Abstract
Epigenetic modification of the mammalian genome by DNA methylation (5-methylcytosine) has a profound impact on chromatin structure, gene expression and maintenance of cellular identity1. The recent demonstration that members of the Ten-eleven translocation (Tet) family of proteins can convert 5-methylcytosine to 5-hydroxymethylcytosine raised the possibility that Tet proteins are capable of establishing a distinct epigenetic state2,3. We have recently demonstrated that Tet1 is specifically expressed in murine embryonic stem (ES) cells and is required for ES cell maintenance2. Using chromatin immunoprecipitation coupled with high-throughput DNA sequencing, here we show in mouse ES cells that Tet1 is preferentially bound to CpG-rich sequences at promoters of both transcriptionally active and Polycomb-repressed genes. Despite an increase in levels of DNA methylation at many Tet1-binding sites, Tet1 depletion does not lead to downregulation of all the Tet1 targets. Interestingly, although Tet1-mediated promoter hypomethylation is required for maintaining the expression of a group of transcriptionally active genes, it is also involved in repression of Polycomb-targeted developmental regulators. Tet1 contributes to silencing of this group of genes by facilitating recruitment of PRC2 to CpG-rich gene promoters. Thus, our study not only establishes a role for Tet1 in modulating DNA methylation levels at CpG-rich promoters, but also reveals a dual function of Tet1 in promoting transcription of pluripotency factors as well as participating in the repression of Polycomb-targeted developmental regulators.
The Tet protein family includes three members (Tet1–3), all of which have the capacity to convert 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) in a 2-oxoglutarate- and Fe(II)-dependent manner2,3. Consistent with the relative enrichment of 5hmC in ES cells, Tet1 is highly expressed in undifferentiated ES cells and Tet1 messenger RNA levels decrease upon ES cell differentiation2,3. Lentiviral-mediated depletion of Tet1 in mouse E14 ES cells cultured under feeder-free conditions leads to phenotypic changes that include partial loss of alkaline phosphatase activity and SSEA1 immunoreactivity, decreased self-renewal capacity and proliferation rate, downregulation of pluripotency factor Nanog and upregulation of differentiation genes (for example, lineage markers for trophectorderm and primitive endoderm in a subset of cells)2. Thus, Tet1 may be required for mouse ES cell maintenance.
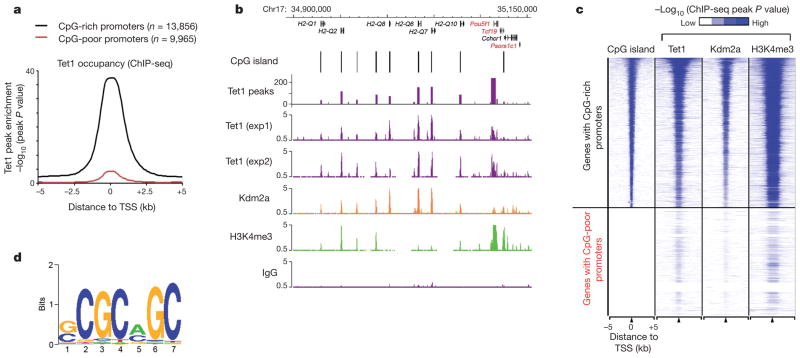
To gain insights into the mechanism by which Tet1 contributes to ES cell function, we investigated the genome-wide distribution of Tet1 in mouse ES cells by chromatin immunoprecipitation coupled with high-throughput DNA sequencing (ChIP-seq) using a highly specific Tet1 antibody (Supplementary Fig. 1a). Analysis of replicate ChIP-seq experiments identified a total of 35,564 binding sites with high confidence (P <10−8, or false discovery rate (FDR) of 0.01) (Supplementary Fig. 1b, c and Supplementary Table 1). In contrast, parallel experiments using rabbit IgG did not yield specific enrichment (Fig. 1b and Supplementary Fig. 1c). Moreover, ChIP-seq analysis also indicated that Tet1 occupancy was generally reduced in fluorescence-activated cell sorting (FACS)-sorted Tet1-depleted ES cells (Supplementary Fig. 2a). ChIP followed by quantitative polymerase chain reaction (qPCR) analysis further confirmed decreased Tet1 occupancy on randomly selected Tet1-binding sites in response to Tet1 depletion (Supplementary Fig. 2b). Most Tet1 binding sites are located in gene-rich euchromatic regions, as 79.8% of all Tet1-bound loci are within intragenic regions or 5 kb intergenic regions up- or downstream of annotated genes (Supplementary Fig. 3a, b). Similar to other CXXC zinc-finger-domain-containing proteins (for example, Cfp1 and Kdm2a)4,5, Tet1 is enriched (86.6%) at CpG islands (Fig. 1a–c). Consistently, de novo motif discovery analysis6 identified a CpG-rich sequence as the highest ranking motif within Tet1-bound regions (Fig. 1d). Quantification of CpG density within Tet1-binding loci indicated that, similar to Kdm2a (Supplementary Fig. 4a, b), Tet1 occupancy positively correlates with CpG density (Supplementary Fig. 4c). Collectively, the above results indicate that Tet1 high-affinity binding sites are generally enriched for CpG-rich sequences.
