Abstract
In this study, bacterial composition of iru produced by natural, uncontrolled fermentation of Parkia biglobosa seeds was assessed using culture-independent method in combination with culture-based genotypic typing techniques. PCR-denaturing gradient gel electrophoresis (DGGE) revealed similarity in DNA fragments with the two DNA extraction methods used and confirmed bacterial diversity in the 16 iru samples from different production regions. DNA sequencing of the highly variable V3 region of the 16S rRNA genes obtained from PCR-DGGE identified species related to Bacillus subtilis as consistent bacterial species in the fermented samples, while other major bands were identified as close relatives of Staphylococcus vitulinus, Morganella morganii, B. thuringiensis, S. saprophyticus, Tetragenococcus halophilus, Ureibacillus thermosphaericus, Brevibacillus parabrevis, Salinicoccus jeotgali, Brevibacterium sp. and uncultured bacteria clones. Bacillus species were cultured as potential starter cultures and clonal relationship of different isolates determined using amplified ribosomal DNA restriction analysis (ARDRA) combined with 16S–23S rRNA gene internal transcribed spacer (ITS) PCR amplification, restriction analysis (ITS-PCR-RFLP), and randomly amplified polymorphic DNA (RAPD-PCR). This further discriminated B. subtilis and its variants from food-borne pathogens such as B. cereus and suggested the need for development of controlled fermentation processes and good manufacturing practices (GMP) for iru production to achieve product consistency, safety quality, and improved shelf life.
Keywords: iru, fermentation, safety, Bacillus, DGGE
Introduction
Fermented vegetable protein seeds used for condiments production in West Africa include African locust bean [Parkia biglobosa (Jacq. Benth)], melon seeds [Citrullus vulgaris (Schrad)], castor oil seeds (Ricinus communis), fluted pumpkin seeds (Telfaria occidentale), African yam bean (Stenophylis stenocarpa), cotton seeds [Gossypium hirsitium (L.)], roselle seeds (Hibiscus sabdariffa), and baobab seeds [Adansonia digitata (L.)] (Odunfa and Oyewole, 1998; Ouoba et al., 2008; Parkouda et al., 2010). Condiments constitute a significant proportion of African diets where they serve as flavorsome and culinary components in various dishes (Achi, 2005). They include iru or dawadawa in Nigeria and Ghana; soumbala, bikalga, and maari in Burkina Faso; afitin and sonru in Benin Republic; nététou in Senegal; kinda in Sierra Leone; dawadawa botso in Niger Republic; datou in Mali; mbuja in Cameroon and furundu in Sudan (N'Dir et al., 1994; Ouoba et al., 2008, 2010; Parkouda et al., 2010).
Though, the vegetable seeds used for the production of these condiments differ considerably from one region to another (Achi, 2005). Nonetheless, the production process is generally characterized by spontaneous solid substrate alkaline fermentation with increase in pH as a result of extensive hydrolysis of the proteins into peptides, amino acids and ammonia, which favors the dominance of Bacillus spp. as the fermenting organisms (Kiers et al., 2000; Sarkar et al., 2002; Ouoba et al., 2003; Achi, 2005). The art is also carried out in exclusive uncontrollable environmental conditions thereby yielding products with variation in quality and organoleptic properties (Sanni, 1993; Steinkraus, 1997; Ouoba et al., 2007). This necessitates the need for the development of appropriate starter cultures to initiate fermentation for the production of consistent products with acceptable qualities.
The processing operations of African locust bean to produce iru involve cleaning and sorting, boiling, dehulling, and fermentation at ambient temperature (Oyeyiola, 1988). Investigations on the microbiology of iru using selective isolation and cultivation based on traditional culture-dependent methods including the biochemical changes that occur during fermentation have been documented (Odunfa, 1981, 1985; Omafuvbe et al., 2004). These techniques often fail to account for minor microbial populations; stressed and injured cells that are present in low number (Fleet, 1999), and have been observed not to give a complete representation of food microbial communities (Kesmen et al., 2012). Also, they are bias, unreliable and lack accurate detection of population dynamics and microbial diversity (Amann et al., 1995; Hugenholtz et al., 1998; Giraffa and Neviani, 2001; Jany and Barbier, 2008).
Culture-independent microbial techniques, such as PCR-denaturing gradient gel electrophoresis (DGGE) has proven much more reliable, fast, and economical in profiling complex bacterial community structure, dynamics and evolution of microbial populations of fermented foods (Giraffa and Neviani, 2001; Randazzo et al., 2002; Cocolin et al., 2007; Jany and Barbier, 2008). PCR-DGGE has been successfully employed to studying the microbial community structures and dynamics of fermented vegetal protein seeds such as doenjang and meju (Cho and Seo, 2007; Kim et al., 2009; Lee et al., 2010) as found in Asia. More recently, barcoded pyrosequencing of V1/V2 region of 16S rRNA gene was used in assessing the microbial community of doenjang (Nam et al., 2012). Meanwhile, there is dearth of information on the application of these techniques to profile microbial community of fermented vegetable foods in Africa.
Studies involving the use of both culture-dependent and culture-independent molecular methods for the identification of dominant bacterial species; accurate determination and detailed investigation of microbial composition of fermented food products have been reported (Pulido et al., 2005; Vilela et al., 2010; Alegría et al., 2012; Kesmen et al., 2012). In this study, we combined both culture-dependent and -independent molecular techniques as a polyphasic strategy to first understand and characterize the microbial community profile of iru produced by natural, uncontrolled fermentation process; secondly, to have a deeper insight into the bacterial diversity, which can be used to locate and identify the dominant bacteria with technological and functional potentials that can be further developed as autochthonous starter cultures to improve industrial production of condiments in W. Africa and guarantee product safety.
Materials and methods
Sample collection
Fermenting P. biglobosa during iru production were obtained from local producers as well as retail markets samples in southwest (Lagos, Ibadan, Abeokuta, Oyo, and Ado-Ekiti) and north central Nigeria (Kaduna and Ilorin). They were transported immediately into the laboratory with the aid of ice pack, stored in the refrigerator at 4°C and analyzed within few days of collection.
