Abstract
The accumulation of the β-amyloid peptide (Aβ) in Alzheimer's disease (AD) is thought to play a causative role in triggering synaptic dysfunction in neurons, leading to their eventual demise through apoptosis. Aβ is produced and secreted upon sequential cleavage of the amyloid precursor protein (APP) by β-secretases and γ-secretases. However, while Aβ levels have been shown to be increased in the brains of AD patients, little is known about how the cleavage of APP and the subsequent generation of Aβ is influenced, or whether the cleavage process changes over time. It has been proposed that Aβ can bind APP and promote amyloidogenic processing of APP, further enhancing Aβ production. Proof of this idea has remained elusive because a clear mechanism has not been identified, and the promiscuous nature of Aβ binding complicates the task of demonstrating the idea. To work around these problems, we used an antibody-mediated approach to bind and cross-link cell-surface APP in cultured rat primary hippocampal neurons. Here we show that cross-linking of APP is sufficient to raise the levels of Aβ in viable neurons with a concomitant increase in the levels of the β-secretase BACE1. This appears to occur as a result of a sorting defect that stems from the caspase-3-mediated inactivation of a key sorting adaptor protein, namely GGA3, which prevents the lysosomal degradation of BACE1. Together, our data suggest the occurrence of a positive pathogenic feedback loop involving Aβ and APP in affected neurons possibly allowing Aβ to spread to nearby healthy neurons.
Introduction
The progressive accumulation and deposition of the β-amyloid peptide (Aβ) leading to senile plaques is an invariant feature of Alzheimer's disease (AD). Processing of the amyloid precursor protein (APP; Kang et al., 1987) by β-secretases and γ-secretases leading to the production of Aβ is a well characterized process (reviewed by Walsh et al., 2007). Although APP is conventionally thought to exist and function as a monomer, biochemical and structural data suggest that APP may exist as functional dimeric or oligomeric complexes (Rossjohn et al., 1999; Scheuermann et al., 2001; Wang and Ha, 2004; Chen et al., 2006).
APP contains three GxxxG motifs at the junction between the juxtamembrane and transmembrane sequences (Liu et al., 2005; Marchesi, 2005; Sato et al., 2006). These motifs mediate sequence-specific dimerization between transmembrane helices by direct glycine–glycine contacts (Bormann et al., 1989; Lemmon et al., 1994) facilitating oligomerization of many transmembrane proteins (Russ and Engelman, 2000), including APP (Munter et al., 2007; Kienlen-Campard et al., 2008). APP homotypic and heterotypic interactions have traditionally been implicated in cellular adhesion (Soba et al., 2005), but APP multimerization has also been proposed to play a factor in the pathogenesis of AD by driving the overproduction of the Aβ (Scheuermann et al., 2001; Kaden et al., 2008). Interestingly, one of the identified mutations (Flemish) in early-onset AD involves an alanine-to-glycine substitution, creating a fourth GxxxG motif in APP (Hendriks et al., 1992). Additionally, inducing dimerization of APP by introducing cysteine residues within the Aβ domain of APP (K28C and L17C) have been found to dramatically increase Aβ production (Scheuermann et al., 2001). Conversely, disrupting dimerization either by mutating a glycine in the GxxxG motifs to an alanine (Munter et al., 2010) or by chemical means (Richter et al., 2010) has been shown to attenuate Aβ production. However, it should be noted that others have reported the opposite results, either with the K28C mutation (Ren et al., 2007) or by using a chimeric APP construct (Eggert et al., 2009).
These conflicting studies led us to revisit the issue in a native system, using endogenous APP in cultured rat primary hippocampal neurons (PHNs). Because a definitive ligand for APP has not been identified, we used a commercially available monoclonal antibody (22C11) raised against the extracellular domain (amino acids 66–81) of APP (Hilbich et al., 1993) to cross-link cell-surface APP and mimic Aβ binding of APP. Here, we show that multimerization of APP triggers the amyloidogenic pathway in viable neurons, resulting in increased Aβ production with a concomitant increase in BACE1 levels. This appears to be the result of decreased lysosomal degradation of BACE1 due to a sorting defect that allows BACE1 to accumulate in recycling endosomes. Our data suggest that aberrant APP signaling in affected neurons may play an important role in not only triggering but also propagating the amyloidogenic pathway to other neurons as well.
Materials and Methods
Materials.
All chemicals used were of the highest grade available. Poly-d-lysine, chloroquine, ammonium chloride (NH4Cl), and drebrin A rabbit antibody (rAb) were purchased from Sigma-Aldrich; APP N-terminal mouse antibody (mAb) 22C11, Aβ mAb 4G8, APP C-terminal mAb, secretory form of APPβ (sAPPβ) mAb, and neprilysin rAb from Millipore; PSD-95 rAb, GGA3 rAb, caspase-3 rAb, cleaved-caspase-3 rAb, and anti-myc-tag rAb from Cell Signaling Technology; BACE1 rAb, insulin-degrading enzyme (IDE) rAb, EEA1 mAb, and GAPDH mAb from Abcam; ERK1 rAb and Aph-1 rAb from Santa Cruz Biotechnology; dendra2 rAb from Evrogen; presenilin-1 (PS1) mAb from Abgent; and βIII-tubulin mAb TUJ1 from Covance.
Hippocampal neuron and cell line cultures.
Hippocampal neuron cultures from both male and female rats pups were prepared following a slightly modified version of the method of Brewer (Brewer et al., 1993). Hippocampal neurons were kept in culture at 37°C with 5% CO2 in Neurobasal medium (Invitrogen) with B27 supplement and Glutamax (Invitrogen) and plated at a density of 2.5 × 105 cell/ml on dishes coated with poly-l-lysine (Sigma-Aldrich). For our experiments, neurons were used after ∼14–21 d in vitro. Human embryonic kidney 293 (HEK293) and murine neuroblastoma B103 cells were cultured and maintained in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 10% FBS plus 5% horse serum (Invitrogen) respectively. For stably transfected B103 cells expressing APP, the media was supplemented with 50 μg/ml Geneticin (Invitrogen).
Diolistic labeling of neurons.
The protocol for diolistic labeling of neurons was adapted from Gan et al. (2000). Briefly, 100 mg of tungsten particles (1.1 μm diameter; Bio-Rad) were thoroughly precipitated with 5.0 mg of lipophilic dye (DiI; Invitrogen) and dissolved in 100 μl of methylene chloride. Dye-coated particles were shot 2–3 times into the cells using the Helios gene gun system (Bio-Rad) and left in 0.1 m PBS overnight to allow dye diffusion along neuronal processes. Cells were postfixed with 4% paraformaldehyde (Pierce) for 1 h to preserve staining, then mounted onto glass slides using Gel Mount (Biomeda) and stored at 4°C in the dark.
Spine imaging, quantification, and statistics.
Labeled neurons were imaged using the Laser Scanning Microscope (LSM) 510 Meta confocal microscope (Zeiss), equipped with 40× 1.3 NA and 100× 1.4 NA oil-immersion objectives. The NIH Image software program ImageJ was used the quantify diolistic-labeled cultured neurons. An average of 12 compressed images (20 μm thick) consisting of pyramidal neurons in the hippocampus were quantified for each treatment. Statistical analysis was performed using the statistics package GraphPad Prism.
Assessment of cell death.
Cell death was assayed using the LIVE/DEAD Cell Viability Assay kit (Invitrogen) following the manufacturer's protocol. After the appropriate treatments, cells were treated with a solution containing 2.0 μm calcein AM and 4.0 μm ethidium homodimer 1 in PBS for 30 min. Live cells are distinguished by the presence of ubiquitous intracellular esterase activity, determined by the enzymatic conversion of the virtually nonfluorescent cell-permeant calcein AM to the intensely fluorescent calcein.
Oligonucleotide delivery into cells.
Oligonucleotide sequences were custom ordered (Dharmacon) with a thiol functionality at the 5′-end (caspase-3 siRNA sequence: 5′Th-AGCCGAAACUCUUCAUCAUUU). Oligonucleotide stocks were solubilized in water at a 10 mm concentration. The delivery peptide penetratin-1 (MP Biomedicals) was cross-linked via a Cys–Cys bond to the desired oligonucleotide as previously described (Davidson et al., 2004).
γ-Secretase activity assay.
γ-Secretase activity in hippocampal neurons was detected as amyloid intracellular domain (AICD)-myc formation using as substrate a C100-myc construct from HEK293 cells transfected with a C100-myc plasmid. CHAPS-solubilized (20 mm HEPES, pH 7.0; 150 mm KCl; 2.0 mm EGTA; 1.0% CHAPS and 2× protease inhibitor mixture) HEK293 cells were incubated together with solubilized 22C11-treated and untreated hippocampal neuron membranes (1:1 ratio) at −20°C or 37°C, with or without the γ-secretase inhibitor, DAPT (1.0 μm), for 2 h. AICD fragments were detected by Western blot analysis using an anti-myc rAb (Hansson et al., 2006).
NucView caspase-3 activity assay.
Caspase-3 activity in hippocampal neurons was assessed with the NucView 488 caspase detection kit (Biotium) following the manufacturer's protocol. Briefly, hippocampal neurons cultured in chamber slides were treated with a 5.0 μm solution of the NucView caspase-3 substrate for 30 min. Cells were then fixed with 4.0% paraformaldehyde (Pierce), mounted, and observed under a fluorescence microscope using a fluorescein isothiocyanate filter.
Immunocytochemistry.
Hippocampal neurons grown in chamber slides were treated with 22C11 (100 ng/ml) only or 22C11 (100 ng/ml) and penetratin-1-linked siCasp3 (80 nm) for 8 h. After fixing with 4.0% paraformaldehyde (Pierce), cells were permeabilized with 0.5% Triton X-100 (Sigma-Aldrich) in PBS and incubated the anti-BACE1 rAb and anti-EEA1 mAb overnight. Anti-mouse and anti-rabbit secondary antibodies conjugated with Alexa Fluor dyes (Invitrogen) were used. Labeled neurons were imaged using the LSM 510 Meta confocal microscope (Zeiss), equipped with 40× 1.3 NA and a 100× 1.4 NA oil-immersion objectives. Scanning used two excitation lines of the argon laser (488 nm for Alexa Fluor-488, 568 nm for Alexa Fluor-568). Z-stacks were collected at 1.0 μm intervals and then compressed into a single image for analysis using Volocity Imaging software (Volocity Acquisition and Volocity Visualization; PerkinElmer).