Figure 1. Tet1 is enriched at genomic regions with high-density CpG dinucleotides.
a, Genome-wide occupancy of Tet1 at all annotated gene promoters in ES cells (black, CpG-rich genes; red, CpG-poor genes). The enrichment of Tet1 binding was determined by ChIP-seq analysis. Average Tet1 binding measured by −log10 (peak P values) in 200-bp bins is shown within genomic regions covering 5 kb up- and downstream of TSSs. b, Enrichment of Tet1 (purple), Kdm2a (orange)4 and H3K4me3 (green)13 measured by ChIP-seq at representative genes in ES cells (black, CpG-rich; red, CpG-poor). ChIP-seq data are shown in reads per million with the y-axis floor set to 0.5 reads per million. Genomic regions with statistically significant enrichment of Tet1 binding (measured by log10 (peak P values); P < 10−8) are also indicated. c, Heatmap representation of genomic regions with high-density CpG sites (CpG islands), binding profiles of Tet1, Kdm2a4 and H3K4me313 in ES cells at all annotated mouse genes promoters (5 kb flanking TSSs of Refseq genes). The heatmap is rank-ordered from genes with CpG islands of longest length to no CpG islands within 5-kb genomic regions flanking TSSs. The presence of CpG islands is shown in colour (blue, present; white, absent). ChIP-seq enrichment was measured by −log10 (peak P values) and is shown by colour scale. The following colour scales (white, no enrichment; blue, high enrichment) were used for Tet1/Kdm2a and H3K4me3 respectively: (0, 50) and (0, 200). d, A DNA motif that is enriched in Tet1-bound loci in ES cells.
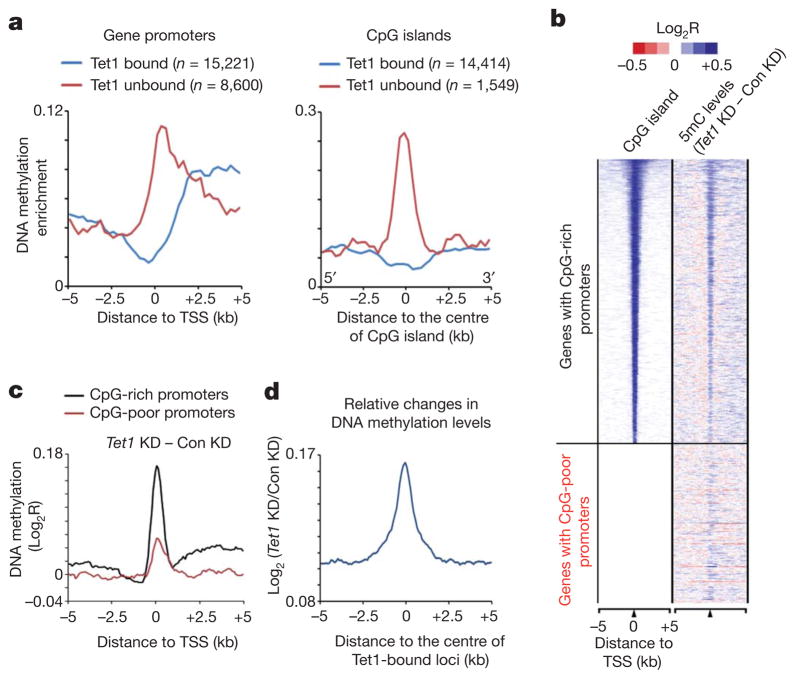
Because Tet proteins are capable of converting 5mC to 5hmC2,3, we investigated the relationship between Tet1 occupancy and DNA methylation in mouse ES cells using methylated DNA immunoprecipitation coupled with mouse whole-genome tiling microarrays (MeDIP-chip). We found that DNA methylation is generally excluded from transcription start sites (TSSs) of Tet1-bound gene promoters (Fig. 2a, blue line in left panel). In contrast, Tet1-unbound gene promoters are frequently DNA methylated (Fig. 2a, red line in left panel). These results are consistent with previous studies demonstrating that CpG-rich gene promoters, where Tet1 is enriched (Fig. 1), are generally hypomethylated7,8. Further analysis indicates that CpG islands not bound by Tet1 are associated with higher 5mC levels compared to Tet1-bound CpG islands (Fig. 2a, right panel). Thus, Tet1 occupancy at gene promoters is inversely correlated to levels of DNA methylation.