Isolation of microorganisms
Samples of fermented iru were homogenized in 90 ml Ringer's solution. Serial dilutions were made and 0.1 ml of appropriate dilutions was spread plated on nutrient agar (Oxoid, Hampshire, UK). Plates were subsequently incubated at 37°C for 18–24 h. Bacterial colonies obtained were purified by repeated streaking to obtain pure cultures.
Culture-dependent analysis
A total of 280 bacterial isolates were isolated from 20 iru samples and Reference bacterial strains obtained from Microbial Type Culture Collection (MTCC), India were subjected to genotypic characterization. Amplified ribosomal DNA restriction analysis (ARDRA) combined with PCR amplification of the 16S–23S rRNA gene internal transcribed spacer (ITS), ITS-PCR-restriction fragment length polymorphism (ITS-PCR-RFLP), and randomly amplified polymorphic DNA (RAPD-PCR) as previously described (Oguntoyinbo et al., 2010) were used for genotypic grouping, clonal relationship, and isolates diversity. The almost complete 16S rRNA genes of representative isolates from the clusters generated from dendrogram of gel fingerprints were amplified and sequenced as described previously (Jeyaram et al., 2010). Closest known identities of the 16S rRNA gene sequences were obtained from NCBI GenBank database.
Culture-independent analysis
DNA extraction and PCR amplification
Two methods of bacterial genomic DNA described previously by Ercolini et al. (2003) and Rantsiou et al. (2005) were used to extract genomic DNA from 16 iru samples (no appreciable DNA was obtained from remaining four samples). The V3 region of the 16S rRNA gene was amplified using the set of primers 338f (5′-ACTCCTACGGGAGGCAGCAG-3′) and 518r (5′-ATTACCGCGGCTGCTGG-3′) (Sigma-Aldrich). A GC-rich clamp was attached to the forward primer as described by Muyzer et al. (1993). Five-microliters PCR amplified product was analysed on 2% agarose (Promega, USA) containing ethidium bromide (0.5 μg/ml) (E1510, Sigma-Aldrich), using 0.5× TBE buffer (45 mM Tris-borate, 1 mM EDTA, pH 8.0) with running conditions of 80 V for about 1 h. DNA fragments of about 240 bp obtained by comparing with PCR 100 bp low ladder (P1473, Sigma-Aldrich) was used for DGGE analysis.
DGGE analysis of fermented iru samples
DGGE was performed using a DCODE™ Universal Mutation Detection System (BIO-RAD, USA) following manufacturer's instructions. PCR amplicons including a DGGE marker designed and used as a reference DNA ladder, were separated in 8% polyacrylamide gels containing 1× TAE with denaturing gradients of 25–55% [100% denaturant corresponds to 7 M urea and 40% (v/v) formamide]. Electrophoresis was run initially at 20 V for 10 min and at a constant temperature of 60°C, and finally at 140 V for 5 h at 60°C. The gels were stained with ethidium bromide, visualized under UV light and documented using Gene Snap (PerkinElmer GELIANCE 200, USA).
PCR-DGGE gel images analysis
The DGGE gel images obtained were converted to densitometric profiles using the software TL120 v2006 (Phoretix 1D Advanced Software, NonLinear Dynamics, Newcastle, UK). The similarity between the DGGE lanes was investigated by constructing non-metric multidimensional scaling (non-metric MDS) scatter plots as described elsewhere (Boon et al., 2002; McOrist et al., 2008) using the software package P A S T (PAleontological STatistics, http://folk.uio.no/ohammer/past/). The Shannon–Weaver index of H diversity and R richness index were determined based on the number presence/absence in the fragments. The importance probability Pi and H were calculated using PAST (Shannon and Weaver, 1963).
Excision of DGGE bands and sequencing of amplicons
DGGE major bands of interest were excised from the polyacrylamide gels, eluted in 50 μl sterile deionized water (Milli Q, Millipore, Bangalore) and incubated overnight at 4°C as described by Cocolin et al. (2001). One microliter of each eluate was used as DNA template for PCR reamplification. PCR products originating from the excised bands were profiled and checked for quality in agarose; presence and position of the bands of interest in DGGE and comparison with parent DGGE profiles. Elution and reamplification were carried out at least twice or until a single band that co-migrate at the same position with the parent DGGE band is obtained. Sequences of major bands obtained from the DGGE gel fragments were compared with the NCBI GenBank database to determine closest relatives.
Results
Culture-based molecular approach
In the cultured analysis, similar colonial morphology and population profile ranging from 106–107 CFU/ml were obtained from fermented samples as well as final product. A total of 280 isolates selected for analysis by ARDRA restriction endonucleases especially CfoI showed differentiation of the isolates into varying species, in particular, B. subtilis and B. cereus phylotypes. Further characterization with ITS-PCR-RFLP revealed intraspecies divergence among the Bacillus isolates. The gel data of ARDRA, ITS-PCR, ITS-PCR-RFLP, and RAPD-PCR were combined to generate polyphasic gel fingerprint. Representative of the major clusters obtained were identified by sequence data of 16S rRNA gene that identified two major bacilli groups; B. subtilis and B. amyloliquefaciens as the dominant, as well as B. cereus group as member of the cultivatable bacteria associated with fermented P. biglobosa cotyledon (Table 1).
Table 1.