Results
Antibody-mediated cross-linking of cell-surface APP triggers neuronal death and loss of dendritic spines in cultured hippocampal neurons
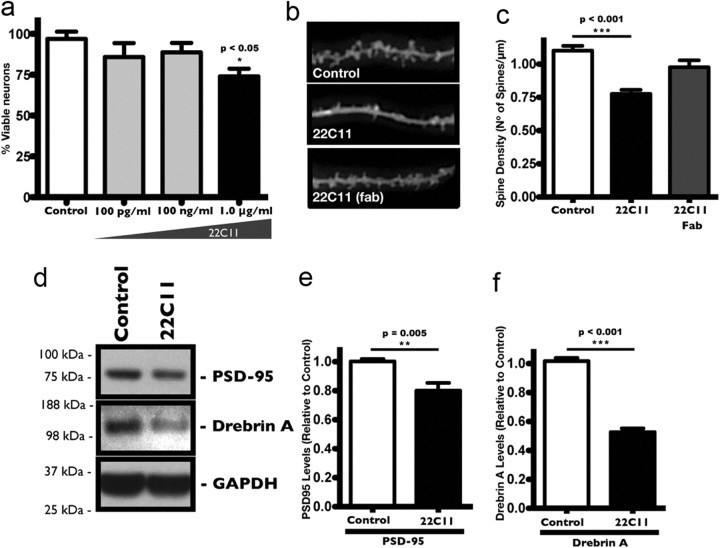
Prior studies using antibodies to cross-link APP have shown a significant amount of apoptotic death in cells exposed to high antibody concentrations (Rohn et al., 2000; Sudo et al., 2000). While neuronal death is invariably an important feature of AD, synaptic dysfunction is thought to be the earliest event in the progression of the disease (Delaère et al., 1989; Terry et al., 1991). We thus wanted to determine a concentration of the antibody sufficient to cause synaptic dysfunction, as measured by the loss of dendritic spines, without affecting their viability. To that end, we treated cultured rat PHNs with various concentrations of the monoclonal antibody, 22C11 (100 pg/ml, 100 ng/ml, and 1.0 μg/ml), and performed a direct count for cell viability after 24 h. We found that 22C11 triggered a significant amount of neuronal death (∼20% cell death) at 1.0 μg/ml, but not at lower concentrations of 100 ng/ml and 100 pg/ml (Fig. 1a). We next investigated the effects of lower concentrations of 22C11 on synaptic function by once again treating PHNs with 100 ng/ml 22C11 for 24 h. A direct count of the number of dendritic spines in diolistically labeled neurons (Fig. 1b) revealed a decrease in density, by as much as ∼40%, in 22C11-treated neurons compared with control neurons treated with PBS (Fig. 1c). The decline in dendritic spine density was accompanied by a corresponding reduction in the protein levels of the dendritic spine markers PSD-95 (Fig. 1d,e) and drebrin A (Fig. 1d,f) after the 24 h period. As a control for cross-linking, PHNs were treated with isolated monovalent Fab fragments of the APP antibody, which had no effect on spine density (Fig. 1c), suggesting that cross-linking of APP is required for its effects in neurons.
Figure 1.
Neurotoxic and synaptotoxic effects of APP cross-linking in cultured rat PHNs. a, Dose–response curve for 22C11 treatment of PHNs >24 h, showing a significant amount of cell death at 1.0 μg/ml compared with control PHNs treated with PBS. Lower concentrations (100 pg/ml and 100 ng/ml) had no significant neurotoxic effects after 24 h. Cell death was assessed with the commercially available LIVE/DEAD cell viability assay kit and quantified from at least three independent experiments and expressed as the mean ± SEM number of viable cells per field (*p < 0.05). b–f, Synaptic changes in PHNs treated for 24 h with a 100 ng/ml solution of intact 22C11 or isolated Fab fragments compared with control PHNs treated with PBS. Synaptic effects were assessed by a direct count of dendritic spine protrusions in diolistically labeled PHNs and by measuring the protein levels of two dendritic spine markers, PSD-95 and drebrin A. b, Representative images of diolistically labeled PHNs showing a decrease in dendritic spine protrusions in 22C11-treated compared with PBS-treated or Fab-treated PHNs. c, Dendritic spines were quantified from an average of 12 neurons taken from at least three independent experiments and expressed as the mean ± SEM number of spines/μm (**p = 0.009). d, Representative Western blots showing a decrease in the synaptic markers PSD-95 and drebrin A in 22C11-treated compared with PBS-treated PHNs. e, f, Densitometric analysis of PSD-95 and drebrin A levels was quantified from three independent experiments and expressed as the mean ± SEM band intensity (**p = 0.005 for PSD-95; ***p < 0.001 for drebrin A).
22C11 treatment increases Aβ levels in cultured hippocampal neurons without affecting cell viability
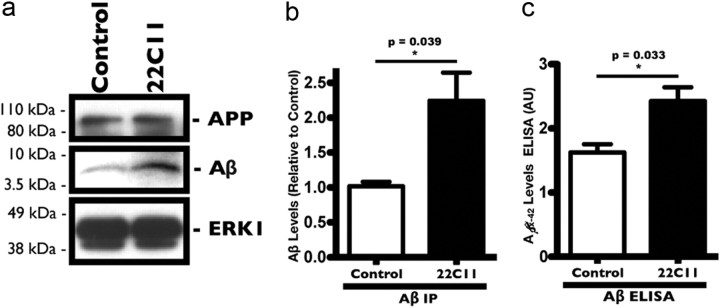
We next examined whether cross-linking of APP with subapoptotic levels of 22C11 increased the levels of Aβ in neurons. PHNs were treated with 100 ng/ml 22C11 for 24 h and harvested to measure the levels of neuronal Aβ by immunoprecipitation (IP) with a monoclonal antibody raised against the Aβ domain of APP, 4G8 (Fig. 2a). We detected a significant increase in Aβ production (∼2-fold) in 22C11-treated neurons compared with PBS-treated control neurons (Fig. 2b). There were no significant changes in the levels of full-length APP, either in 22C11-treated or in PBS-treated control neurons (Fig. 2a), suggesting that 22C11 treatment increases Aβ levels by directly promoting its production or by affecting its clearance. In a second approach, the levels of Aβ in lysates were measured using a commercially available ELISA kit, which measures the levels of Aβ(x-42). Consistent with our immunoprecipitation results, we found a significant increase in the levels of intraneuronal Aβ(x-42) in the 22C11-treated cells compared with PBS-treated control cells (Fig. 2c). Interestingly, we did not detect a significant change in the levels of secreted Aβ in the culture medium of either 22C11-treated neurons or of PBS-treated control cells, as measured by ELISA or IP (data not shown), suggesting that Aβ is accumulating intracellularly as a result of 22C11 treatment.
Figure 2.
Treatment with 22C11 increases Aβ levels in cultured rat PHNs. Treatment of PHNs for 24 h with a 100 ng/ml solution of 22C11 resulted in a significant increase in Aβ levels compared with control PHNs treated with PBS. Intracellular Aβ levels from PHN lysates were assessed by IP with the commercially available anti-Aβ antibody 4G8 and by ELISA. a, Representative Western blots from IP of Aβ showing an increase in Aβ levels in 22C11-treated compared with PBS-treated PHNs. b, Densitometric analysis of the levels of Aβ was quantified from at least three independent experiments and expressed as the mean ± SEM band intensity (**p = 0.039). c, Aβ(x-42) levels from 22C11-treated and PBS-treated PHN lysates measured by ELISA were quantified from four independent experiments, expressed as the mean ± SEM signal intensity in arbitrary units (AU) (*p = 0.033).
Increased Aβ levels after 22C11 treatment correlate with increased BACE1 levels and activity but not with increased γ-secretase activity
Our next objective was to determine the mechanism by which Aβ levels were increased in 22C11-treated neurons. Since the levels of APP remained unchanged in treated cells, we hypothesized that the changes in Aβ levels were due to a perturbation in the equilibrium between its clearance and production rates. Several studies have implicated the metalloproteases neprilysin and IDE in the degradation of Aβ (for review, see Miners et al., 2008). The levels of these two proteins have also shown to be reduced in several AD mouse models and in AD patients (Kurochkin and Goto, 1994; Yasojima et al., 2001; Apelt et al., 2003; Cook et al., 2003). However, we did not detect any significant changes in the protein levels of either IDE or neprilysin (data not shown) in 22C11-treated neurons after 8 h, indicating that increased β-secretase and/or γ-secretase processing of APP was the likely cause of the elevated Aβ levels.
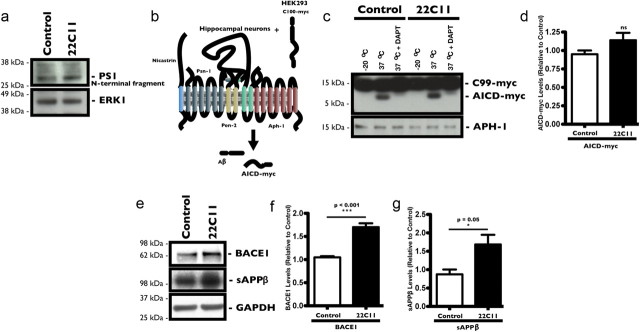
We used two different approaches to evaluate γ-secretase processing of APP. Because active endogenous PS exists mainly as a heterodimeric complex formed from the endoproteolytically processed N-terminal and C-terminal fragments of PS (Thinakaran et al., 1996), we first measured the levels of N-terminal fragments (∼28 kDa) by Western blot using a commercially available N-terminal antibody for PS1. There was no statistical difference in the levels of cleaved PS1 N-terminal fragments between neurons treated with 22C11 and those treated with PBS (Fig. 3a). To assess whether 22C11 could enhance the activity of γ-secretase, we used a previously described cell-free assay (Sastre et al., 2001; Hansson et al., 2006) designed to directly measure cleavage of C-terminal fragment β (CTFβ; C100) by γ-secretase (Fig. 3b). Briefly, HEK293 cells were transiently transfected with a C100-myc construct and membrane fractions were prepared from these cells 48 h post-transfection. These membrane fractions, enriched with APP C100-myc fragments (data not shown), were combined with membrane extracts (γ-secretase complex source) from 22C11-treated and PBS-treated PHNs and incubated for 2 h at 37°C to allow γ-secretase cleavage of C100-myc. The AICD-myc fragments generated were then detected by Western blot using a commercially available polyclonal anti-myc antibody (Fig. 3c). We did not observe a significant difference across the samples (Fig. 3d). γ-Secretase-mediated cleavage of C100-myc was confirmed by the addition of the γ-secretase inhibitor DAPT (Dovey et al., 2001), which completely abolished generation of AICD-myc fragments (Fig. 3c).
Figure 3.