Figure 2. Tet1 maintains a DNA hypomethylated state at Tet1-bound regions.
a, The distribution frequency of regions enriched with DNA methylation is shown for Tet1-bound (blue) and unbound (red) gene promoters (left) or CpG islands (right) in mouse ES cells. b, Heatmap representation of CpG islands and the changes in DNA methylation (5mC) in response to Tet1 depletion. The DNA methylation gained after Tet1 depletion was calculated by deduction of 5mC levels in control knockdown (Con KD) from that in Tet1 knockdown (Tet1 KD). c, Changes in 5mC levels in response to Tet1 knockdown are shown for both CpG-rich and CpG-poor gene promoters. Note that proximal promoters and 5′ intragenic regions of CpG-rich genes are associated with a higher increase in 5mC levels as compared to those of CpG-poor genes in response to Tet1 depletion. d, An increase in 5mC levels in response to Tet1 depletion is specifically enriched at the centre of Tet1-binding loci. Changes in 5mC levels between control knockdown and Tet1 knockdown ES cells were determined by co-hybridizing and analysing genomic DNA from control knockdown and Tet1 knockdown cells on the whole-genome tiling microarrays.
To investigate whether Tet1 is required for maintaining the hypomethylated state at Tet1-bound regions, we analysed DNA methylation profiles in Tet1-depleted ES cells and demonstrated that Tet1 deficiency led to a general increase in 5mC levels at both TSSs and genomic regions flanking the proximal promoters of CpG-rich genes (Fig. 2b, c and Supplementary Fig. 5a, b). An increase in 5mC levels was also detected within proximal promoter regions of a subset of CpG-poor gene promoters (Fig. 2b, c and Supplementary Fig. 5a, b). The observed 5mC changes in Tet1-depleted cells were not due to interarray variations as a co-hybridization strategy analysing biologically independent replicates also revealed that the increase in 5mC levels induced by Tet1 deficiency was generally enriched at Tet1-binding sites (Fig. 2d and Supplementary Table 2). Locus-specific bisulphite sequencing confirmed that Tet1-binding sites and their surrounding regions became more DNA methylated in response to Tet1 depletion (Supplementary Fig. 6). Collectively, these data suggest that Tet1 binding is required for maintaining a DNA hypomethylated state at a large cohort of CpG-rich gene promoters.
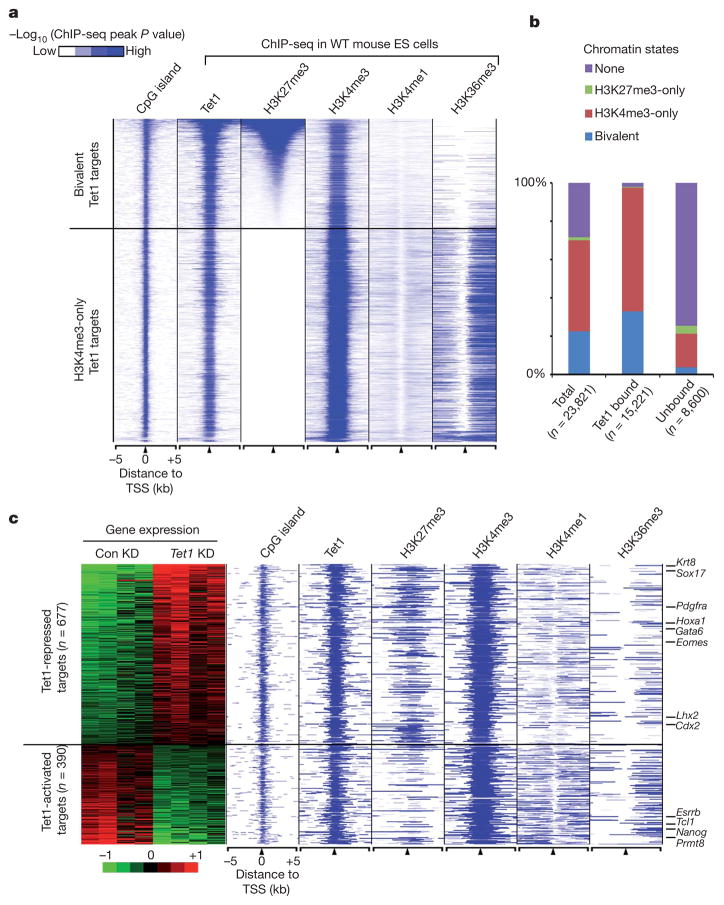
Previous studies have established a link between DNA methylation and histone methylation9–11. To explore a potential relationship between Tet1 occupancy and histone modifications, we compared the binding profile of Tet1 with that of major histone modifications in mouse ES cells previously determined by ChIP-seq (Supplementary Table 3)12,13. We found that histone H3 lysine 4 trimethylation (H3K4me3) is positively correlated to Tet1 binding at gene promoters, as 71.3% of all Tet1-binding sites (n = 25,359) overlapped with H3K4me3 peaks (Fig. 1c). Analysis of the histone modification profiles that flank TSSs of Tet1-bound genes revealed two categories of Tet1 targets (Fig. 3a, b and Supplementary Table 4). The first group is associated with bivalent domains, a chromatin state characterized by the presence of both H3K4me3 and H3K27me314. Interestingly, bivalent gene promoters in ES cells are generally hypomethylated15. In contrast, the second group is associated with active histone marks, including H3K4me3, H3K4me1 and H3K36me3 (Fig. 3a). These data indicate that Tet1 can associate with both actively transcribed as well as repressed target genes. Gene ontology analysis indicated that genes related to development and cell differentiation are highly enriched in the first group of Tet1 targets, whereas genes involved in housekeeping functions are enriched in the second group of Tet1 targets (Supplementary Fig. 7).