Genomic characterization of Bacillus species isolated from iru, traditional fermented Parkia biglobosa.
| ARDRA groups | 16S rRNA-RFLP profile (sizes in bp) | ITS-PCR sub- groups (sizes in bp) | ITS-PCR-RFLP sub-groups (sizes in bp) | No. of isolates | Representative strains | 16S rRNA gene sequencing | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HaeIII | CfoI | DdeI | HinfI | TaqI | RsaI | Closest known relative | Similarity | NCBI acc. no. | |||||
| I | a627, 473 | 908 | 508 | 581 | 573 | 525 | IA. | IA1. | 93 | U4E BA | B. subtilis | 99% | JN255703 |
| 318 | 451 | 465 | 355 | 458 | 477 | 1065, 945, | 502, 417, 339, 278, 260, 214, | ||||||
| 156 | 248 | 237 | 317 | 349 | 429 | 869, | 143, 136 | ||||||
| 150 | 177 | 214 | 149 | IA2. | 26 | U104 | B. subtilis | 98% | JN165753.1 | ||||
| 122 | 506, 339, 280, 141 | ||||||||||||
| II | 622, 467 | 904 | 504 | 592 | 590 | 515 | IIA. | IIA1. | 1 | U154B | B. subtilis | 100% | JN255715 |
| 314, 151 | 445, | 381 | 362 | 412 | 470 | 1036, 845 | 485, 401, 322, 263, 247, 218, | ||||||
| 243 | 280 | 324 | 361 | 417 | 138, 129 | ||||||||
| 147 | 183 | 221 | 146 | IIA2. | 3 | U191A | B. subtilis | 97% | JN255725 | ||||
| 117 | 489, 329, 271, 231, 137 | ||||||||||||
| IIA3. | 1 | U233 | B. subtilis | 100% | JN255727 | ||||||||
| 494, 406, 330, 269, 255, | |||||||||||||
| 225, 127, 120 | |||||||||||||
| IIA4. | 1 | U122A | B. subtilis | 99% | JN255710 | ||||||||
| 494, 329, 268, 223, 134 | |||||||||||||
| IIA5. | 2 | U213A | B. pumilus | 99% | JN255726 | ||||||||
| 490, 339 259, 149 | U158 | B. pumilus | 100% | JN255717 | |||||||||
| III | 627, 472, | 912 | 510 | 609 | 722 | 515 | IIIA. | IIIAl. | 62 | U184B | B.amylolique-faciens | 99% | JN255722 |
| 318, 156 | 451 | 392 | 374 | 375 | 424 | 1057, 939, 863 | 504, 420, 343, 274, | ||||||
| 247 | 286 | 335 | 232 | 147 | 259, 145, 137 | ||||||||
| 236 | 190 | IIIA2. | 37 | U79BA | B.amyloliquefaciens | 98% | JN255707 | ||||||
| 122 | 506, 424, 384, 369, 343, | ||||||||||||
| 274, 260, 147, 138 | |||||||||||||
| IV | 622, 475 | 414 | 518 | 581 | 570 | 498 | IVA. | IVAl. | 3 | U186 | B. licheniformis | 98% | JN255724 |
| 324, 156 | 326 | 288 | 344 | 395 | 459 | 1024, 845 | 496, 336, 272, 229, 142 | ||||||
| 228 | 277 | 307 | 345 | 409 | IVB. | IVB1. | 1 | U126 | B. licheniformis | 100% | JN255711 | ||
| 172 | 237 | 166 | 211 | 142 | 1044, 926, | 489, 435, 414, 335, 271, 129 | |||||||
| 132 | 842, 339 | ||||||||||||
| 115 | |||||||||||||
| V | 595, 477 | 567 | 516 | 943 | 600 | 494 | VA. | VA1. | 49 | U175 | B. cereus | 96% | JN255721 |
| 322, 161 | 417 | 287 | 312 | 481 | 411 | 1078, 827 | 506, 311, 168, 136 | U243 | B. cereus | 92% | JN255728 | ||
| 332 | 275 | 170 | 134 | 355 | |||||||||
| 175 | 172 | 142 | |||||||||||
| 150 | |||||||||||||
| 115 | |||||||||||||
| VI | 627, 477, | 412 | 546 | 973 | 575 | 452 | VIA. | VIA1. | 1 | U185B | Brevibacillus formosus | 99% | JN255723 |
| 228, 157 | 324 | 441 | 304 | 389 | 411 | 1359, 1161, | 461, 355, 294, 262, 225, 205, | ||||||
| 218 | 238 | 234 | 365 | 353 | 1103, 1020, | 198, 193, 167, 157, 142, 133 | |||||||
| 172 | 167 | 167 | 196 | 131 | 910 | ||||||||
| 120 | 107 | ||||||||||||
DNA fragment sizes.
Analysis of bacterial communities and diversity of iru using PCR-DGGE
The two genomic DNA extraction protocols- enzymatic and chemical proved to be efficient in terms of quantity, purity, and amplifiabiltiy, and thus produced DNA suitable for PCR amplification. The results of the PCR-DGGE gel fingerprints of the V3 region of amplified 16S rRNA gene showed identical DGGE patterns and high degree of similarity in DNA fragments with the two DNA extraction methods. The PCR-DGGE gel profiles were also used to assess the bacterial community profile; richness, biodiversity, and dominance indexes of naturally fermented iru samples obtained from different geographical locations in Nigeria. The species richness index (R) determined was highest for DGGE profiles of iru samples from Ilorin (R
![]() 19), thus exhibiting the highest number of bands/bacterial species than those of Lagos (16 < R < 18); Ado-Ekiti (R
19), thus exhibiting the highest number of bands/bacterial species than those of Lagos (16 < R < 18); Ado-Ekiti (R
![]() 16); Ibadan (11 < R < 14); Oyo (R
16); Ibadan (11 < R < 14); Oyo (R
![]() 10) and Abeokuta (9 < R < 17) (Table 2). Bacterial diversity index (H) calculated on the basis of number of bands on a gel track was also highest for iru samples from Ilorin (H
10) and Abeokuta (9 < R < 17) (Table 2). Bacterial diversity index (H) calculated on the basis of number of bands on a gel track was also highest for iru samples from Ilorin (H
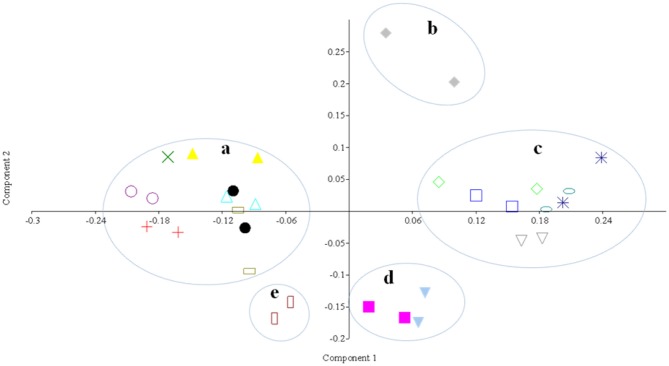
![]() 2.92) with the lowest being Abeokuta (2.19 < H < 2.80) (Table 2). Similarity and variation among DGGE gel patterns of the various fermented condiments were established based on combined analysis of Dice similarity coefficient and nMDS. The results obtained showed that the 16 iru samples analysed clustered into five groups—a, b, c, d, and e (Figure 1). Noticeable similarity can be said to exist in the bacterial community structure of the different samples of iru obtained in the two geographical zones under study (southwest and north central Nigeria). Apart from two iru samples from Lagos south western Nigeria, that clustered into the same group “a” variation was observed among samples within the same geographical location especially southwest Nigerian samples.