Increased Aβ levels in 22C11-treated cultured rat PHNs correlate with increased BACE1 protein levels and not with increased γ-secretase activity. Treatment of PHNs for 8 h with a 100 ng/ml solution of 22C11 did not significantly affect PS1 cleavage compared with PBS-treated PHNs. a, Representative Western blots showing no significant changes in the levels of the N-terminal fragment of PS1 in 22C11-treated PHNs compared with PBS-treated PHNs. b–d, Treatment of PHNs for 8 h with a 100 ng/ml solution of 22C11 did not significantly affect γ-secretase activity in 22C11-treated PHNs compared with PBS-treated PHNs. b, Scheme depicting the in vitro γ-secretase activity assay: solubilized membranes from C100-myc overexpressing HEK293 cells were mixed with solubilized 22C11-treated (100 ng/ml; 8 h) or PBS-treated PHNs and incubated for 2 h, resulting in the formation of AICD-myc fragments detectable by Western blot. c, Representative Western blots showing no observable difference in AICD-myc production in 22C11-treated compared with PBS-treated PHNs. d, Densitometric analysis of the AICD-myc levels was quantified from three independent experiments, expressed as the relative mean ± SEM signal intensity (ns). e–g, Treatment of PHNs for 8 h with a 100 ng/ml solution of 22C11 resulted a significant increase in BACE1 protein levels with a concomitant increase in secreted APPβ fragments. e, Representative Western blots showing increased levels of BACE1 and sAPPβ in 22C11-treated compared with PBS-treated PHNs. f, g, Densitometric analysis of BACE1 levels was quantified from at least three independent experiments and expressed as the mean ± SEM band intensity (***p < 0.001 for BACE1; *p = 0.05 for sAPPβ).
To assess β-secretase cleavage of APP, we measured the total levels of cellular BACE1 protein by Western blot in PHNs after an 8 h treatment with 100 ng/ml 22C11 (Fig. 3e). BACE1 protein levels were significantly increased in 22C11-treated PHNs compared with PBS-treated control PHNs (Fig. 3f). To correlate increased BACE1 levels with increased activity, we also measured the levels of sAPPβ in the culture medium by Western blot (Fig. 3e). Concomitant with the increase in BACE1, sAPPβ levels were significantly higher in the culture medium from 22C11-treated PHNs compared with that of control PHNs (Fig. 3g), consistent with increased activity of BACE1 and increased processing of APP.
Together, these results suggest that the observed increases in the levels of intracellular Aβ in 22C11-treated PHNs is unlikely to be caused by a deficiency in the clearance mechanism of Aβ. This is supported by the unchanged levels of IDE and neprilysin in 22C11-treated neurons. Instead, our results point toward increased processing of APP by BACE1 at the β-secretase cleavage site of APP as the main element driving the elevated Aβ levels.
Increased BACE1 protein levels are not caused by increased synthesis but rather are due to enhanced protein stability
Having demonstrated elevated levels of BACE1 in 22C11-treated PHNs, we turned our focus on determining the cause. We first investigated whether 22C11 activated transcription of BACE1 in hippocampal neurons. qRT-PCR analysis of BACE1 mRNA levels in 22C11-treated neurons did not reveal any significant changes over control neurons after 8 h (data not shown), ruling out the possibility that increased transcription was the likely cause of increased BACE1 levels. This is in line with several findings that failed to show a significant increase in BACE1 mRNA levels in AD brains and in AD mouse models, despite elevated BACE1 protein levels (Holsinger et al., 2002; Zhao et al., 2007; Hébert et al., 2008).
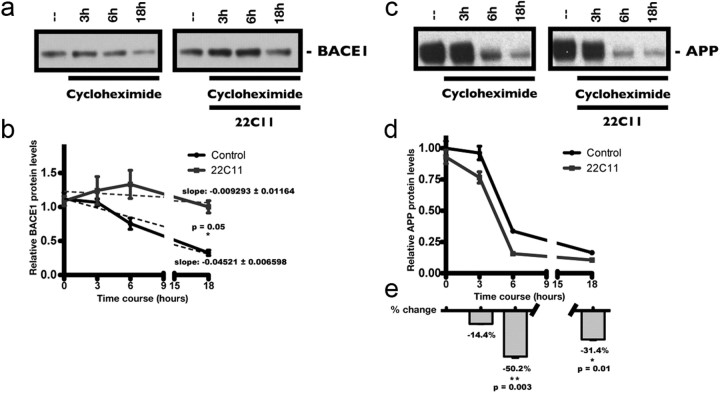
We therefore reasoned that the increase in BACE1 protein levels was due to increased stability of the protein. To test this hypothesis, we performed a cycloheximide (CHX) degradation assay to directly examine changes in the turnover rate of BACE1. PHNs were treated either with CHX (150 μm) alone, or with both CHX and 22C11 (100 ng/ml) over the course of 18 h. BACE1 protein levels were evaluated by Western blot (Fig. 4a). When we compared the rate of degradation of BACE1, we found a significant delay in its clearance in 22C11-treated PHNs compared with PHN treated with CHX alone (Fig. 4b), consistent with the idea of increased protein stability. The apparent increase in BACE1 in CHX/22C11-treated PHNs was likely due to the fact that all the samples were normalized for equal amounts of protein. Since BACE1 is stabilized following 22C11 treatment while most other proteins continue to be degraded at their normal rates, the levels of BACE1 in relation to the total amount of proteins are seemingly elevated. We also compared the turnover rate of APP between CHX alone and CHX/22C11-treated PHNs (Fig. 4c) and found a significant decrease in the levels of APP, suggesting that APP was processed at a faster rate in 22C11-treated PHNs (Fig. 4d). APP levels were decreased by ∼50 and 31% after 6 and 18 h respectively in PHNs treated with 22C11 (Fig. 4e).
Figure 4.
Increased BACE1 levels in 22C11-treated cultured rat PHNs occur through increased stability of the protein, not through increased synthesis. Cycloheximide (CHX) degradation time course: treatment of PHNs with a 100 ng/ml solution of 22C11 significantly delayed the degradation of BACE1 following CHX treatment, while increasing APP processing. a, Representative Western blots showing the degradation of BACE1 in PHNs at various time points (3, 6, and 18 h) after the addition of CHX (150 μm) alone or CHX (150 μm) plus 22C11 (100 ng/μl). b, Densitometric analysis of BACE1 levels was quantified from at least three independent experiments and expressed as the mean ± SEM signal intensity (*p = 0.05). c, Representative Western blots showing the processing of APP in PHNs at various time points (3, 6, and 18 h) after the addition of CHX alone (150 μm) or CHX (150 μm) plus 22C11 (100 ng/μl). d, Densitometric analysis of the APP levels was quantified from at least three independent experiments and expressed as the mean ± SEM signal intensity. e, Quantification of APP levels expressed as the percentage change after treatment with CHX alone (150 μm) or CHX (150 μm) plus 22C11 (100 ng/μl), indicating increased APP turnover by 22C11 treatment (**p = 0.003 and *p = 0.01 for 6 and 18 h time points, respectively).
These results are consistent with our initial hypothesis that 22C11 treatment affects the turnover rate of BACE1, increasing the half-life of endogenous BACE1 protein and argues against the possibility that increased synthesis is the cause of elevated BACE1 levels.
BACE1 is primarily degraded through the lysosomal pathway in hippocampal neurons
Collectively, the above data suggested that 22C11 drove Aβ production by altering the steady-state levels of BACE1, the rate-limiting enzyme in the biogenesis of Aβ. It also indicated that the increase in BACE1 levels might be, at least in part, the result of increased stabilization of BACE1. At least three different mechanisms have been reported to control the degradation of BACE1: (1) endoproteolysis within its catalytic domain (Huse et al., 2003), (2) the lysosomal pathway (Koh et al., 2005), and (3) the ubiquitin-proteasomal pathway (Qing et al., 2004). The latter study by Qing and colleagues was largely based on overexpression of BACE1 in several cell lines and it remains unclear whether endogenous BACE1 would normally be degraded through the proteasomal pathway. In addition, because the effects of lysosomal inhibitors were not studied, any potential contribution from lysosomal degradation of BACE1 in unclear. Moreover, the interpretation of experiments involving proteasome inhibitors can be problematic because many proteasome inhibitors can also inhibit lysosomal cathepsins (Kisselev and Goldberg, 2001; Kozlowski et al., 2001).
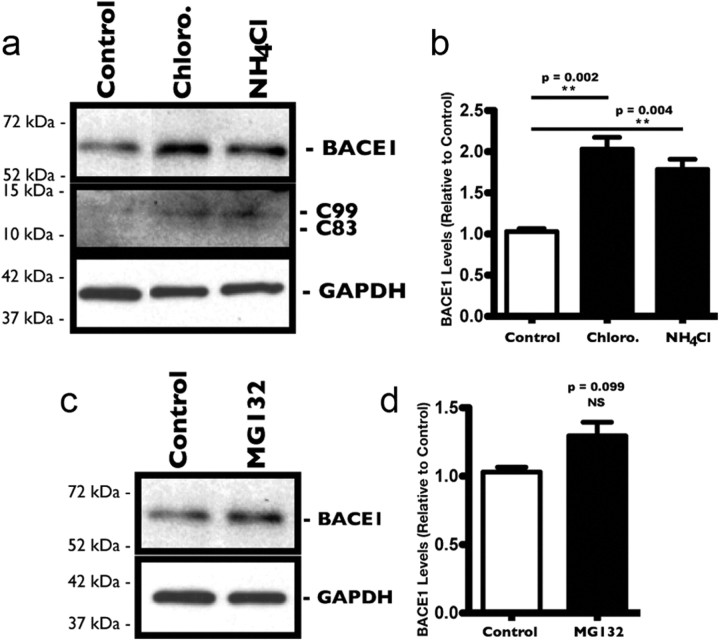
To overcome these shortcomings, we evaluated the respective contributions of proteasomal and lysosomal degradative pathways in regulating the levels of endogenous BACE1 in our model system, namely cultured rat PHNs. To assess lysosomal degradation of endogenous BACE1, PHNs were treated for 8 h with NH4Cl (500 μm) and chloroquine (100 μm), weak bases known to inhibit lysosomal hydrolases by reducing the acidification of the endosomal/lysosomal compartments (Ohkuma and Poole, 1978). After treatments, neurons were harvested and the levels of BACE1 protein were evaluated by Western blot analysis (Fig. 5a). Both chloroquine and NH4Cl treatments induced a marked increase in endogenous BACE1 protein levels after 8 h (Fig. 5b) compared with control PHNs. Increasing the concentrations of each inhibitor did not appear to significantly affect the buildup of BACE1. However, longer treatments did lead to higher BACE1 levels (data not shown). These results were in line with previous studies that demonstrated a similar effect in several cell lines, including Chinese hamster ovary and SY5Y cells and in cortical neurons (Koh et al., 2005). To assess proteasomal degradation, a similar protocol was followed and PHNs were treated with the proteasomal inhibitor MG132 (1.0 μm) for 8 h. Western blot analysis (Fig. 5c) showed a slight increase in BACE1 levels after 8 h (Fig. 5d) that did not reach statistical significance. Increasing the concentration of MG132, or extending the treatment over a longer time frame proved problematic due to toxicity. Comparable results were also obtained with another proteasomal inhibitor, lactacystin (data not shown).
Figure 5.