Figure 3. Tet1 binds to and functions in both repressed (bivalent) and actively transcribed (H3K4me3-only) genes.
a, Heatmap representation of genomic regions with high-density CpG sites (CpG islands), binding profile of Tet1, and major histone modifications (H3K4me1 (ref. 12), H3K4me3, H3K27me3 and H3K36me3 (ref. 13)) in mouse ES cells at indicated Tet1 target genes (5 kb flanking TSSs). The heatmap is rank-ordered from genes with highest H3K27me3 enrichment to no H3K27me3 within 5-kb genomic regions flanking TSSs. The following colour scales (white, no enrichment; blue, high enrichment) were used for Tet1/H3K27me3/H3K4me1/ H3K36me3 and H3K4me3 respectively: (0, 50) and (0, 200). WT, wild type. b, Relative percentage of genes with different chromatin states shown for all genes, Tet1-bound and unbound genes. c, Heatmap representation of differentially expressed Tet1 targets between control knockdown and Tet1 knockdown mouse ES cells. Note that Tet1-repressed targets are preferentially associated with bivalent chromatin states, whereas Tet1-activated targets are generally H3K4me3-only genes.
The fact that Tet1 occupies the promoters of actively transcribed as well as repressed genes suggests that Tet1 might have a dual function in transcription regulation. Microarray analysis comparing the gene expression of control and Tet1-depleted mouse ES cells identified a total of 1,332 genes that are differentially expressed (788 upregulated and 544 downregulated in Tet1 knockdown cells) (Supplementary Fig. 8). Of these differentially expressed genes, a significant percentage (80%) are associated with Tet1 occupancy within 5 kb up- or downstream of their TSSs (1,067 out of 1,332) (Fig. 3c and Supplementary Table 5). Interestingly, despite the fact that DNA methylation has been primarily associated with transcriptional repression, more Tet1 targets are upregulated rather than downregulated in response to Tet1 depletion (677 targets are upregulated, P = 2.0 × 10−45, compared with 390 targets downregulated, P = 4.1 × 10−5, Fisher’s exact test) (Fig. 3c and Supplementary Fig. 8a), indicating that Tet1 may also be involved in gene repression in mouse ES cells. Notably, genes with known functions in development and differentiation, for example, Cdx2 (trophectoderm), Sox17 (endoderm) and Krt8 (ectoderm), are among the upregulated Tet1 targets (Fig. 3c and Supplementary Fig. 8b). In contrast, genes related to pluripotency and ES cell functions (for example, Nanog, Tcl1 and Esrrb) are among the downregulated Tet1 targets (Fig. 3c and Supplementary Fig. 8b). Consistent with the notion that changes in gene expression in response to Tet1 depletion are mainly due to Tet1-occupancy-mediated effects, instead of a secondary effect due to Nanog downregulation, overexpression of Nanog in Tet1-depleted targets (Supplementary Fig. 9a, b). Notably, the rescued targets include pluripotency-related genes such as Tcl1 and Esrrb. Gene expression profiling and qPCR with reverse transcription (RT–qPCR) analysis demonstrated that overexpression of Nanog rescued a subset of genes through direct (Nanog bound) or indirect (Nanog unbound) regulation (Supplementary Fig. 9a–c and Supplementary Table 6). Collectively, these results indicate that Tet1 is not only required for maintaining the expression of a subset of genes important for ES cell pluripotency, but also required for the repression of a cohort of developmental regulators.
Because many developmental regulators are repressed by Polycomb repressive complexes PRC1 and PRC2 (refs 16, 17), we sought to determine whether Tet1 might facilitate silencing of developmental regulators by promoting Polycomb repression. Comparison of our expression data sets to a published data set16 revealed that 43% of Tet1-repressed genes were also in the upregulated gene list of Eed-deficient ES cells, which is significantly higher than that expected by chance (43% versus 9.5%, P = 3.59 × 10−144, Fisher’s exact test), supporting a potential role for Tet1 in PRC2-mediated repression of developmental regulators. Indeed, analysis of the histone modification states of Tet1-regulated genes in wild-type ES cells indicated that Tet1-represssed genes were preferentially associated with H3K27me3 (Fig. 3c), a mark deposited by PRC218,19. In contrast, Tet1-activated targets were preferentially associated with H3K36me3, a mark associated with transcriptional elongation20 (Fig. 3c), supporting the notion that Tet1-mediated DNA hypomethylation at these gene promoters may facilitate their expression.