2.92) with the lowest being Abeokuta (2.19 < H < 2.80) (Table 2). Similarity and variation among DGGE gel patterns of the various fermented condiments were established based on combined analysis of Dice similarity coefficient and nMDS. The results obtained showed that the 16 iru samples analysed clustered into five groups—a, b, c, d, and e (Figure 1). Noticeable similarity can be said to exist in the bacterial community structure of the different samples of iru obtained in the two geographical zones under study (southwest and north central Nigeria). Apart from two iru samples from Lagos south western Nigeria, that clustered into the same group “a” variation was observed among samples within the same geographical location especially southwest Nigerian samples.
Table 2.
Species richness estimates (R) and Shannon's index of diversity (H) of DGGE profiles of fermented iru samples obtained from various geographical locations in Nigeria.
| Samples | R | H |
|---|---|---|
| iru Abeokuta 1 | 12 | 2.44 |
| iru Abeokuta 2 | 13 | 2.57 |
| iru Abeokuta 3 | 17 | 2.80 |
| iru Abeokuta 4 | 12 | 2.49 |
| iru Abeokuta 5 | 11 | 2.34 |
| iru Abeokuta 6 | 11 | 2.35 |
| iru Abeokuta 7 | 15 | 2.70 |
| iru Abeokuta 8 | 9 | 2.19 |
| iru Lagos 1 | 18 | 2.88 |
| iru Lagos 2 | 16 | 2.74 |
| iru Ilorin 1 | 19 | 2.92 |
| iru Ilorin 2 | 19 | 2.92 |
| iru Ibadan 1 | 11 | 2.40 |
| iru Ibadan 2 | 14 | 2.60 |
| iru Oyo | 10 | 2.25 |
| iru Ado-Ekiti | 16 | 2.77 |
| Mean iru Abeokuta (±STDEV) | 12.50 ± 2.35 | 2.49 ± 0.19 |
| Mean iru Lagos (±STDEV) | 17.00 ± 1.00 | 2.81 ± 0.05 |
| Mean iru Ilorin (±STDEV) | 19.00 ± 0.00 | 2.92 ± 0.00 |
| Mean iru Ibadan (±STDEV) | 12.50 ± 1.50 | 2.50 ± 0.10 |
Figure 1.
Non-metric Multidimensional Scaling analysis of DGGE data Group (a): iru Abeokuta 2 southwest Nigeria ( ); iru Abeokuta 3 southwest Nigeria (
); iru Abeokuta 3 southwest Nigeria ( ); iru Lagos 1 southwest Nigeria (
); iru Lagos 1 southwest Nigeria ( ); iru Lagos 2 southwest Nigeria (•); iru Ilorin 1 northcentral Nigeria (
); iru Lagos 2 southwest Nigeria (•); iru Ilorin 1 northcentral Nigeria ( ); iru Ilori n 2 northcentral Nigeria (
); iru Ilori n 2 northcentral Nigeria ( ); iru Ibadan 1 southwest Nigeria (
); iru Ibadan 1 southwest Nigeria ( ). Group (b): iru Abeokuta 4 southwest Nigeria (
). Group (b): iru Abeokuta 4 southwest Nigeria ( ). Group (c): iru Abeokuta 1 southwest Nigeria (
). Group (c): iru Abeokuta 1 southwest Nigeria ( ); iru Abeokuta 5 southwest Nigeria (
); iru Abeokuta 5 southwest Nigeria ( ); iru Abeokuta 8 southwest Nigeria (
); iru Abeokuta 8 southwest Nigeria ( ); iru Oyo southwest Nigeria (
); iru Oyo southwest Nigeria ( ); iru Ado-Ekiti southwest Nigeria (
); iru Ado-Ekiti southwest Nigeria ( ). Group (d): iru Abeokuta 6 southwest Nigeria (
). Group (d): iru Abeokuta 6 southwest Nigeria ( ); iru lbadan 2 southwest Nigeria (
); iru lbadan 2 southwest Nigeria ( ). Group (e): iru Abeokuta 7 southwest Nigeria (
). Group (e): iru Abeokuta 7 southwest Nigeria ( ).
).
Identification of major bacterial PCR-DGGE bands
DNA sequencing was carried out on the major bacterial bands. The closest known identities, percentage similarity and frequency of occurrence of each band in the fermented condiments are as shown in Table 3. Among them all, B. subtilis occurred most frequently (band 10), Figure 2). It was found to be present in 94% of the iru samples, which confirms its consistency, dominance, and viability in the fermented condiments. B. licheniformis and Brevibacillus species previously isolated in the culture-dependent study were also found present in at least a sample of the fermented condiment (Table 3). Other bands identified include close relatives of S. vitulinus, Morganella morganii, B. thuringiensis, S. saprophyticus, T. halophilus, Ureibacillus thermosphaericus, Salinicoccus jeotgali, Brevibacterium sp., and uncultured bacteria clones.
Table 3.