Inhibition of lysosomal hydrolase leads to increased levels of endogenous BACE1 in cultured rat PHNs. Treatment of PHNs for 8 h with various protease inhibitors (chloroquine and NH4Cl for lysosome; MG132 for proteasome) resulted in a significant increase in BACE1 protein levels in PHNs in which lysosomal degradation was blocked, suggesting that lysosomal degradation is the preferred pathway for BACE1 in PHNs. a, Representative Western blots showing increased levels of BACE1 and APP fragment C99 in PHNs treated for 8 h with the lysosomal inhibitors chloroquine (100 μm) and NH4Cl (500 μm). b, Densitometric analysis of BACE1 levels was quantified from three independent experiments and expressed as the mean ± SEM band intensity (**p = 0.002 for chloroquine; **p = 0.004 for NH4Cl). c, Representative Western blots showing BACE1 levels in PHNs treated for 8 h with the proteasomal inhibitor MG132 (1.0 μm). d, Densitometric analysis of BACE1 levels was quantified from three independent experiments and expressed as the mean ± SEM band intensity (NS).
22C11 treatment interferes with the sorting properties of BACE1 by GGA3 in hippocampal neurons
The results above indicated that endogenous BACE1 degradation in PHNs was primarily regulated by the activity of lysosomal hydrolases. We hypothesized that 22C11 treatment was interfering with lysosomal degradation of BACE1. Because levels of BACE1 protein rise when leucine residues are mutated to leucine residues, the dileucine (DXXLL) motif of BACE1 is believed to play a critical role in its degradation (Pastorino et al., 2002). Mutagenesis of the dileucine motif has been shown to interfere with the lysosomal degradation of BACE1 by blocking its transport from endosomes to lysosomes (Koh et al., 2005). This trafficking pathway is mediated by the Vps-27, Hrs, and STAM (VHS) domain of the Golgi-localized gamma-ear-containing ARF-binding (GGA) proteins (GGA1, GGA2, and GGA3), which binds the dileucine motif of BACE1 (for review, see Bonifacino, 2004). Depletion of GGA proteins by RNAi increases accumulation of BACE1 in early endosomes (He et al., 2005; Wahle et al., 2005; Tesco et al., 2007) and increases Aβ secretion in neurons (Wahle et al., 2006). Meanwhile, overexpression of GGA proteins reduces CTFβ generation and Aβ production (Wahle et al., 2006). Interestingly, of the three GGA family member proteins, GGA3 was highlighted in a recent report by Tesco et al. (2007) for its potential role in AD pathology. GGA3 protein levels were found to be significantly decreased in AD brains and inversely correlated with increased levels of BACE1.
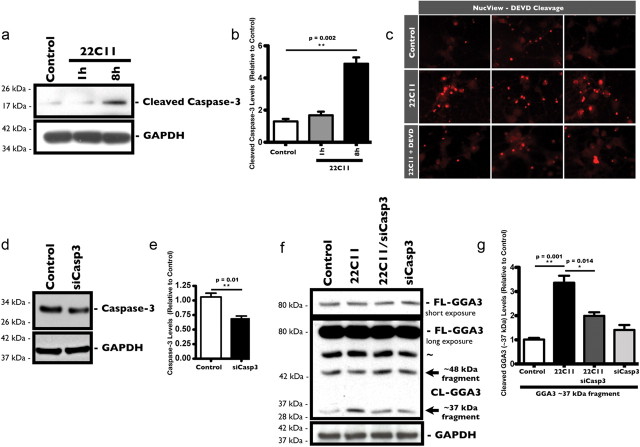
Because our data suggested that BACE1 was predominantly degraded through the lysosomal pathway in PHNs, we investigated the fate of GGA3 in response to 22C11 treatment. Tesco et al. (2007) demonstrated that GGA3 was a substrate for caspase-3, which led us to determine whether treatment with 22C11 could lead to the activation of caspase-3 in PHNs. To that end, we used an antibody that recognizes endogenous levels of the large fragment (17/19 kDa) of cleaved caspase-3 to analyze the levels of cleaved caspase-3 in PHNs treated with sublethal levels of 22C11 (100 ng/ml) for 1 and 8 h by Western blot (Fig. 6a). We found a significant increase in cleaved caspase-3 levels after 8 h in PHNs treated 22C11 compared with PBS-treated PHNs (Fig. 6b).
Figure 6.
22C11 treatment of cultured rat PHNs triggers the caspase-3-dependent cleavage of GGA3. a–c, Treatment of PHNs with a 100 ng/ml solution of 22C11 resulted in increased cleavage and activation of caspase-3 after 8 h. a, Representative Western blots showing an increase in cleaved caspase-3 levels after 8 h in 22C11-treated PHNs compared with PBS-treated PHNs. b, Densitometric analysis of cleaved caspase-3 levels was quantified from three independent experiments and expressed as the mean ± SEM (**p = 0.002) band intensity. c, NucView assay for in vivo caspase-3 activity: PHNs in chamber slides were treated with 22C11 (100 ng/ml) for 8 h before incubating with the DEVD-NucView caspase substrate. Upon cleavage of this substrate by caspase-3, the NucView moiety enters the nucleus and fluoresces upon binding with DNA. Treated PHNs showed a marked increase in fluorescence compared with PBS-treated neurons, indicative of increased caspase-3 activity. Pretreatment for 1 h with the caspase-3 inhibitor Z-DEVD-FMK blocked increased NucView fluorescence, confirming that fluorescence was a result of caspase activity. d, g, Treatment of PHNs with a 100 ng/ml solution of 22C11 resulted in increased cleavage of GGA3 compared with PBS-treated neurons in a caspase-3-dependent manner. d, Representative Western blots showing the downregulation of caspase-3 in PHNs after an 8 h treatment with penetratin-1-linked caspase-3 siRNA (siCasp3; 80 nm). e, Densitometric analysis of caspase-3 levels was quantified from three independent experiments and expressed as the mean ± SEM (*p = 0.01) band intensity. f, Representative Western blots with a C-terminal-specific GGA3 antibody showing increased production of the ∼37 kDa fragment in 22C11-treated compared with PBS-treated PHNs. This effect was blocked by downregulation of caspase-3. g, Densitometric analysis of cleaved GGA3 (∼37 kDa fragment) levels was quantified from three independent experiments and expressed as the mean ± SEM (**p = 0.001 for Control vs 22C11; *p = 0.014 for 22C11 vs 22C11 plus siCasp3) band intensity.
It is often assumed that increased caspase-3 cleavage equates to increased caspase-3 activity. However, inhibitors of aptosis protein (IAPs), such as xIAP, c-IAP1/2, and survivin, can bind processed caspase-3 and block its activity (for review, see Liston et al., 2003). To determine whether increased caspase-3 cleavage indeed equates to increased caspase-3 activity, we directly measured the activity of caspase-3 in live PHNs using a commercially available fluorescent caspase-3 substrate, DEVD (acetyl-aspartyl-glutamyl-valyl)-NucView. The DEVD peptide is attached to a DNA-binding dye, which produces fluorescence only in the presence of DNA. Upon entering the cell cytoplasm, it is cleaved by active caspase-3 to release the high-affinity DNA dye. The released dye migrates to the nucleus and brightly stains it. Consistent with the observed increased in cleaved caspase-3 levels, we found that PHNs treated with 22C11 had notably higher fluorescence compared with control PHNs (Fig. 6c), indicative of increased DEVD cleavage by active caspase-3. Fluorescence was significantly attenuated by pretreating PHNs for 1 h with the competing nonfluorescent caspase- 3 inhibitor Z-DEVD-FMK (N-benzyloxycarbonyl-Asp(OMe)-Gly(OMe)-Val-Asp-(OMe)-fluoro-methylketone; 25 μm; Fig. 6c).
Because caspase-3 activity was increased in 22C11-treated PHNs, we decided to look at the cleavage of endogenous GGA3 protein in these cells. As described earlier, PHNs were treated with 22C11 (100 ng/ml) for 8 h and cleavage of endogenous GGA3 was assessed by Western blot. Cleavage of recombinant human GGA3 by recombinant active caspase-3 occurs at three major sites within its hinge domain, generating three C- terminal fragments of ∼50 kDa, ∼48 kDa, and ∼37 kDa, as well as an N-terminal fragment, which also functions as a dominant negative form of GGA3 (Tesco et al., 2007). We used a commercially available antibody directed against the C terminus of GGA3, which recognizes the full-length protein as well as C-terminal fragments (Fig. 6f). PHNs treated with 22C11 had increased levels of cleaved GGA3 (∼37 kDa fragment) compared with control PHNs (Fig. 6g). We did not detect the larger ∼50 kDa fragment of GGA3 possibly because the levels were below the limit of detection. This was consistent with the findings of Tesco et al. (2007), who were unsuccessful at detecting endogenous levels of the larger fragment (∼50 kDa) in lysates from the H4 human glioblastoma cell line. Interestingly, no significant change in the levels of the ∼48 kDa fragment was observed in 22C11-treated PHNs, perhaps due to a preference for the third cleavage site to produce the smaller fragment.
To confirm the role of caspase-3 in the cleavage of GGA3 PHNs, we used a siRNA to deplete the levels of endogenous caspase-3 in PHNs before treating with 22C11. The siRNA oligonucleotide was delivered to PHNs by linking it to the cell-penetrating peptide penetratin-1, previously used as an effective method for delivering siRNA sequences to primary neurons (Davidson et al., 2004). We achieved ∼40% knockdown of endogenous caspase-3 protein in hippocampal neurons after 8 h (Fig. 6d,e), which was sufficient to significantly abrogate the cleavage of GGA3 in 22C11-treated PHNs, as demonstrated by the reduced levels of the ∼37 kDa fragment (Fig. 6g).
Increased BACE1 and Aβ levels in 22C11-treated neurons are caspase-3 and GGA3 dependent
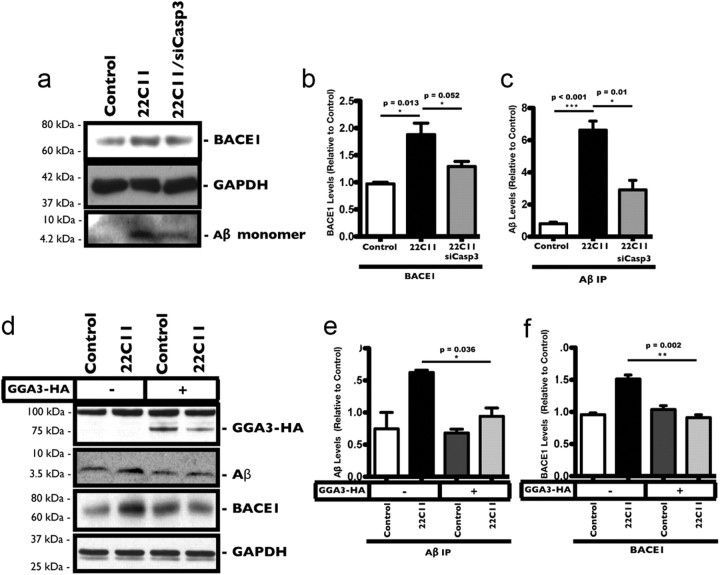
The results above suggested a mechanism whereby 22C11 increased caspase-3 activity in PHNs, resulting in the cleavage and blocking of GGA3 function. We predicted that depletion of caspase-3 by siRNA as shown should reverse the effects of 22C11 on both BACE1 and Aβ levels. To test this, we treated PHNs with 22C11 (100 ng/ml) in the presence or absence of the caspase-3 siRNA and analyzed the levels of BACE1 by Western blot and of Aβ by IP (Fig. 7a). The increases in BACE1 and Aβ levels in 22C11-treated PHNs were completely blocked in PHNs treated with siCasp3 (Fig. 7b,c).
