pettus JR, Froehlich HM, Memoli VA, Morganelli PM, Swinnen JV, Timmerman LA, Chaychi L, Fricano CJ, Eisenberg BL, Coleman WB, Kinlaw WB. Lipoprotein Lipase Links Dietary Fat to Solid Tumor Cell Proliferation. Mol Cancer Ther. 2011;10:427–436. doi: 10.1158/1535-7163.MCT-10-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhajda FP, Jenner K, Wood FD, Hennigar RA, Jacobs LB, Dick JD, Pasternack GR. Fatty-Acid Synthesis - a Potential Selective Target for Antineoplastic Therapy. P Natl Acad Sci USA. 1994;91:6379–6383. doi: 10.1073/pnas.91.14.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, Vander Heiden MG. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature genetics. 2011a;43:869–874. doi: 10.1038/ng.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW, Grassian AR, Melman T, Lyssiotis CA, Mattaini KR, Bass AJ, Heffron G, Metallo CM, Muranen T, Sharfi H, Sasaki AT, Anastasiou D, Mullarky E, Vokes NI, Sasaki M, Beroukhim R, Stephanopoulos G, Ligon AH, Meyerson M, Richardson AL, Chin L, Wagner G, Asara JM, Brugge JS, Cantley LC, Vander Heiden MG. Phosphoglycerate dehydrogenase diverts glycolytic flux and contributes to oncogenesis. Nature genetics. 2011b;43 doi: 10.1038/ng.890. 869-U879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JZ, Li W, Booker L, Burston JJ, Kinsey SG, Schlosburg JE, Pavon FJ, Serrano AM, Selley DE, Parsons LH, Lichtman AH, Cravatt BF. Selective blockade of 2-arachidonoylglycerol hydrolysis produces cannabinoid behavioral effects. Nature chemical biology. 2009;5:37–44. doi: 10.1038/nchembio.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O'Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483 doi: 10.1038/nature10860. 474-U130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annual review of cell and developmental biology. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Luo WB, Hu HX, Chang R, Zhong J, Knabel M, O'Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate Kinase M2 Is a PHD3-Stimulated Coactivator for Hypoxia-Inducible Factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Li D, Zhao D, Lin RT, Chu YJ, Zhang H, Zha ZY, Liu Y, Li Z, Xu YP, Wang G, Huang YR, Xiong Y, Guan KL, Lei QY. Acetylation Targets the M2 Isoform of Pyruvate Kinase for Degradation through Chaperone-Mediated Autophagy and Promotes Tumor Growth. Mol Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du FY, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ, Schindler JK, Pohl CS, Wallis JW, Shi XQ, Lin L, Schmidt H, Tang YZ, Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD, Nagarajan R, Link DC, Walter MJ, Graubert TA, DiPersio JF, Wilson RK, Ley TJ. Recurring Mutations Found by Sequencing an Acute Myeloid Leukemia Genome. New Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nature reviews. Cancer. 2007a;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nature Reviews Cancer. 2007b;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, Jewell CM, Johnson ZR, Irvine DJ, Guarente L, Kelleher JK, Vander Heiden MG, Iliopoulos O, Stephanopoulos G. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migita T, Ruiz S, Fornari A, Fiorentino M, Priolo C, Zadra G, Inazuka F, Grisanzio C, Palescandolo E, Shin E, Fiore C, Xie W, Kung AL, Febbo PG, Subramanian A, Mucci L, Ma J, Signoretti S, Stampfer M, Hahn WC, Finn S, Loda M. Fatty acid synthase: a metabolic enzyme and candidate oncogene in prostate cancer. Journal of the National Cancer Institute. 2009;101:519–532. doi: 10.1093/jnci/djp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills GB, Moolenaar WH. The emerging role of lysophosphatidic acid in cancer. Nature Reviews Cancer. 2003;3:582–591. doi: 10.1038/nrc1143. [DOI] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman KM, Kenny HA, Penicka CV, Ladanyi A, Buell-Gutbrod R, Zillhardt MR, Romero IL, Carey MS, Mills GB, Hotamisligil GS, Yamada SD, Peter ME, Gwin K, Lengyel E. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17 doi: 10.1038/nm.2492. 1498-U1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nature reviews. Cancer. 2010a;10:630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Lombardi DP, Chang JW, Niessen S, Ward AM, Long JZ, Hoover HH, Cravatt BF. Monoacylglycerol lipase exerts dual control over endocannabinoid and fatty acid pathways to support prostate cancer. Chemistry & biology. 2011;18:846–856. doi: 10.1016/j.chembiol.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, Cravatt BF. Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell. 2010b;140:49–61. doi: 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RGW, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K, Network CGAR. Identification of a CpG Island Methylator Phenotype that Defines a Distinct Subgroup of Glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Jones S, Zhang XS, Lin JCH, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, Olivi A, McLendon R, Rasheed BA, Keir S, Nikolskaya T, Nikolsky Y, Busam DA, Tekleab H, Diaz LA, Hartigan J, Smith DR, Strausberg RL, Marie SKN, Shinjo SMO, Yan H, Riggins GJ, Bigner DD, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. An integrated genomic analysis of human glioblastoma Multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and Molecular Heterogeneity of Breast Cancers Correlates with Their Cancer Stem Cell Content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Possemato R, Marks KM, Shaul YD, Pacold ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, Chen WW, Barrett FG, Stransky N, Tsun ZY, Cowley GS, Barretina J, Kalaany NY, Hsu PP, Ottina K, Chan AM, Yuan B, Garraway LA, Root DE, Mino-Kenudson M, Brachtel EF, Driggers EM, Sabatini DM. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature. 2011;476 doi: 10.1038/nature10350. 346-U119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne NJ, Pyne S. Sphingosine 1-phosphate and cancer. Nature Reviews Cancer. 2010;10:489–503. doi: 10.1038/nrc2875. [DOI] [PubMed] [Google Scholar]