Identities of major bacterial bands excised from PCR-DGGE gels of fermented Parkia biglobosa seeds.
| Bands | Source/locationsa | 16S rRNA gene closest known relative | % Similarity |
|---|---|---|---|
| 1 | Oyo (1), Abeokuta (4), Ado-Ekiti (1), Lagos (1), Ibadan (1), Ilorin (2) | Staphylococcus vitulinus | 98 |
| 2 | Ado-Ekiti (1), Ibadan (2), Abeokuta (4), Lagos (2), Ilorin (2) | Morganella morganii | 98 |
| 3 | Abeokuta (2), Ibadan (1), Lagos (1) | Bacillus thuringiensis | 99 |
| 4 | Abeokuta (2), Ibadan (1), Ilorin (1) | B. licheniformis | 98 |
| 5 | Oyo (1), Abeokuta (1), Lagos (1), Ilorin (1) | S. saprophyticus | 99 |
| 6 | Oyo (1), Abeokuta (7), Ado-Ekiti (1), Lagos (2), Ilorin (2) | Uncultured bacterium clone | 97 |
| 7 | Abeokuta (3), Ibadan (1), Lagos (1) | Tetragenococcus halophilus | 95 |
| 8 | Oyo (2), Ibadan (2), Abeokuta (1), Lagos (1) | Ureibacillus thermosphaericus | 100 |
| 9 | Ado-Ekiti (1), Ibadan (1), Abeokuta (1), Lagos (1) | Bacillus sp. | 98 |
| 10 | Oyo (1), Abeokuta (7), Ado-Ekiti (1), Ibadan (2), Lagos (2), Ilorin (2) | B. subtilis | 99 |
| 11 | Abeokuta (1) | Brevibacillus parabrevis | 97 |
| 12 | Ibadan (1), Abeokuta (1) | Salinicoccus jeotgali | 99 |
| 13 | Ado-Ekiti (1), Lagos (1) | Brevibacterium sp. | 97 |
Bands are as shown in Figure 2.
Values in parenthesis represent number of fermented iru samples, with total number of iru sampled being 16.
Figure 2.
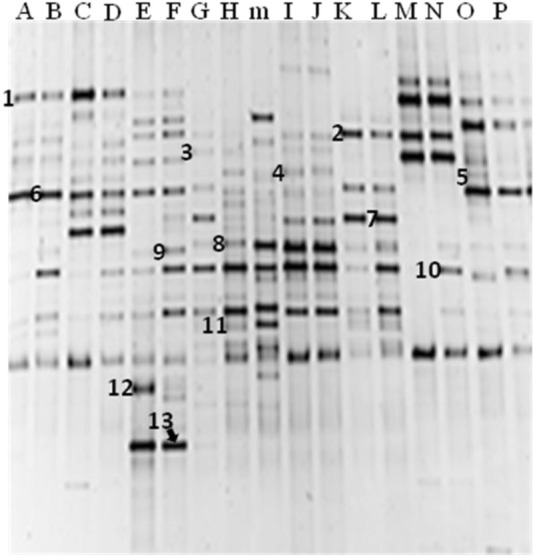
DGGE profiles of PCR-amplified 16S rRNA gene fragments of sixteen (16) fermented iru samples showing major bacterial amplicons. Samples A: iru Oyo; B: iru Lagos 1; C: iru Ilorin 2; D: iru Ilorin 1; E: iru Ibadan 1; F: iru Ado-Ekiti; G: iru Abeokuta 2; H: iru Abeokuta 1; I: iru Ibadan 2; J: iru Abeokuta 3; K: iru Abeokuta 4; L: iru Abeokuta 5; M: iru Abeokuta 6; N: iru Lagos 2; Î: iru Abeokuta 7; P: iru Abeokuta 8; “m” is DGGE reference DNA ladder. Bands 1: Staphylococcus vitulinus; 2: Morganella morganii; 3: Bacillus thuringiensis; 4: B. licheniformis; 5: S. saprophyticus; 6: Uncultured bacterium clone; 7: Tetragenococcus halophilus; 8: Ureibacillus thermosphaericus; 9: Bacillus sp.; 10: B. subtilis; 11: Brevibacillus parabrevis; 12: Salinicoccus jeotgali; 13: Brevibacterium sp.
Discussion
In W. Africa traditional fermented condiments form an important component of the dietary protein requirement of the people, where they are added generously to various dishes to supplement protein intake, improve nutritive value, and impart sensory attributes. This study combined molecular techniques to reveal microbiota of traditional fermented condiments sample tested at different fermentation time regimes with consideration for geographical spread in Nigeria. This is aimed at future objectives of developing predictable process through the use of identified bacteria that can be applied as starter cultures to dominate the fermentation, impact desirable biochemical changes and improve safety quality.
The results of culture-dependent analysis and 16S rRNA gene sequencing showed that iru is composed of clonally related Bacillus species identified as B. subtilis, B. amyloliquefaciens, B. cereus, B. licheniformis, and Brevibacillus formosus in order of frequency of incidence. Also, the combination of ARDRA, ITS-PCR, ITS-PCR-RFLP, and RAPD-PCR revealed high degree of diversity among the Bacillus isolates and identified the dominant species to be B. subtilis and B. amyloliquefaciens. The results clearly showed sub-typing of autochthonous clonally related species of bacilli and genetically identified dominant groups, this is in agreement with previous reports in W. Africa and India during fermentation of similar vegetable proteins studies (Sarkar et al., 2002; Ouoba et al., 2004; Jeyaram et al., 2008; Kim et al., 2010; Oguntoyinbo et al., 2010; Parkouda et al., 2010).