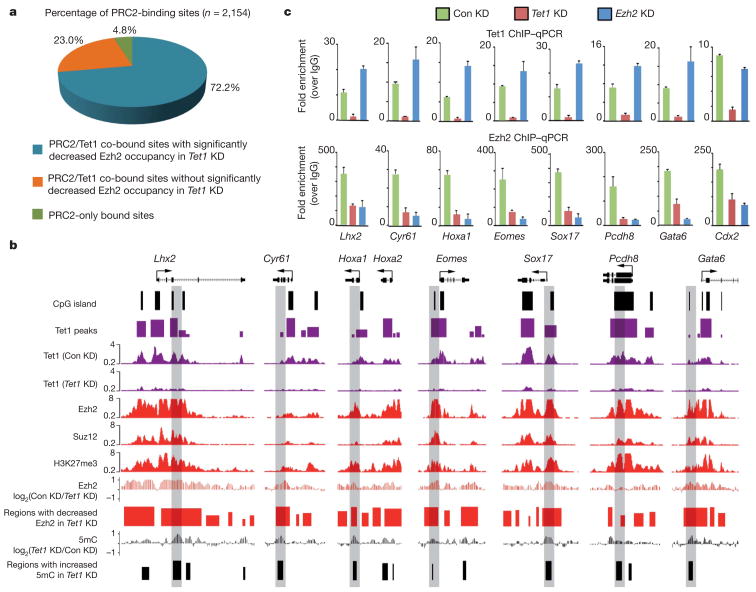
The fact that genes upregulated in response to Tet1 knockdown significantly overlap with those upregulated by Eed deficiency indicates that Tet1 may cooperate with PRC2 in silencing this group of genes. Given that the protein levels of PRC2 subunits are not significantly altered in response to Tet1 depletion (Supplementary Fig. 10), Tet1 is unlikely to affect PRC2 expression or stability. As 95.2% of PRC2-binding sites (defined as Ezh2/Suz12 co-bound21) overlapped with Tet1-bound loci (Fig. 4a), we next evaluated the effect of Tet1 depletion on the chromatin-binding ability of PRC2. ChIP coupled with whole genome tiling microarrays (ChIP-chip) in control and Tet1 knockdown cells revealed that Tet1 depletion impaired the binding of Ezh2, a core subunit of PRC2, to a large fraction (72.2%) of PRC2-binding sites (Fig. 4a, b, Supplementary Figs 11, 12a and Supplementary Table 7). ChIP–qPCR further confirmed the effect of Tet1 knockdown on Ezh2/ Suz12 recruitment (Fig. 4c and Supplementary Fig. 12b). Interestingly, depletion of Ezh2 did not affect Tet1 binding to chromatin (Fig. 4c), indicating that Tet1 may function upstream of PRC2. Furthermore, overexpression of Nanog in Tet1-depleted cells also failed to fully rescue the Ezh2 binding to Tet1/PRC2 co-bound targets (Supplementary Fig. 9d). Given that previous purification of the PRC2 complex did not uncover Tet1 as an associated component18,19,22 and the fact that a stable interaction between Tet1 and PRC2 could not be demonstrated (unpublished observation), we favour a model in which Tet1 may indirectly contribute to PRC2 recruitment by maintaining a DNA hypomethylated state at PRC2-bound loci. This model is supported by a recent study demonstrating that DNA methylation impedes binding of PRC2 to chromatin23.
Figure 4. Tet1 is required for chromatin binding of PRC2 in mouse ES cells.
a, Tet1 depletion affects the binding of PRC2 to the majority of its targets. PRC2-binding sites are divided into three groups (Tet1/PRC2 co-bound Tet1 dependent, Tet1 independent and PRC2-only bound). b, Shown are Tet1, Ezh2 and Suz12 (ref. 21), and H3K27me3 (ref. 13) occupancy, and the effect of Tet1 depletion on Ezh2 occupancy and 5mC levels at seven representative Tet1-repressed bivalent targets. Regions associated with significant changes in Ezh2 occupancy between control and Tet1-depleted ES cells were measured by whole-genome tiling microarrays. Genomic regions that are further examined by locus-specific ChIP–qPCR in c are shaded. c, ChIP–qPCR analysis of Tet1 (top panels) and Ezh2 (bottom panels) occupancy at the promoters of eight representative Tet1-repressed targets in control (Con KD), Tet1-depleted (Tet1 KD) and Ezh2-depleted (Ezh2 KD) ES cells. Error bars represents standard deviation determined from duplicate experiments.