Figure 7.
Increased BACE1 and Aβ levels in 22C11-treated neurons is caspase-3 and GGA3 dependent. a–c, Cultured rat PHNs were treated with 22C11 (100 ng/ml) only or with 22C11 plus siCasp3 for 8 h. a, Representative Western blots and IP analysis showing increased levels of BACE1 and Aβ in 22C11-treated compared with PBS-treated PHNs. Increases in both BACE1 and Aβ levels were blocked by downregulation of caspase-3. b, c, Densitometric analysis of BACE1 and Aβ levels was quantified from three independent experiments and expressed as the mean ± SEM band intensity (BACE1: *p = 0.013 for Control vs 22C11; *p = 0.052 for 22C11 vs 22C11 plus siCasp3; Aβ: ***p < 0.0001 for Control vs 22C11; *p = 0.01 for 22C11 vs 22C11 plus siCasp3). d--f, Mouse neuroblastoma cells (B103) stably expressing APP (B103APP) were transfected with 4.0 μg of GGA3 plasmid or with 4.0 μg of empty vector plasmid and treated with 500 ng/ml of a 22C11 solution 48 h post-transfection. d, Representative Western blots showing increased BACE1 levels in mock-transfected cells, as well as increased Aβ production (determined by IP). Overexpression of human GGA3 in the cells blocked both BACE1 increase and Aβ production. e, f, Densitometric analysis of BACE1 and Aβ (IP) levels was quantified from three independent experiments and expressed as the mean ± SEM band intensity (Aβ: *p = 0.036 for 22C11 vs 22C11 plus GGA3; BACE1: **p = 0.002 for 22C11 vs 22C11 plus GGA3).
Our model also predicted that flooding the neurons with GGA3 by overexpression should block the increase in BACE1 and Aβ production triggered by 22C11. Because of the low transfection efficiency in PHNs, we used the B103 murine neuroblastoma cell line (Schubert et al., 1974), stably expressing APP. These cells were treated with 500 ng/ml 22C11 for 8 h and the levels of neuronal Aβ and BACE1 were measured (Fig. 7d). Similar to PHNs, B103 cells treated with 22C11 showed a significant increase in both BACE1 levels and Aβ production (Fig. 7e,f). To test our prediction, B103 cells were transfected with a plasmid expressing full-length human GGA3 and treated after 48 h with 22C11 as above. Consistent with our proposed model, B103 cells overexpressing GGA3 did not exhibit an increase in BACE1 levels or Aβ production in response to 22C11 treatment (Fig. 7e,f).
Taken collectively, these results provide consistent support for a mechanism in which 22C11 promotes BACE1 accumulation in hippocampal neurons by interfering with its normal degradation process. Activation of caspase-3 by 22C11 leads to increased cleavage and inactivation of GGA3, delaying the degradation of BACE1 and allowing it to accumulate.
Treatment with 22C11 leads to accumulation of BACE1 in endosomes in hippocampal neurons
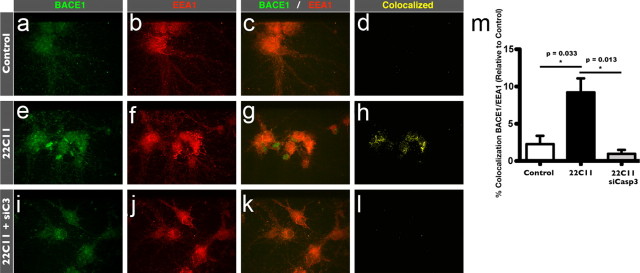
As mentioned above, depletion of GGA3 by siRNA impairs its normal function of targeting BACE1 to lysosomes for degradation, leading to the accumulation of BACE1 in early endosomes (He et al., 2005; Wahle et al., 2005; Tesco et al., 2007). Since 22C11 treatment promoted GGA3 cleavage, we further reasoned that 22C11 treatment should also increase BACE1 in the endosomal compartments of PHNs. To test this final prediction, PHNs were again treated with 22C11 for 8 h, after which they were fixed with paraformaldehyde and costained with antibodies against the early endosome marker EEA1 (Fig. 8b,f,j) and endogenous BACE1 (Fig. 8a,e,i). Confocal analysis of the obtained images (Fig. 8c,g,k) revealed a marked increase in the colocalization of BACE1 and EEA1 in 22C11-treated PHNs compared with PBS-treated PHNs (Fig. 8d,h,m), indicative of BACE1 accumulation in early endosomal compartments, in agreement with our proposed model.
Figure 8.
Treatment of cultured rat PHNs with 22C11 induces the accumulation of BACE1 in early endosomal compartments. a–l, PHNs in chamber slides (Control or pretreated with siCasp3) were treated with a 100 ng/ml solution of 22C11 for 8 h. Following fixation and permeabilization, PHNs were stained with antibodies for BACE1 (a, e, i, green) and the subcellular early endosome marker EEA1 (b, f, j, red) and imaged and analyzed by confocal microscopy (c, g, k). Colocalization of BACE1 with EEA1 (d, h, l) was markedly increased in PHNs treated with 22C11-treated (d) compared with PBS-treated PHNs (h), consistent with an endosome-to-lysosome trafficking defect. Downregulation of caspase-3 by siRNA completely blocked colocalization of BACE1 with EEA1 (l). m, Colocalization of BACE1 and EEA1 was quantified from an average of 10 neurons from three independent experiments using the Volocity software and expressed as the mean ± SEM percentage of BACE1 colocalized with EEA1 (*p = 0.033 for Control vs 22C11; *p = 0.013 for Control vs 22C11 plus siCasp3).
We also tested whether caspase-3 downregulation could reverse the 22C11-induced accumulation of BACE1 in endosomes. Indeed, consistent with our model, downregulation of caspase-3 in 22C11-treated PHNs resulted in a notable reduction in the colocalization of BACE1 with EEA1 (Fig. 8h,l,m), suggesting that BACE1 is prevented from accumulating in early endosomes.
Discussion
Much progress has been made toward deciphering some of the biological functions of APP, particularly its role in facilitating cell–cell adhesion through homodimerization and heterodimerization with the other APP family members, namely APLP1 and APLP2. However, the focus has remained mostly on its pathogenic role, as the source of the Aβ peptide in the context of AD. The identification and characterization of APP-cleaving enzymes, such as secretases (BACE1 and the γ-secretase complex) and caspases, have provided a valuable insight into the complex steps involved in the proteolytic processing of APP and the production of Aβ. Whether the Aβ peptide itself is the cause of AD remains a subject for debate. The identification of several mutations in the APP and PS genes in early-onset familial AD seem to support this view, as these mutations have been shown promote the production of Aβ. In late-onset AD, however, the picture becomes a little more muddled. While many risk-factor genes have been identified, age remains the greatest risk factor for AD. This raises several important questions: Does the cleavage process of APP change with aging or in AD, and, if so, what triggers these changes? Do elevated BACE1 levels in the brain play a role in AD pathogenesis, and, if so, what causes BACE1 to become elevated? Could dimerization of APP provide some answers?
The exact consequences of homodimerization of APP on the processing of APP are not fully understood. Introduction of a cysteine mutation in the juxtamembrane region of APP has been reported to enhance Aβ production through the formation of stable disulfide-linked APP dimers (Scheuermann et al., 2001), consistent with the observation that stable Aβ dimers can be found intracellularly in vitro and in vivo in brains (Walsh et al., 2000). However, other laboratories have reported the opposite effect, where enhanced dimerization of APP leads to decreased APP processing and decreased Aβ levels (Struhl and Adachi, 2000; Eggert et al., 2009). Reconciling this dichotomy remains difficult, but it could simply be explained by differences in the manner through which APP dimerization is promoted in each model system.
Here we show that cross-linking of endogenous APP through the use of a divalent antibody triggers the amyloidogenic pathway in cultured hippocampal neurons, resulting in a rise in the levels of intracellular Aβ. Interestingly, this increase in Aβ is observed under nonapoptotic conditions, at least within 48 h of treatment, but is accompanied by a significant loss of dendritic spine protrusions as well as a decrease in synaptic markers, PSD-95 and drebrin A. This is an important consideration because neuronal and non-neuronal cells undergoing apoptosis have been shown to overproduce and secrete Aβ, whether triggered by staurosporine or by trophic factor withdrawal (LeBlanc, 1995; Barnes et al., 1998; Galli et al., 1998; Gervais et al., 1999; Guo et al., 2001; Tesco et al., 2003; Sodhi et al., 2004; Matrone et al., 2008a,b). This wide array of conditions and cell types raises questions as to whether the observed Aβ overproduction is a specific process or simply a general response to apoptotic stimuli. Our result supports the former view, but it is possible that both APP signaling and apoptotic stimuli share common pathways.
Whether driven by apoptosis or by APP signaling, enhanced Aβ production seems to correlate with high levels of BACE1. We show that the observed Aβ overproduction in treated neurons is due primarily to increased processing of APP by BACE1, and not by γ-secretase. This is reflected by a significant increase in the production of sAPPβ fragments correlating with elevated BACE1 protein levels. BACE1 levels rise in response to physiological stress or injury, such as oxidative stress (Tamagno et al., 2002), traumatic brain injury (Blasko et al., 2004), ischemia (Wen et al., 2004), hypoxia (Zhang and Xu, 2007), and energy impairment (Velliquette et al., 2005). BACE1 is also increased in brains from late-onset and early-onset AD patients compared with cognitively normal individuals (Fukumoto et al., 2002; Holsinger et al., 2002; Tyler et al., 2002; Yang et al., 2003; Li et al., 2004). Our results imply that in addition to age-related stress, aberrant signaling triggered by APP oligomerization may also enhance levels of BACE1 and Aβ in the brain, and drive AD pathogenesis.