Culture-independent techniques have been previously used to describe microbiota of fermented foods in Africa (Humblot and Guyot, 2009; Oguntoyinbo et al., 2011). Although none of this studies reported microbiota of fermented vegetable protein used as condiments. In this study, during profiling of bacterial community of fermented iru, two methods of genomic DNA extraction were employed. No significant differences in the DNA yields, fragments and DGGE band patterns were observed. Mafra et al. (2008) reported similar findings in a study involving comparison of four DNA extraction methods for downstream processing, comparison such as this guarantees better microbiome profiling. Results of PCR-DGGE gel analysis of iru sampled from Ilorin, north central Nigeria had the highest number of bacterial species than others, including the highest bacterial diversity index; the variable production process could possibly be responsible for this observation. The intraspecific variation observed in the fermented iru samples as predicated by Dice similarity coefficient and nMDS is an indication that the bacterial community profile of this condiment is diverse; having grouped into distinct clusters with varying diversity indexes. Iru from different geographical zones were produced from the same substrate P. biglobosa seeds but variable fermentation periods. Therefore, diversity observed in the microbial communities could be due to difference in fermentation conditions, which is also spontaneous and uncontrolled, without the use of any known specific isolate as starter culture. Other workers have also reported similar variation in the microbial composition of naturally fermented food materials produced using same substrate (Ampe and Miambi, 2000; Oguntoyinbo et al., 2011).
PCR-DGGE bands sequencing identified B. subtilis as the major bacterial species associated with iru, which is in agreement with the result of culture-based PCR analysis. Earlier studies using traditional culture-dependent methods and biochemical characterization have also reported the dominance of B. subtilis during iru fermentation (Antai and Ibrahim, 1986; Odunfa and Oyewole, 1986). Other DGGE bands corresponded to potential food-borne pathogens and contaminants such as S. vitulinus, S. saprophyticus, B. thuringiensis, Tetragenococcus halophilus, etc. Of interest is T. halophilus, a halophilic lactic acid bacterium, predominantly found in fermented foods with high salt concentration such as soysauce, miso and doenjang (Tanasupawat et al., 2002; Onda et al., 2003; Kim et al., 2009). The source of this bacterium in iru can somewhat be attributed to the salt added as a preservative at the end of the fermentation process. The detection of B.cereus, B. thuringiensis, and Staphylococcus species clearly raises doubt regarding the microbiological safety of this fermented condiment. Information on the toxigenic potential of food-borne pathogen in traditional African fermented foods exist (Oguntoyinbo and Oni, 2004; Abriouel et al., 2007; Oguntoyinbo et al., 2010).
In conclusion, the results indicated that combination of culture-dependent and culture-independent techniques can be used to profile bacterial microbiota, examine dynamics and diversity of naturally fermented food. On the overall, this study showed that bacterial composition of iru is significantly more diverse than earlier reported and justified the need to develop process across production sites to facilitate safety quality and consistency. This study confirmed dominance of Bacillus species and occurrence of other bacteria with pathogenic trait and unknown functions. Condiments can support dietary need of the people if better understanding of the functional roles of all members of the community during fermentation can be determined, this will support development of predictable process, eliminate variation and inconsistency.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This project was funded by the Department of Biotechnology (DBT), Government of India and the academy of Sciences for the Developing World (TWAS), Italy through the award of a DBT-TWAS Postgraduate Sandwich fellowship to Gbenga A. Adewumi and SfAM New Lecturer Grant, UK to Folarin A. Oguntoyinbo. The leave of absence also granted to Gbenga A. Adewumi by Bells University of Technology, Ota, Nigeria that facilitated his collaboration with IBSD, Imphal, India is gratefully acknowledged.
References
- Abriouel H., Ben Omar N., López R. L., Cañamero M. M., Ortega E., Gálvez A. (2007). Differentiation and characterization by molecular techniques of Bacillus cereus group isolates from poto poto and degue, two traditional cereal-based fermented foods of Burkina Faso and Republic of Congo. J. Food Prot. 70, 1165–1173 [DOI] [PubMed] [Google Scholar]
- Achi O. K. (2005). Traditional fermented protein condiments in Nigeria. Afr. J. Biotechnol. 4, 1612–1621 [Google Scholar]
- Alegría A., Szczesny P., Mayo B., Bardowski J., Kowalczyk M. (2012). Biodiversity in oscypek, a traditional Polish cheese determined by culture-dependent and in-dependent approaches. Appl. Environ. Microbiol. 78, 1890–1898 10.1128/AEM.06081-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann R. I., Ludwig W., Schleifer K.-H. (1995). Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59, 143–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampe F., Miambi E. (2000). Cluster analysis, richness and biodiversity indexes derived from denaturing gradient gel electrophoresis fingerprints of bacterial communities demonstrate that traditional maize fermentations are driven by transformation process. Int. J. Food Microbiol. 60, 91–97 10.1016/S0168-1605(00)00358-5 [DOI] [PubMed] [Google Scholar]
- Antai S. P., Ibrahim M. H. (1986). Microorganisms associated with African locust bean (Parkia biglobosa Welw) fermentation for dawadawa production. J. Appl. Bacteriol. 61, 145–148 [Google Scholar]