- Reitman ZJ, Jin GL, Karoly ED, Spasojevic I, Yang JA, Kinzler KW, He YP, Bigner DD, Vogelstein B, Yan H. Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. P Natl Acad Sci USA. 2011;108:3270–3275. doi: 10.1073/pnas.1019393108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah M, Emami S, Redeuilh G, Julien S, Prevost G, Zimber A, Ouelaa R, Bracke M, De Wever O, Gespach C. Molecular signature and therapeutic perspective of the epithelial-to-mesenchymal transitions in epithelial cancers. Drug Resist Update. 2008;11:123–151. doi: 10.1016/j.drup.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Scott DA, Richardson AD, Filipp FV, Knutzen CA, Chiang GG, Ronai ZA, Osterman AL, Smith JW. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. The Journal of biological chemistry. 2011;286:42626–42634. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical chemistry. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- Snyder F, Wood R. Alkyl and alk-1-enyl ethers of glycerol in lipids from normal and neoplastic human tissues. Cancer research. 1969;29:251–257. [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AWM, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LGT, Huse JT, Mellinghoff IK, Chan TA. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483 doi: 10.1038/nature10866. 479-U137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez MJ, Leavens W, Liu RG, Rodriguez B, Read M, Richards S, Winegar D, Dominguez JM. Discovery of GSK837149A, an inhibitor of human fatty acid synthase targeting the beta-ketoacyl reductase reaction. Febs J. 2008;275:1556–1567. doi: 10.1111/j.1742-4658.2008.06314.x. [DOI] [PubMed] [Google Scholar]
- Wahrheit J, Nicolae A, Heinzle E. Eukaryotic metabolism: measuring compartment fluxes. Biotechnology journal. 2011;6:1071–1085. doi: 10.1002/biot.201100032. [DOI] [PubMed] [Google Scholar]
- Walsh MJ, Brimacombe KR, Veith H, Bougie JM, Daniel T, Leister W, Cantley LC, Israelsen WJ, Vander Heiden MG, Shen M, Auld DS, Thomas CJ, Boxer MB. 2-Oxo-N-aryl-1,2,3,4-tetrahydroquinoline–6-sulfonamides as activators of the tumor cell specific M2 isoform of pyruvate kinase. Bioorg Med Chem Lett. 2011;21:6322–6327. doi: 10.1016/j.bmcl.2011.08.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther JL, Metallo CM, Zhang J, Stephanopoulos G. Optimization of C-13 isotopic tracers for metabolic flux analysis in mammalian cells. Metab Eng. 2012;14:162–171. doi: 10.1016/j.ymben.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang DZ, Dubois RN. Eicosanoids and cancer. Nature Reviews Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, Dematteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. P Natl Acad Sci USA. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood R, Snyder F. Characterization and identification of glyceryl ether diesters present in tumor cells. Journal of lipid research. 1967;8:494–500. [PubMed] [Google Scholar]
- Wu M, Singh SB, Wang J, Chung CC, Salituro G, Karanam BV, Lee SH, Powles M, Ellsworth KP, Lassman ME, Miller C, Myers RW, Tota MR, Zhang BB, Li C. Antidiabetic and antisteatotic effects of the selective fatty acid synthase (FAS) inhibitor platensimycin in mouse models of diabetes. P Natl Acad Sci USA. 2011;108:5378–5383. doi: 10.1073/pnas.1002588108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymann MP, Schneiter R. Lipid signalling in disease. Nature reviews. Molecular cell biology. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu LX, Jiang WQ, Liu J, Zhang JY, Wang B, Frye S, Zhang Y, Xu YH, Lei QY, Guan KL, Zhao SM, Xiong Y. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of alpha-Ketoglutarate-Dependent Dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WW, Xia Y, Ji HT, Zheng YH, Liang J, Huang WH, Gao X, Aldape K, Lu ZM. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480 doi: 10.1038/nature10598. 118-U289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying HQ, Kimmelman AC, Lyssiotis CA, Hua SJ, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang HL, Coloff JL, Yan HY, Wang W, Chen SJ, Viale A, Zheng HW, Paik JH, Lim C, Guimaraes AR, Martin ES, Chang J, Hezel AF, Perry SR, Hu J, Gan BY, Xiao YH, Asara JM, Weissleder R, Wang YA, Chin L, Cantley LC, DePinho RA. Oncogenic Kras Maintains Pancreatic Tumors through Regulation of Anabolic Glucose Metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni N. 13C metabolic flux analysis in complex systems. Current opinion in biotechnology. 2011;22:103–108. doi: 10.1016/j.copbio.2010.08.009. [DOI] [PubMed] [Google Scholar]
- Zielske SP, Spalding AC, Lawrence TS. Loss of tumor-initiating cell activity in cyclophosphamide-treated breast xenografts. Transl Oncol. 2010;3:149–152. doi: 10.1593/tlo.09307. [DOI] [PMC free article] [PubMed] [Google Scholar]