The exact mechanism of this upregulation is not fully understood and hypotheses vary from transcriptional, posttranscriptional, translational, and posttranslational modifications of BACE1 (Holsinger et al., 2002; Zhao et al., 2007; Faghihi et al., 2008; Hébert et al., 2008; Wen et al., 2008). Our results indicate that the BACE1 increase may be due, at least in part, to enhanced protein stabilization and accumulation. This is reflected by the fact that no significant changes are observed in BACE1 mRNA in treated hippocampal neurons. Alternatively, BACE1 half-life is significantly prolonged in treated neurons, persisting to near control levels after 18 h of CHX treatment. These results suggest that oligomerization of APP may trigger a signaling cascade that directly interferes with the normal degradation of BACE1 protein, allowing BACE1 to accumulate in treated neurons. Our data show that activation of this pathway may lead to the loss of function of the GGA3 protein. GGA family proteins are known to be involved in the trafficking of proteins, such as BACE1, which contain the DXXLL signal between different compartments (e.g., Golgi complex, endosomes, and lysosomes) (for review, see Bonifacino, 2004). We demonstrated that activation of caspase-3 in treated neurons promotes cleavage of GGA3, perhaps generating increased amounts of the dominant negative N-terminal fragment (Tesco et al., 2007). The accumulation of BACE1 in early endosomal compartments in treated neurons was consistent with a loss of function of the GGA3 protein. Furthermore, downregulation of caspase-3 by siRNA prevented the accumulation of BACE1 in endosomes as well as an increase in intracellular Aβ. A similar effect was observed in cells overexpressing human GGA3, which did not exhibit an accumulation of BACE1 or overproduction of Aβ.
The mechanism by which GGA3 targets its cargo to lysosomes has been shown to be ubiquitin dependent (Puertollano and Bonifacino, 2004). While there is some evidence that BACE1 is ubiquitinated (Qing et al., 2004), future studies will be required to determine whether GGA3-dependent degradation of BACE1 requires ubiquitination or whether it occurs via another mechanism (e.g., binding the VHS domain of GGA3). Additionally, RNAi silencing of GGA1 and GGA2 has also been shown to lead to the accumulation of BACE1 in endosomes. However, unlike GGA3, which shuttles BACE1 from endosomes to lysosomes, GGA1 and GGA2 appear to regulate retrograde transport of BACE1 from endosomes to the trans-Golgi network (TGN) (Wahle et al., 2005). Further studies will be required to determine whether loss of function or depletion of GGA1 and GGA2 contributes to BACE1 accumulation in our model. Finally, phosphorylation of BACE1 at Serine498 facilitates its binding to GGA proteins (He et al., 2002, 2003; Shiba et al., 2004; von Arnim et al., 2004). However, we did not observe any changes in the phosphorylation status of BACE1 in treated neurons (data not shown).
In summary, our results argue for a well defined mechanism through which aberrant APP signaling can trigger amyloidogenic processing of APP, without affecting neuronal survival, as depicted in Figure 9. As we discussed earlier, alterations in synaptic density occur early in AD and strongly correlate with the cognitive decline observed in the disease (for review, see Scheff and Price, 2006). Our results suggest that aberrant signaling through APP oligomerization is sufficient to drive synaptic dysfunction as well as promote Aβ production in hippocampal neurons. Whether, these effects are dependent on each other remains unclear. It is interesting to note, however, that in our model system, while Aβ production is dependent on caspase-3, downregulation of caspase-3 did not protect neurons against the synaptotoxic effects of 22C11, suggesting on the surface that the effects we observed are independent of Aβ production. While somewhat puzzling, a simple explanation could be that signaling through APP results in the activation of a parallel pathway, which itself drives synaptic dysfunction and perhaps even neuronal death. Indeed, another consequence of APP cross-linking is the production of another toxic 31 aa cytosolic fragment, termed C31 (Bertrand et al., 2001; Lu et al., 2003; Shaked et al., 2006), which exerts its effects independently of Aβ (Park et al., 2009). This raises the interesting possibility that abnormal oligomerization of APP initiates a positive feedback loop in an affected neuronal population, resulting in local synaptic dysfunction while simultaneously raising the levels of Aβ, which can spread and “infect” healthy neurons in a manner similar to Prion's disease. This may also suggest that the development of drugs aimed at disrupting APP dimerization may be a viable therapeutic approach worth exploring.
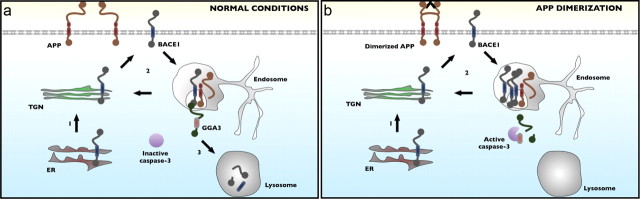
Figure 9.
Schematic illustration of 22C11-induced BACE1 accumulation and Aβ production in neurons. Following synthesis in the endoplasmic reticulum, pro-BACE1 traffics through the trans-Golgi network (TGN), where it matures before localizing to the plasma membrane. a, Under normal conditions, surface BACE1 is reinternalized through the endosomal pathway and may be recycled from endosomes to the TGN back to the plasma membrane. Alternatively, BACE1 can interact with GGA3, which targets BACE1 to lysosomes for degradation. b, Dimerization of APP and subsequent activation of caspase-3 results in the cleavage of GGA3, preventing GGA3 from shuttling BACE1 to lysosomes. This allows BACE1 to accumulate in endosomes, resulting in increased cleavage of APP and increased Aβ production.
Footnotes
This work was supported in part by National Institutes of Health Grant P50AG008702 (to M.S.). We thank Dr. Edward Koo (University of California San Diego) for providing B103 cells. We are thankful to Dr. Carol Troy (Columbia University) for helpful discussions and advice. We are grateful to Dr. Soline Aubry and Andreina Dominguez for critically reading and carefully editing this manuscript.
References
- Apelt J, Ach K, Schliebs R. Aging-related down-regulation of neprilysin, a putative beta-amyloid-degrading enzyme, in transgenic Tg2576 Alzheimer-like mouse brain is accompanied by an astroglial upregulation in the vicinity of beta-amyloid plaques. Neurosci Lett. 2003;339:183–186. doi: 10.1016/s0304-3940(03)00030-2. [DOI] [PubMed] [Google Scholar]
- Barnes NY, Li L, Yoshikawa K, Schwartz LM, Oppenheim RW, Milligan CE. Increased production of amyloid precursor protein provides a substrate for caspase-3 in dying motoneurons. J Neurosci. 1998;18:5869–5880. doi: 10.1523/JNEUROSCI.18-15-05869.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand E, Brouillet E, Caillé I, Bouillot C, Cole GM, Prochiantz A, Allinquant B. A short cytoplasmic domain of the amyloid precursor protein induces apoptosis in vitro and in vivo. Mol Cell Neurosci. 2001;18:503–511. doi: 10.1006/mcne.2001.1030. [DOI] [PubMed] [Google Scholar]
- Blasko I, Beer R, Bigl M, Apelt J, Franz G, Rudzki D, Ransmayr G, Kampfl A, Schliebs R. Experimental traumatic brain injury in rats stimulates the expression, production and activity of Alzheimer's disease beta-secretase (BACE-1) J Neural Transm. 2004;111:523–536. doi: 10.1007/s00702-003-0095-6. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS. The GGA proteins: adaptors on the move. Nat Rev Mol Cell Biol. 2004;5:23–32. doi: 10.1038/nrm1279. [DOI] [PubMed] [Google Scholar]
- Bormann BJ, Knowles WJ, Marchesi VT. Synthetic peptides mimic the assembly of transmembrane glycoproteins. J Biol Chem. 1989;264:4033–4037. [PubMed] [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Chen CD, Oh SY, Hinman JD, Abraham CR. Visualization of APP dimerization and APP-Notch2 heterodimerization in living cells using bimolecular fluorescence complementation. J Neurochem. 2006;97:30–43. doi: 10.1111/j.1471-4159.2006.03705.x. [DOI] [PubMed] [Google Scholar]
- Cook DG, Leverenz JB, McMillan PJ, Kulstad JJ, Ericksen S, Roth RA, Schellenberg GD, Jin LW, Kovacina KS, Craft S. Reduced hippocampal insulin-degrading enzyme in late-onset Alzheimer's disease is associated with the apolipoprotein E-epsilon4 allele. Am J Pathol. 2003;162:313–319. doi: 10.1016/s0002-9440(10)63822-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson TJ, Harel S, Arboleda VA, Prunell GF, Shelanski ML, Greene LA, Troy CM. Highly efficient small interfering RNA delivery to primary mammalian neurons induces MicroRNA-like effects before mRNA degradation. J Neurosci. 2004;24:10040–10046. doi: 10.1523/JNEUROSCI.3643-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaère P, Duyckaerts C, Brion JP, Poulain V, Hauw JJ. Tau, paired helical filaments and amyloid in the neocortex: a morphometric study of 15 cases with graded intellectual status in aging and senile dementia of Alzheimer type. Acta Neuropathol. 1989;77:645–653. doi: 10.1007/BF00687893. [DOI] [PubMed] [Google Scholar]
- Dovey HF, John V, Anderson JP, Chen LZ, de Saint Andrieu P, Fang LY, Freedman SB, Folmer B, Goldbach E, Holsztynska EJ, Hu KL, Johnson-Wood KL, Kennedy SL, Kholodenko D, Knops JE, Latimer LH, Lee M, Liao Z, Lieberburg IM, Motter RN, et al. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. J Neurochem. 2001;76:173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- Eggert S, Midthune B, Cottrell B, Koo EH. Induced dimerization of the amyloid precursor protein leads to decreased amyloid-beta protein production. J Biol Chem. 2009;284:28943–28952. doi: 10.1074/jbc.M109.038646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feedforward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto H, Cheung BS, Hyman BT, Irizarry MC. Beta-secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch Neurol. 2002;59:1381–1389. doi: 10.1001/archneur.59.9.1381. [DOI] [PubMed] [Google Scholar]