In summary, we demonstrate that Tet1 is preferentially enriched in CpG-island-containing gene promoters in mouse ES cells. This result is consistent with the presence of a CXXC domain in Tet1 and the demonstration that the CXXC domain is preferentially bound to CpG-rich sequences4,5. The nonrandom genomic distribution of Tet1 suggests that genes with CpG-rich promoters are selectively regulated by a Tet1-dependent epigenetic state (that is, 5hmC) or active demethylation process. The convergence of CpG-binding proteins at CpG islands, including Cfp1, Kdm2a and Tet1, cooperatively contributes to the establishment of a specialized chromatin/epigenetic state at CpG-rich gene promoters. Specifically, Cfp1 confers H3K4me3 by recruiting the H3K4me3 methyltransferase Setd1 (ref. 5); Kdm2a leads to depletion of H3K36me2 (ref. 4), and Tet1 maintains DNA at a hypomethylation state at CpG islands (Fig. 2).
In addition to binding to gene promoters with CpG islands, Tet1 also binds to a subset of actively transcribed CpG-poor gene promoters, such as Nanog, Tcl1 and Esrrb, whose gene products have an important role in ES cell maintenance. In this scenario, Tet1 has an important role in promoting the transcriptionally active state of these genes by maintaining a hypomethylated promoter state2. Interestingly, Tet1 also contributes to the silencing of a group of developmental regulators and somatic lineage differentiation genes that are silenced by Polycomb group proteins (Fig. 3c). Depletion of Tet1 leads to a decrease in Ezh2 occupancy at many PRC2 targets, indicating that Tet1 contributes to PRC2 recruitment. Therefore, our study reveals a novel function for Tet1 in the recruitment of PRC2 and silencing of developmental regulators, which also contributes to the role of Tet1 in mouse ES cell maintenance. We note that, in contrast to our results, a recent study has shown that knockdown of Tet1 alone is not sufficient to confer any noticeable phenotype in mouse ES cells24. This difference is probably due to the use of different ES cell lines, culture conditions and knockdown efficiency (see Supplementary Information for details). Collectively, our study establishes a dual function for Tet1 in transcriptional regulation in mouse ES cells.
METHODS
Constructs and antibodies
All the constructs and antibodies used in this study have been described previously2,18 or were purchased from the following sources: EZH2 (Cell Signaling; catalogue no. 4905); EED (Santa Cruz; sc-133537); Jarid2 (Abcam; ab48137); AEBP2 (Proteintech group; 11232-2-AP), EZH1 (Abcam; ab64850) and actin (Sigma; AC-40).
Mouse ES cell cultures and lentiviral knockdown
Mouse E14Tg2A ES cells were cultured in feeder-free conditions2. For Tet1 knockdown, mouse ES cells were infected with lentiviruses expressing both the GFP reporter and short-hairpin RNA (shRNA) specific for Tet1 (5′-GCAGATGGCCGTGACACAAAT-3′). For Ezh2 knockdown, mouse ES cells were infected with lentiviruses expressing both the GFP reporter and shRNA specific for Ezh2 (5′-GTATGTGGGCATCGAACGA-3′) as previously described25. All analyses were performed usingTet1-orEzh2-depleted ES cells that were purified on the basis of GFP fluorescence by FACS 8 days after lentiviral transduction. Lentiviruses expressing GFP alone was used as a control.
RNA isolation, qPCR and expression microarray analysis
Total RNA from cultured cells was isolated using RNeasy Mini Kit (Qiagen), and cDNA was generated with Improm-IITM Reverse Transcription System (Promega). Real-time qPCR reactions were performed on an ABI PRISM 7700 Sequence Detection System (Applied Biosystems) using SYBR Green (Invitrogen). cDNA levels of target genes were analysed using comparative CT methods, where CT is the cycle threshold number and normalized to GAPDH. RT–qPCR primers are listed in Supplementary Table 8.
For expression microarray analysis comparing control and Tet1 knockdown ES cells, 2 μg of total RNA purified from GFP sorted cells were reverse-transcribed into cDNA with a T7-(dT)24 primer from a custom kit (Life Technologies). Biotinylated cRNA was then generated from the cDNA reaction using the BioArray High Yield RNA Transcript Kit. The cRNA was then fragmented in fragmentation buffer (40 mM Tris-acetate, pH 8.1, 100 mM KOAc and 150 mM MgOAc) at 94 °C for 35 min before microarray hybridization. Fifteen micrograms of fragmented cRNA was then added to a hybridization cocktail (0.05 mg ml−1 fragmented cRNA, 50 pM control oligonucleotide B2, BioB, BioC, BioD and cre hybridization controls, 0.1 mg ml−1 herring sperm DNA, 0.5 mg ml−1 acetylated BSA, 100 mM MES, 1 M Na+, 20 mM EDTA, 0.01% Tween 20). Ten micrograms of cRNA were used for hybridization to Affymetrix GeneChip Mouse Genome 430 2.0 Array. Hybridization was carried out at 45 °C for 16 h. The arrays were then washed and stained with R-phycoerythrin streptavidin, before scanning. Washing, scanning and basic analysis was carried out using Affymetrix GeneChip Microarray Suite 5.0 software. Raw signal intensity (.cel files) was RMA normalized using affy (R/bioconductor). For identification of differentially expressed genes, we used NIA array analysis tool (http://lgsun.grc.nia.nih.gov/ANOVA). Of all the probes present on the microarray, signal intensity of redundant probes was averaged before analysis. The following parameters were used for analysing statistically significant differential expression: threshold z-value to remove outliers, 10,000; Error Model, Max (Average, Bayesian); error variance averaging window, 200; proportion of highest error variances to be removed, 0.05; Bayesian degrees of freedom, 20; the FDR threshold was set at 0.05.