- Boon N., De Windt W., Verstraete W., Top E. M. (2002). Evaluation of nested PCR-DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol. Ecol. 39, 101–112 10.1111/j.1574-6941.2002.tb00911.x [DOI] [PubMed] [Google Scholar]
- Cho K. M., Seo W. T. (2007). Bacterial diversity in a Korean traditional soybean fermented foods (doenjang and ganjang) by 16S rRNA gene sequence analysis. Food Sci. Biotechnol. 16, 320–324 [Google Scholar]
- Cocolin L., Diez A., Urso R., Rantsiou K., Comi G., Bergmaier I., et al. (2007). Optimization of conditions for profiling bacterial populations of food by culture-independent methods. Int. J. Food Microbiol. 120, 100–109 10.1016/j.ijfoodmicro.2007.06.015 [DOI] [PubMed] [Google Scholar]
- Cocolin L., Manzano M., Cantoni C., Comi G. (2001). Denaturing gradient gel electrophoresis analysis of the 16S rRNA gene V1 region to monitor dynamic changes in the bacterial population during fermentation of Italian sausages. Appl. Environ. Microbiol. 67, 5113–5121 10.1128/AEM.67.11.5113-5121.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercolini D., Hill P. J., Dodd C. E. R. (2003). Bacterial community structure and location in stilton cheese. Appl. Environ. Microbiol. 69, 3540–3548 10.1128/AEM.69.6.3540-3548.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet G. H. (1999). Microorganisms in food ecosystems. Int. J. Food Microbiol. 50, 101–117 10.1016/S0168-1605(99)00080-X [DOI] [PubMed] [Google Scholar]
- Giraffa G., Neviani E. (2001). DNA-based, culture-independent strategies for evaluating microbial communities in food-associated ecosystems. Int. J. Food Microbiol. 67, 19–34 [DOI] [PubMed] [Google Scholar]
- Hugenholtz P., Goebbel B. M., Pace N. R. (1998). Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180, 4765–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humblot C., Guyot J. P. (2009). Pyrosequencing of tagged 16S rRNA gene amplicons for rapid deciphering of the microbiomes of fermented foods such as pearly millet slurries. Appl. Environ. Microbiol. 75, 4354–4361 10.1128/AEM.00451-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jany J. L., Barbier G. (2008). Culture-independent methods for identifying microbial communities in cheese. Food Microbiol. 25, 839–848 10.1016/j.fm.2008.06.003 [DOI] [PubMed] [Google Scholar]
- Jeyaram K., Romi W., Singh T. A., Devi A. R., Devi S. (2010). Bacterial species associated with traditional starter cultures used for fermented bamboo shoot production in Manipur state of India. Int. J. Food Microbiol. 143, 1–8 10.1016/j.ijfoodmicro.2010.07.008 [DOI] [PubMed] [Google Scholar]
- Jeyaram K., Singh W. M., Premarani T., Devi A. R., Chanu K. S., Talukdar N. C., et al. (2008). Molecular identification of dominant microflora associated with hawaijar – a traditional fermented soybean [Glycine max (L.)] food of Manipur of India. Int. J. Food Microbiol. 122, 259–268 [DOI] [PubMed] [Google Scholar]
- Kesmen Z., Yetiman A. E., Gulluce A., Kacmaz N., Sagdic O., Cetin B., et al. (2012). Combination of culture-dependent and culture-independent molecular methods for the determination of lactic microbiota in sucuk. Int. J. Food Microbiol. 153, 428–435 10.1016/j.ijfoodmicro.2011.12.008 [DOI] [PubMed] [Google Scholar]
- Kiers J. L., Van Laeken A. E. A., Rombouts F. M., Nout M. J. R. (2000). In vitro digestibility of Bacillus fermented soya bean. Int. J. Food Microbiol. 60, 163–169 10.1016/S0168-1605(00)00308-1 [DOI] [PubMed] [Google Scholar]
- Kim T.-W., Kim Y.-H., Kim S.-E., Lee J.-H., Park C.-S., Kim H.-Y. (2010). Identification and distribution of Bacillus species in doenjang by whole-cell protein patterns and 16S rRNA gene sequence analysis. J. Microbiol. Biotechnol. 20, 1210–1214 [DOI] [PubMed] [Google Scholar]
- Kim T.-W., Lee J.-H., Kim S.-E., Park M.-H., Chang H. C., Kim H.-Y. (2009). Analysis of microbial communities in doenjang, a Korean fermented soybean paste, using nested PCR-denaturing gradient gel electrophoresis. Int. J. Food Microbiol. 131, 265–271 10.1016/j.ijfoodmicro.2009.03.001 [DOI] [PubMed] [Google Scholar]
- Lee J.-H., Kim T.-W., Lee H., Chang H. C., Kim H.-Y. (2010). Determination of microbial diversity in meju, a fermented cooked soya beans, using nested PCR-denaturing gradient gel electrophoresis. Lett. Appl. Microbiol. 51, 388–394 10.1111/j.1472-765X.2010.02906.x [DOI] [PubMed] [Google Scholar]
- Mafra I., Silva S. A., Moreira E. J. M. O., da Silva C. S., Beatriz M., Oliveira P. P. (2008). Comparative study of DNA extraction methods for soybean derived food products. Food Control 19, 1183–1190 10.1002/rcm.1899 [DOI] [PubMed] [Google Scholar]
- McOrist A. L., Abell G. C. J., Cooke C., Nyland K. (2008). Bacterial population dynamics and faecal short-chain fatty acid (SCFA) concentrations in healthy humans. Br. J. Nutr. 100, 138–146 10.1017/S0007114507886351 [DOI] [PubMed] [Google Scholar]
- Muyzer G., De Waal E. D., Uitterlinden A. G. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam Y.-D., Lee S.-Y., Lim S.-I. (2012). Microbial community analysis of Korean soybean pastes by next-generation sequencing. Int. J. Food Microbiol. 155, 36–42 10.1016/j.ijfoodmicro.2012.01.013 [DOI] [PubMed] [Google Scholar]
- N'Dir B., Hbid C. L., Cornelius C., Roblain D., Jacques P., Vanhentenryck F., et al. (1994). Proptiétés antifongiques de la microflora sporulée du nététu. Cahiers Agric. 3, 23–30 19042653 [Google Scholar]
- Odunfa S. A. (1981). Microorganisms associated with fermentation of African locust bean (Parkia filicoidea) during iru preparation. J. Plant Foods 3, 245–250 [Google Scholar]
- Odunfa S. A. (1985). Biochemical changes in fermenting African locust bean (Parkia biglobosa) during iru fermentation. J. Food Technol. 20, 295–303 [Google Scholar]