- Galli C, Piccini A, Ciotti MT, Castellani L, Calissano P, Zaccheo D, Tabaton M. Increased amyloidogenic secretion in cerebellar granule cells undergoing apoptosis. Proc Natl Acad Sci U S A. 1998;95:1247–1252. doi: 10.1073/pnas.95.3.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan WB, Grutzendler J, Wong WT, Wong RO, Lichtman JW. Multicolor “DiOlistic” labeling of the nervous system using lipophilic dye combinations. Neuron. 2000;27:219–225. doi: 10.1016/s0896-6273(00)00031-3. [DOI] [PubMed] [Google Scholar]
- Gervais FG, Xu D, Robertson GS, Vaillancourt JP, Zhu Y, Huang J, LeBlanc A, Smith D, Rigby M, Shearman MS, Clarke EE, Zheng H, Van Der Ploeg LH, Ruffolo SC, Thornberry NA, Xanthoudakis S, Zamboni RJ, Roy S, Nicholson DW. Involvement of caspases in proteolytic cleavage of Alzheimer's amyloid-beta precursor protein and amyloidogenic A beta peptide formation. Cell. 1999;97:395–406. doi: 10.1016/s0092-8674(00)80748-5. [DOI] [PubMed] [Google Scholar]
- Guo Q, Xie J, Chang X, Du H. Prostate apoptosis response-4 enhances secretion of amyloid beta peptide 1–42 in human neuroblastoma IMR-32 cells by a caspase-dependent pathway. J Biol Chem. 2001;276:16040–16044. doi: 10.1074/jbc.M010996200. [DOI] [PubMed] [Google Scholar]
- Hansson CA, Popescu BO, Laudon H, Cedazo-Minguez A, Popescu LM, Winblad B, Ankarcrona M. Caspase cleaved presenilin-1 is part of active gamma-secretase complexes. J Neurochem. 2006;97:356–364. doi: 10.1111/j.1471-4159.2006.03735.x. [DOI] [PubMed] [Google Scholar]
- He X, Chang WP, Koelsch G, Tang J. Memapsin 2 (beta-secretase) cytosolic domain binds to the VHS domains of GGA1 and GGA2: implications on the endocytosis mechanism of memapsin 2. FEBS Lett. 2002;524:183–187. doi: 10.1016/s0014-5793(02)03052-1. [DOI] [PubMed] [Google Scholar]
- He X, Zhu G, Koelsch G, Rodgers KK, Zhang XC, Tang J. Biochemical and structural characterization of the interaction of memapsin 2 (beta-secretase) cytosolic domain with the VHS domain of GGA proteins. Biochemistry. 2003;42:12174–12180. doi: 10.1021/bi035199h. [DOI] [PubMed] [Google Scholar]
- He X, Li F, Chang WP, Tang J. GGA proteins mediate the recycling pathway of memapsin 2 (BACE) J Biol Chem. 2005;280:11696–11703. doi: 10.1074/jbc.M411296200. [DOI] [PubMed] [Google Scholar]
- Hébert SS, Horré K, Nicolaï L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks L, van Duijn CM, Cras P, Cruts M, Van Hul W, van Harskamp F, Warren A, McInnis MG, Antonarakis SE, Martin JJ. Presenile dementia and cerebral haemorrhage linked to a mutation at codon 692 of the beta-amyloid precursor protein gene. Nat Genet. 1992;1:218–221. doi: 10.1038/ng0692-218. [DOI] [PubMed] [Google Scholar]
- Hilbich C, Mönning U, Grund C, Masters CL, Beyreuther K. Amyloid-like properties of peptides flanking the epitope of amyloid precursor protein-specific monoclonal antibody 22C11. J Biol Chem. 1993;268:26571–26577. [PubMed] [Google Scholar]
- Holsinger RM, McLean CA, Beyreuther K, Masters CL, Evin G. Increased expression of the amyloid precursor beta-secretase in Alzheimer's disease. Ann Neurol. 2002;51:783–786. doi: 10.1002/ana.10208. [DOI] [PubMed] [Google Scholar]
- Huse JT, Byant D, Yang Y, Pijak DS, D'Souza I, Lah JJ, Lee VM, Doms RW, Cook DG. Endoproteolysis of beta-secretase (beta-site amyloid precursor protein-cleaving enzyme) within its catalytic domain. A potential mechanism for regulation. J Biol Chem. 2003;278:17141–17149. doi: 10.1074/jbc.M213303200. [DOI] [PubMed] [Google Scholar]
- Kaden D, Munter LM, Joshi M, Treiber C, Weise C, Bethge T, Voigt P, Schaefer M, Beyermann M, Reif B, Multhaup G. Homophilic interactions of the amyloid precursor protein (APP) ectodomain are regulated by the loop region and affect beta-secretase cleavage of APP. J Biol Chem. 2008;283:7271–7279. doi: 10.1074/jbc.M708046200. [DOI] [PubMed] [Google Scholar]
- Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Müller-Hill B. The precursor of Alzheimer's disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- Kienlen-Campard P, Tasiaux B, Van Hees J, Li M, Huysseune S, Sato T, Fei JZ, Aimoto S, Courtoy PJ, Smith SO, Constantinescu SN, Octave JN. Amyloidogenic processing but not amyloid precursor protein (APP) intracellular C-terminal domain production requires a precisely oriented APP dimer assembled by transmembrane GXXXG motifs. J Biol Chem. 2008;283:7733–7744. doi: 10.1074/jbc.M707142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev AF, Goldberg AL. Proteasome inhibitors: from research tools to drug candidates. Chem Biol. 2001;8:739–758. doi: 10.1016/s1074-5521(01)00056-4. [DOI] [PubMed] [Google Scholar]
- Koh YH, von Arnim CA, Hyman BT, Tanzi RE, Tesco G. BACE is degraded via the lysosomal pathway. J Biol Chem. 2005;280:32499–32504. doi: 10.1074/jbc.M506199200. [DOI] [PubMed] [Google Scholar]
- Kozlowski L, Stoklosa T, Omura S, Wójcik C, Wojtukiewicz MZ, Worowski K, Ostrowska H. Lactacystin inhibits cathepsin A activity in melanoma cell lines. Tumour Biol. 2001;22:211–215. doi: 10.1159/000050618. [DOI] [PubMed] [Google Scholar]
- Kurochkin IV, Goto S. Alzheimer's beta-amyloid peptide specifically interacts with and is degraded by insulin degrading enzyme. FEBS Lett. 1994;345:33–37. doi: 10.1016/0014-5793(94)00387-4. [DOI] [PubMed] [Google Scholar]
- LeBlanc A. Increased production of 4 kDa amyloid beta peptide in serum deprived human primary neuron cultures: possible involvement of apoptosis. J Neurosci. 1995;15:7837–7846. doi: 10.1523/JNEUROSCI.15-12-07837.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Treutlein HR, Adams PD, Brünger AT, Engelman DM. A dimerization motif for transmembrane alpha-helices. Nat Struct Biol. 1994;1:157–163. doi: 10.1038/nsb0394-157. [DOI] [PubMed] [Google Scholar]
- Li R, Lindholm K, Yang LB, Yue X, Citron M, Yan R, Beach T, Sue L, Sabbagh M, Cai H, Wong P, Price D, Shen Y. Amyloid beta peptide load is correlated with increased beta-secretase activity in sporadic Alzheimer's disease patients. Proc Natl Acad Sci U S A. 2004;101:3632–3637. doi: 10.1073/pnas.0205689101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston P, Fong WG, Korneluk RG. The inhibitors of apoptosis: there is more to life than Bcl2. Oncogene. 2003;22:8568–8580. doi: 10.1038/sj.onc.1207101. [DOI] [PubMed] [Google Scholar]
- Liu W, Crocker E, Zhang W, Elliott JI, Luy B, Li H, Aimoto S, Smith SO. Structural role of glycine in amyloid fibrils formed from transmembrane alpha-helices. Biochemistry. 2005;44:3591–3597. doi: 10.1021/bi047827g. [DOI] [PubMed] [Google Scholar]
- Lu DC, Shaked GM, Masliah E, Bredesen DE, Koo EH. Amyloid beta protein toxicity mediated by the formation of amyloid-beta protein precursor complexes. Ann Neurol. 2003;54:781–789. doi: 10.1002/ana.10761. [DOI] [PubMed] [Google Scholar]
- Marchesi VT. An alternative interpretation of the amyloid Abeta hypothesis with regard to the pathogenesis of Alzheimer's disease. Proc Natl Acad Sci U S A. 2005;102:9093–9098. doi: 10.1073/pnas.0503181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrone C, Di Luzio A, Meli G, D'Aguanno S, Severini C, Ciotti MT, Cattaneo A, Calissano P. Activation of the amyloidogenic route by NGF deprivation induces apoptotic death in PC12 cells. J Alzheimers Dis. 2008b;13:81–96. doi: 10.3233/jad-2008-13109. [DOI] [PubMed] [Google Scholar]
- Matrone C, Ciotti MT, Mercanti D, Marolda R, Calissano P. NGF and BDNF signaling control amyloidogenic route and Abeta production in hippocampal neurons. Proc Natl Acad Sci U S A. 2008a;105:13139–13144. doi: 10.1073/pnas.0806133105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miners JS, Baig S, Palmer J, Palmer LE, Kehoe PG, Love S. Abeta-degrading enzymes in Alzheimer's disease. Brain Pathol. 2008;18:240–252. doi: 10.1111/j.1750-3639.2008.00132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munter LM, Voigt P, Harmeier A, Kaden D, Gottschalk KE, Weise C, Pipkorn R, Schaefer M, Langosch D, Multhaup G. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Abeta42. EMBO J. 2007;26:1702–1712. doi: 10.1038/sj.emboj.7601616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munter LM, Botev A, Richter L, Hildebrand PW, Althoff V, Weise C, Kaden D, Multhaup G. Aberrant amyloid precursor protein (APP) processing in hereditary forms of Alzheimer disease caused by APP familial Alzheimer disease mutations can be rescued by mutations in the APP GxxxG motif. J Biol Chem. 2010;285:21636–21643. doi: 10.1074/jbc.M109.088005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuma S, Poole B. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc Natl Acad Sci U S A. 1978;75:3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SA, Shaked GM, Bredesen DE, Koo EH. Mechanism of cytotoxicity mediated by the C31 fragment of the amyloid precursor protein. Biochem Biophys Res Commun. 2009;388:450–455. doi: 10.1016/j.bbrc.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino L, Ikin AF, Nairn AC, Pursnani A, Buxbaum JD. The carboxyl-terminus of BACE contains a sorting signal that regulates BACE trafficking but not the formation of total A(beta) Mol Cell Neurosci. 2002;19:175–185. doi: 10.1006/mcne.2001.1065. [DOI] [PubMed] [Google Scholar]
- Puertollano R, Bonifacino JS. Interactions of GGA3 with the ubiquitin sorting machinery. Nat Cell Biol. 2004;6:244–251. doi: 10.1038/ncb1106. [DOI] [PubMed] [Google Scholar]
- Qing H, Zhou W, Christensen MA, Sun X, Tong Y, Song W. Degradation of BACE by the ubiquitin-proteasome pathway. FASEB J. 2004;18:1571–1573. doi: 10.1096/fj.04-1994fje. [DOI] [PubMed] [Google Scholar]