For heatmap display, RMA-normalized signal intensity was log2 transformed and median-centred. Heatmaps were generated using Cluster3 and Java Treeview.
ChIP-seq
ChIP-seq experiments were performed as described26. Briefly, cells were cross-linked with 1% formaldehyde at 25 °C for 10 min and sonicated to generate chromatin fragments of 200–500 bp. Chromatin fragments from 10–20×106 cells were immunoprecipitated using 8 μg of the Tet1 antibody2 or IgG control from two biologically independent samples. ChIP-seq library construction and Illumina sequencing were performed as described previously26. All sequencing reads were mapped to the mouse genome (mm8). Sequencing reads from both Tet1 ChIP-seq experiments were combined and Tet1-enriched regions were determined by the MACS program (version1.3.7.1). Sequencing reads from IgG control experiments were used as negative controls in MACS. Only uniquely mapped reads were retained and redundant reads were filtered out. The statistical cutoff used for identifying Tet1-binding sites was P value <10−8 (or FDR <1%) and fold enrichment (over IgG control) > 10. ChIP-seq data sets of H3K4me1 (ref. 12), H3K4me3, H3K27me3, H3K36me3 (ref. 13), Ezh2, Suz12 (ref. 21), Kdm2a (ref. 4) and RNA pol II (ref. 28) were obtained from previous publications and reanalysed in MACS using identical parameters (except statistical cutoff was set to P value <10−5). A summary of all ChIP-seq experiments used in this study (generated by this work and by previous publications) is provided in Supplementary Table 3. ChIP-seq sequencing read counts for each ChIP-seq experiments were binned into 400-bp windows at 100-bp steps along the genome and visualized in the Cisgenome browser29. To assign ChIP-seq enriched regions to genes, a complete set of Refseq genes was downloaded from the UCSC table browser (accessed May,2010). For all data sets, genes with enriched regions within 5 kb of their TSSs were called bound.
Gene ontology analysis
Functional enrichment analysis of bivalent and H3K4me3-only Tet1 were calculated by hypergeometric distribution followed by Benjamini correction in DAVID.
Genome-wide DNA methylation (5mC) analysis
Methylated DNA immunoprecipitation (MeDIP) was performed as described previously with minor modifications8. Briefly, genomic DNA was sequentially digested with proteinase K and RNase A, and purified by phenol/chloroform extraction. Purified genomic DNA was sonicated and heat denatured (95 °C, 10 min). An aliquot of sonicated genomic DNA was saved as input. Five micrograms of fragmented genomic DNA was immunoprecipitated with 5 μl of a monoclonal antibody against 5-methylcytidine (Eurogentec) at 4 °C overnight in a final volume of 500 μl of IP buffer (10 mM sodium phosphate (pH 7.0), 140 mM NaCl, 0.05% Triton X-100). We incubated the DNA–antibody mixture with 30 μl protein G Dynabeads (Invitrogen) for 2 h at 4 °C and washed it three times with 1 ml IP buffer. We then treated the beads with proteinase K for at least 3 h at 55 °C and purified the methylated DNA by phenol-chloroform extraction followed by ethanol precipitation. For whole-genome DNA tiling microarray analysis, immunoprecipitated DNA prepared from both control and Tet1-depleted ES cells were co-hybridized to mouse whole-genome tiling microarrays (NimbleGen).
Whole-genome tiling microarray analysis
For whole-genome DNA tiling microarray analysis of relative changes in 5mC levels or Ezh2 occupancy, immunoprecipitated DNA was prepared from both control and Tet1-depleted ES cells and amplified using whole genome amplification kit (Sigma). Amplified DNA was labelled (5′ Cy5- or Cy3-random nonamers, TriLink Biotechnologies) using the standard protocol (NimbleGen Arrays User’s Guide for ChIP-chip analysis). Hybridization of labelled samples to whole genome HD2 microarrays 4-array set (Roche/NimbleGen, ~2.1 million tiling probes per array, covering the entire non-repetitive portion of mouse genome) was carried out for 16–20 h at 42 °C using NimbleGen hybridization System 4. After stringent washes, microarrays were subsequently scanned using an Agilent scanner at 5-μm resolution. Data were extracted and analysed using NimbleScan v2.5 (Roche/NimbleGen).