- Odunfa S. A., Oyewole O. B. (1986). Identification of Bacillus species from iru, a fermented African locust bean product. J. Basic Microbiol. 2, 101–108 [Google Scholar]
- Odunfa S. A., Oyewole O. B. (1998). African fermented foods, in Microbiology of Fermented Foods, 2nd Edn, Vol. 2, ed Wood B. J. B. (London, UK: Blackie Academic and Professional; ), 712–752 [Google Scholar]
- Oguntoyinbo F. A., Huch M., Cho G.-S., Schillinger U., Holzapfel W., Sanni A. I., et al. (2010). Diversity of Bacillus species isolated from okpehe, a traditional fermented soup condiment of Nigeria. J. Food Prot. 73, 870–878 [DOI] [PubMed] [Google Scholar]
- Oguntoyinbo F. A., Oni O. M. (2004). Incidence and characterization of Bacillus cereus isolated from traditional fermented meals in Nigeria. J. Food Prot. 67, 2805–2808 [DOI] [PubMed] [Google Scholar]
- Oguntoyinbo F. A., Tourlomousis P., Gasson M., Narbad A. (2011). Analysis of bacterial communities of traditional fermented West African cereal foods using culture-independent methods. Int. J. Food Microbiol. 145, 205–210 10.1016/j.ijfoodmicro.2010.12.025 [DOI] [PubMed] [Google Scholar]
- Omafuvbe B. O., Falade O. S., Osuntogun B. A., Adewusi S. R. A. (2004). Chemical and biochemical changes in African locust bean (Parkia biglobosa) and melon (Citrullus vulgaris) seeds during fermentation to condiments. Pak. J. Nutr. 3, 140–145 [Google Scholar]
- Onda T., Yanagida F., Uchimura T., Tsuji M., Ogino S., Shinohara T., et al. (2003). Analysis of lactic acid bacterial flora during miso fermentation. Food Sci. Technol. Res. 9, 17–24 10.1146/annurev.food.102308.124134 [DOI] [PubMed] [Google Scholar]
- Ouoba L. I. I., Diawara B., Amoa-Awua W. K., Traoré A. S., Møller P. L. (2004). Genotyping of starter cultures of Bacillus subtilis and Bacillus pumilus for fermentation of African locust bean (Parkia biglobosa) to produce soumbala. Int. J. Food Microbiol. 90, 197–205 10.1016/S0168-1605(03)00302-7 [DOI] [PubMed] [Google Scholar]
- Ouoba L. I. I., Diawara B., Jespersen L., Jakobsen M. (2007). Antimicrobial activity of Bacillus subtilis and Bacillus pumilus during the fermentation of African locust bean (Parkia biglobosa) for soumbala production. J. Appl. Microbiol. 102, 963–970 10.1111/j.1365-2672.2006.03156.x [DOI] [PubMed] [Google Scholar]
- Ouoba L. I. I., Nyanga-Koumou C. A. G., Parkouda C., Sawadogo H., Kobawila S. C., Keleke S., et al. (2010). Genotypic diversity of lactic acid bacteria isolated from African traditional alkaline-fermented foods. J. Appl. Microbiol. 108, 2019–2029 10.1111/j.1365-2672.2009.04603.x [DOI] [PubMed] [Google Scholar]
- Ouoba L. I. I., Parkouda C., Diawara B., Scotti C., Varnam A. H. (2008). Identification of Bacillus spp. from bikalga, fermented seeds of Hibiscus sabdariffa: phenotypic and genotypic characterization. J. Appl. Microbiol. 104, 122–131 10.1111/j.1365-2672.2007.03550.x [DOI] [PubMed] [Google Scholar]
- Ouoba L. I. I., Rechinger K. B., Barkholt V., Diawara B., Traore A. S., Jakobsen M. (2003). Degradation of proteins during the fermentation of African locust bean (Parkia biglobosa) by strains of Bacillus subtilis and Bacillus pumilus for production of soumbala. J. Appl. Microbiol. 94, 396–402 10.1046/j.1365-2672.2003.01845.x [DOI] [PubMed] [Google Scholar]
- Oyeyiola G. P. (1988). Microbiology of iru pete and iru woro obtained from local producers. MIRCEN J. 4, 439–445 [Google Scholar]
- Parkouda C., Thorsen L., Compaore C. S., Nielsen D. S., Tano-Debrah K., Jensen J. S., et al. (2010). Microorganisms associated with maari, a baobab seed fermented product. Int. J. Food Microbiol. 142, 292–301 10.1016/j.ijfoodmicro.2010.07.004 [DOI] [PubMed] [Google Scholar]
- Pulido R. P., Ben Omar N., Abriouel H., López R. L., Cañamero M. M., Gálvez A. (2005). Microbiological study of lactic fermentation of caper berries by molecular and culture-dependent methods. Appl. Environ. Microbiol. 71, 7872–7879 10.1128/AEM.71.12.7872-7879.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo C. L., Torriani S., Akkermans A. L. D., de Vos W. M., Vaughan E. E. (2002). Diversity, dynamics and activity of bacterial communities during production of an artisanal Sicilian cheese as evaluated by 16S rRNA analysis. Appl. Environ. Microbiol. 68, 1882–1892 10.1128/AEM.68.4.1882-1892.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rantsiou K., Urso R., Iacumin L., Cantoni C., Cattaneo P., Comi G., et al. (2005). Culture-dependent and -independent methods to investigate the microbial ecology of Italian fermented sausages. Appl. Environ. Microbiol. 71, 1977–1986 10.1128/AEM.71.4.1977-1986.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanni A. I. (1993). The need for process optimization of African fermented foods and beverages. Int. J. Food Microbiol. 18, 85–95 [DOI] [PubMed] [Google Scholar]
- Sarkar P. K., Hasenack B., Nout M. J. R. (2002). Diversity and functionality of Bacillus and related genera isolated from spontaneously fermented soybeans (Indian kinema) and locust beans (soumbala). Int. J. Food Microbiol. 77, 175–186 [DOI] [PubMed] [Google Scholar]
- Shannon C. E., Weaver W. (1963). The Mathematical Theory of Communication. Urbana, IL: The University of Illinois Press [Google Scholar]
- Steinkraus K. H. (1997). Classification of fermented foods: worldwide review of household fermentation techniques. Food Control 8, 311–317 [Google Scholar]
- Tanasupawat S., Thongsanit J., Okada S., Komagata K. (2002). Lactic acid bacteria isolated from soy sauce mash in Thailand. J. Gen. Appl. Microbiol. 48, 201–209 [DOI] [PubMed] [Google Scholar]
- Vilela D. M., Pereira G. B. M., Silva C. F., Batista L. B., Schwan R. F. (2010). Molecular ecology and polyphasic characterization of the microbiota associated with semi-dry processed coffee (Coffee arabica L.). Food Microbiol. 27, 1128–1135 10.1016/j.fm.2010.07.024 [DOI] [PubMed] [Google Scholar]