- Ren Z, Schenk D, Basi GS, Shapiro IP. Amyloid beta-protein precursor juxtamembrane domain regulates specificity of gamma-secretase-dependent cleavages. J Biol Chem. 2007;282:35350–35360. doi: 10.1074/jbc.M702739200. [DOI] [PubMed] [Google Scholar]
- Richter L, Munter LM, Ness J, Hildebrand PW, Dasari M, Unterreitmeier S, Bulic B, Beyermann M, Gust R, Reif B, Weggen S, Langosch D, Multhaup G. Amyloid beta 42 peptide (Abeta42)-lowering compounds directly bind to Abeta and interfere with amyloid precursor protein (APP) transmembrane dimerization. Proc Natl Acad Sci U S A. 2010;107:14597–14602. doi: 10.1073/pnas.1003026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohn TT, Ivins KJ, Bahr BA, Cotman CW, Cribbs DH. A monoclonal antibody to amyloid precursor protein induces neuronal apoptosis. J Neurochem. 2000;74:2331–2342. doi: 10.1046/j.1471-4159.2000.0742331.x. [DOI] [PubMed] [Google Scholar]
- Rossjohn J, Cappai R, Feil SC, Henry A, McKinstry WJ, Galatis D, Hesse L, Multhaup G, Beyreuther K, Masters CL, Parker MW. Crystal structure of the N-terminal, growth factor-like domain of Alzheimer amyloid precursor protein. Nat Struct Biol. 1999;6:327–331. doi: 10.1038/7562. [DOI] [PubMed] [Google Scholar]
- Russ WP, Engelman DM. The GxxxG motif: a framework for transmembrane helix-helix association. J Mol Biol. 2000;296:911–919. doi: 10.1006/jmbi.1999.3489. [DOI] [PubMed] [Google Scholar]
- Sastre M, Steiner H, Fuchs K, Capell A, Multhaup G, Condron MM, Teplow DB, Haass C. Presenilin-dependent gamma-secretase processing of beta-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep. 2001;2:835–841. doi: 10.1093/embo-reports/kve180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Kienlen-Campard P, Ahmed M, Liu W, Li H, Elliott JI, Aimoto S, Constantinescu SN, Octave JN, Smith SO. Inhibitors of amyloid toxicity based on beta-sheet packing of Abeta40 and Abeta42. Biochemistry. 2006;45:5503–5516. doi: 10.1021/bi052485f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheff SW, Price DA. Alzheimer's disease-related alterations in synaptic density: neocortex and hippocampus. J Alzheimers Dis. 2006;9:101–115. doi: 10.3233/jad-2006-9s312. [DOI] [PubMed] [Google Scholar]
- Scheuermann S, Hambsch B, Hesse L, Stumm J, Schmidt C, Beher D, Bayer TA, Beyreuther K, Multhaup G. Homodimerization of amyloid precursor protein and its implication in the amyloidogenic pathway of Alzheimer's disease. J Biol Chem. 2001;276:33923–33929. doi: 10.1074/jbc.M105410200. [DOI] [PubMed] [Google Scholar]
- Schubert D, Heinemann S, Carlisle W, Tarikas H, Kimes B, Patrick J, Steinbach JH, Culp W, Brandt BL. Clonal cell lines from the rat central nervous system. Nature. 1974;249:224–227. doi: 10.1038/249224a0. [DOI] [PubMed] [Google Scholar]
- Shaked GM, Kummer MP, Lu DC, Galvan V, Bredesen DE, Koo EH. Abeta induces cell death by direct interaction with its cognate extracellular domain on APP (APP 597–624) FASEB J. 2006;20:1254–1256. doi: 10.1096/fj.05-5032fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba T, Kametaka S, Kawasaki M, Shibata M, Waguri S, Uchiyama Y, Wakatsuki S. Insights into the phosphoregulation of beta-secretase sorting signal by the VHS domain of GGA1. Traffic. 2004;5:437–448. doi: 10.1111/j.1600-0854.2004.00188.x. [DOI] [PubMed] [Google Scholar]
- Soba P, Eggert S, Wagner K, Zentgraf H, Siehl K, Kreger S, Löwer A, Langer A, Merdes G, Paro R, Masters CL, Müller U, Kins S, Beyreuther K. Homo- and heterodimerization of APP family members promotes intercellular adhesion. EMBO J. 2005;24:3624–3634. doi: 10.1038/sj.emboj.7600824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodhi CP, Rampalli S, Perez RG, Koo EH, Quinn B, Gottardi-Littell NR. The endocytotic pathway is required for increased A beta 42 secretion during apoptosis. Brain Res Mol Brain Res. 2004;128:201–211. doi: 10.1016/j.molbrainres.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Struhl G, Adachi A. Requirements for presenilin-dependent cleavage of notch and other transmembrane proteins. Mol Cell. 2000;6:625–636. doi: 10.1016/s1097-2765(00)00061-7. [DOI] [PubMed] [Google Scholar]
- Sudo H, Jiang H, Yasukawa T, Hashimoto Y, Niikura T, Kawasumi M, Matsuda S, Takeuchi Y, Aiso S, Matsuoka M, Murayama Y, Nishimoto I. Antibody-regulated neurotoxic function of cell-surface beta-amyloid precursor protein. Mol Cell Neurosci. 2000;16:708–723. doi: 10.1006/mcne.2000.0910. [DOI] [PubMed] [Google Scholar]
- Tamagno E, Bardini P, Obbili A, Vitali A, Borghi R, Zaccheo D, Pronzato MA, Danni O, Smith MA, Perry G, Tabaton M. Oxidative stress increases expression and activity of BACE in NT2 neurons. Neurobiol Dis. 2002;10:279–288. doi: 10.1006/nbdi.2002.0515. [DOI] [PubMed] [Google Scholar]
- Terry RD, Masliah E, Salmon DP, Butters N, DeTeresa R, Hill R, Hansen LA, Katzman R. Physical basis of cognitive alterations in Alzheimer's disease: synapse loss is the major correlate of cognitive impairment. Ann Neurol. 1991;30:572–580. doi: 10.1002/ana.410300410. [DOI] [PubMed] [Google Scholar]
- Tesco G, Koh YH, Tanzi RE. Caspase activation increases beta-amyloid generation independently of caspase cleavage of the beta-amyloid precursor protein (APP) J Biol Chem. 2003;278:46074–46080. doi: 10.1074/jbc.M307809200. [DOI] [PubMed] [Google Scholar]
- Tesco G, Koh YH, Kang EL, Cameron AN, Das S, Sena-Esteves M, Hiltunen M, Yang SH, Zhong Z, Shen Y, Simpkins JW, Tanzi RE. Depletion of GGA3 stabilizes BACE and enhances beta-secretase activity. Neuron. 2007;54:721–737. doi: 10.1016/j.neuron.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thinakaran G, Borchelt DR, Lee MK, Slunt HH, Spitzer L, Kim G, Ratovitsky T, Davenport F, Nordstedt C, Seeger M, Hardy J, Levey AI, Gandy SE, Jenkins NA, Copeland NG, Price DL, Sisodia SS. Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron. 1996;17:181–190. doi: 10.1016/s0896-6273(00)80291-3. [DOI] [PubMed] [Google Scholar]
- Tyler SJ, Dawbarn D, Wilcock GK, Allen SJ. alpha- and beta-secretase: profound changes in Alzheimer's disease. Biochem Biophys Res Commun. 2002;299:373–376. doi: 10.1016/s0006-291x(02)02635-9. [DOI] [PubMed] [Google Scholar]
- Velliquette RA, O'Connor T, Vassar R. Energy inhibition elevates β-secretase levels and activity and is potentially amyloidogenic in APP transgenic mice: possible early events in Alzheimer's disease pathogenesis. J Neurosci. 2005;25:10874–10883. doi: 10.1523/JNEUROSCI.2350-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim CA, Tangredi MM, Peltan ID, Lee BM, Irizarry MC, Kinoshita A, Hyman BT. Demonstration of BACE (beta-secretase) phosphorylation and its interaction with GGA1 in cells by fluorescence-lifetime imaging microscopy. J Cell Sci. 2004;117:5437–5445. doi: 10.1242/jcs.01422. [DOI] [PubMed] [Google Scholar]
- Wahle T, Prager K, Raffler N, Haass C, Famulok M, Walter J. GGA proteins regulate retrograde transport of BACE1 from endosomes to the trans-Golgi network. Mol Cell Neurosci. 2005;29:453–461. doi: 10.1016/j.mcn.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Wahle T, Thal DR, Sastre M, Rentmeister A, Bogdanovic N, Famulok M, Heneka MT, Walter J. GGA1 is expressed in the human brain and affects the generation of amyloid β-peptide. J Neurosci. 2006;26:12838–12846. doi: 10.1523/JNEUROSCI.1982-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DM, Tseng BP, Rydel RE, Podlisny MB, Selkoe DJ. The oligomerization of amyloid beta-protein begins intracellularly in cells derived from human brain. Biochemistry. 2000;39:10831–10839. doi: 10.1021/bi001048s. [DOI] [PubMed] [Google Scholar]
- Walsh DM, Minogue AM, Sala Frigerio C, Fadeeva JV, Wasco W, Selkoe DJ. The APP family of proteins: similarities and differences. Biochem Soc Trans. 2007;35:416–420. doi: 10.1042/BST0350416. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ha Y. The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain. Mol Cell. 2004;15:343–353. doi: 10.1016/j.molcel.2004.06.037. [DOI] [PubMed] [Google Scholar]
- Wen Y, Onyewuchi O, Yang S, Liu R, Simpkins JW. Increased beta-secretase activity and expression in rats following transient cerebral ischemia. Brain Res. 2004;1009:1–8. doi: 10.1016/j.brainres.2003.09.086. [DOI] [PubMed] [Google Scholar]
- Wen Y, Yu WH, Maloney B, Bailey J, Ma J, Marié I, Maurin T, Wang L, Figueroa H, Herman M, Krishnamurthy P, Liu L, Planel E, Lau LF, Lahiri DK, Duff K. Transcriptional regulation of beta-secretase by p25/cdk5 leads to enhanced amyloidogenic processing. Neuron. 2008;57:680–690. doi: 10.1016/j.neuron.2008.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LB, Lindholm K, Yan R, Citron M, Xia W, Yang XL, Beach T, Sue L, Wong P, Price D, Li R, Shen Y. Elevated beta-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat Med. 2003;9:3–4. doi: 10.1038/nm0103-3. [DOI] [PubMed] [Google Scholar]
- Yasojima K, Akiyama H, McGeer EG, McGeer PL. Reduced neprilysin in high plaque areas of Alzheimer brain: a possible relationship to deficient degradation of beta-amyloid peptide. Neurosci Lett. 2001;297:97–100. doi: 10.1016/s0304-3940(00)01675-x. [DOI] [PubMed] [Google Scholar]
- Zhang YW, Xu H. Molecular and cellular mechanisms for Alzheimer's disease: understanding APP metabolism. Curr Mol Med. 2007;7:687–696. doi: 10.2174/156652407782564462. [DOI] [PubMed] [Google Scholar]
- Zhao J, Fu Y, Yasvoina M, Shao P, Hitt B, O'Connor T, Logan S, Maus E, Citron M, Berry R, Binder L, Vassar R. β-Site amyloid precursor protein cleaving enzyme 1 levels become elevated in neurons around amyloid plaques: implications for Alzheimer's disease pathogenesis. J Neurosci. 2007;27:3639–3649. doi: 10.1523/JNEUROSCI.4396-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]