For identification of probes associated with significant increase in 5mC levels or decrease in Ezh2 occupancy in response to Tet1 depletion in microarray experiments with the IP/IP configuration (DNA from control knockdown and Tet1 knockdown were co-hybridized to the same microarrays), a non-parametric one-sided Kolmogorov–Smirno (KS) test was used (KS score). Briefly, from the scaled log2-ratio data, a fixed-length window (750 bp) is placed around each consecutive probe and the one-sided KS test is applied to determine whether the probes are drawn from a significantly more positive distribution of intensity log-ratios than those in the rest of the array. The resulting score for each probe is the −log10 P value from the windowed KS test around that probe. Using NimbleScan v2.5, peak data files are generated from the P-value data files. NimbleScan software detects peaks by searching for at least 2 probes above a P-value minimum cutoff (−log10) of 2. Peaks within 500 bp of each other are merged. For calculating the absolute 5mC levels in control knockdown and Tet1 knockdown ES cells (Supplementary Fig. 5a), the MEDME program30 was used to correct the nonlinear relationship between MeDIP-chip signals (measured by microarray experiments with the IP/input configuration) and genomic CpG density.
For visualizing raw microarray signal intensity in the genome browser, probe level smoothing (log2 ratios of probes within 1 kb are averaged) was performed for each probe. For calculating the peak distribution, regions associated with significant changes in 5mC levels or Ezh2 occupancy were binned to 500-bp intervals using a 250-bp sliding window within genomic regions 5-kb up- and downstream of TSSs of annotated Refseq genes. Heatmaps were generated and visualized using Cluster3 and Java TreeView, respectively.
Locus-specific ChIP assays and bisulphite sequencing
Cells were fixed in a final concentration of 1% formaldehyde. After incubation at 25 °C for 10 min, the reaction was stopped by the addition of 125 mM glycine. ChIP assays were performed using a protocol associated with the ChIP assay kit (Upstate Biotechnology). After extensive washing, ChIPed DNA was eluted from the beads, and analysed on an ABI 7300 Real Time PCR System (Applied Biosystems) using SYBR Green (Invitrogen). Primer sequences are listed in Supplementary Table 9.
Bisulphite sequencing was performed as described previously with minor modifications2. Five micrograms of sodium-bisulphite-treated DNA samples was subjected to PCR amplification using the first set of primers; PCR products were used as templates for a subsequent PCR reaction using nested primers. The PCR products of the second reaction were then subcloned using the Invitrogen TA cloning Kit following the manufacturer’s instructions. PCRs and subcloning were performed in duplicate for each sample. The clones were sequenced using the M13 reverse primer. Primers for bisulphite sequencing are listed in Supplementary Table 10.
Supplementary Material
Acknowledgments
We thank B. Abraham and I. Chepelev for Illumina sequencing and data transfer; J. He and A. T. Nguyen for FACS sorting; O. Taranova for discussion; S. Wu for critical reading of the manuscript. This work was supported by NIH grants GM68804 (to Y.Z.), R56MH082068 (to Y.E.S.) and support from the Division of Intramural Research Program of National Heart, Lung and Blood Institute, NIH (K.Z.). S.I. is a research fellow of the Japan Society for the Promotion of Science. Y.Z. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Y.Z. conceived the project; H.W., A.C.D’A. and Y.Z. designed the experiments; H.W., A.C.D’A., S.I., Z.W. and K.C. performed the experiments; H.W. and K.X. analysed the data; H.W., A.C.D’A., K.Z., Y.E.S. and Y.Z. interpreted the data; H.W. and Y.Z. wrote the manuscript.
ChIP-seq and microarray data have been deposited in the Gene Expression Omnibus under accession number GSE26833. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nature Rev Genet. 2008;2008:129–140. doi: 10.1038/nrg2295. [DOI] [PubMed] [Google Scholar]
- 2.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackledge NP, et al. CpG islands recruit a histone H3 lysine 36 demethylase. Mol Cell. 2010;38:179–190. doi: 10.1016/j.molcel.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomson JP, et al. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature. 2010;464:1082–1086. doi: 10.1038/nature08924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein–DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nature Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 7.Fouse SD, et al. Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, PcG complex, and histone H3 K4/K27 trimethylation. Cell Stem Cell. 2008;2:160–169. doi: 10.1016/j.stem.2007.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 9.Mohn F, et al. Lineage-specific polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell. 2008;30:755–766. doi: 10.1016/j.molcel.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlesinger Y, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nature Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 12.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 15.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 17.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 19.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xiao T, et al. Phosphorylation of RNA polymerase II CTD regulates H3 methylation in yeast. Genes Dev. 2003;17:654–663. doi: 10.1101/gad.1055503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasini D, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464:306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 23.Wu H, et al. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu SC, Kallin EM, Zhang Y. Role of H3K27 methylation in the regulation of lncRNA expression. Cell Res. 2010;20:1109–1116. doi: 10.1038/cr.2010.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, et al. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji H, et al. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nature Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelizzola M, et al. MEDME: an experimental and analytical methodology for the estimation of DNA methylation levels based on microarray derived MeDIP-enrichment. Genome Res. 2008;18:1652–1659. doi: 10.1101/gr.080721.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.