Highlights
► MHV nsp15 is distributed throughout the cytoplasm of virus-infected cells. ► MHV but not SARS-CoV or TGEV nsp15 formed speckles when ectopically expressed in cell. ► Two domains (I and III) within nsp15 are responsible for the speckle formation. ► A novel protein-interaction domain is identified in the C-terminus of MHV nsp15. ► nsp15s from the 3 coronaviruses have differential effects on MHV replication.
Keywords: Coronavirus, Mouse hepatitis virus, SARS coronavirus, Transmissible gastroenteritis virus, Nonstructural protein 15
Abstract
The purpose of this study was to compare the biochemical and biological properties of nonstructural protein (nsp) 15 among mouse hepatitis virus (MHV), severe acute respiratory syndrome coronavirus (SARS-CoV) and transmissible gastroenteritis virus (TGEV) in virus-infected and ectopically expressed cells. In virus-infected cells, MHV nsp15 distributed unevenly throughout the cytoplasm but predominantly in the perinuclear region. When expressed as N-terminal enhanced green fluorescence protein (EGFP) fusion, it predominantly formed speckles in the cytoplasm. In contrast, SARS-CoV and TGEV EGFP-nsp15s distributed smoothly in the whole cell and did not form speckles. Deletion mapping experiments identified two domains responsible for the speckle formation in MHV EGFP-nsp15: Domain I (aa101–150) and Domain III (aa301–374). Interestingly, Domain II (aa151–250) had an inhibitory effect on Domain III- but not on Domain I-mediated speckle formation. Expression of a small (35aa) sequence in Domain III alone was sufficient to form speckles for all 3 viral nsp15s. However, addition of surrounding sequences in Domain III abolished the speckle formation for TGEV nsp15 but not for MHV and SARS-CoV nsp15s. Further domain swapping experiments uncovered additional speckle-inducing and -suppressive elements in nsp15s of SARS-CoV and TGEV. Homotypic interaction involving Domain III of MHV nsp15 was further demonstrated biochemically. Moreover, the biological functions of the expressed nsp15s were assessed in MHV-infected cells. It was found that the effects of EGFP-nsp15s on MHV replication were both virus species- and nsp15 domain-dependent. Collectively these results thus underscore the differential biochemical and biological functions among the nsp15s of MHV, TGEV and SARS-CoV in host cells.
1. Introduction
Coronavirus (CoV) is an enveloped RNA virus that belongs to the family Coronaviridae in the order of Nidovirales. It is highly prevalent among humans and diverse species of domestic and wild animals, causing diseases ranging from respiratory, digestive, neurological to immune-mediated diseases. Based on genetic and antigenic relatedness, members of the Coronaviridae can be classified into 3 groups, as exemplified by the transmissible gastroenteritis virus (TGEV) in pigs (group I), the mouse hepatitis virus (MHV) in rodents (group II), and the infectious bronchitis virus (IBV) in chicken (group III). While the recently emerged severe acute respiratory syndrome (SARS)-CoV in humans is quite divergent from all existing coronaviruses, it is relatively more closely related to group II than to other groups of coronaviruses (Snijder et al., 2003). Despite their huge variations in genome sequence, antigenicity, host species specificity and disease spectrum, all coronaviruses share a common “crown”-like virion morphology and a single-strand, positive-sense RNA genome of 26–32 kilobase (kb) in length, the largest among all RNA viruses.
Upon infection, the viral genomic RNA, which is capped at the 5′-end and polyadenylated at the 3′-end, serves as a mRNA for translation of the two overlapping open reading frames (ORF) (gene 1a/1b) at the 5′ two-third of the genome via the ribosomal frame-shifting translation mechanism (Bredenbeek et al., 1990, Brierley et al., 1987, Brierley et al., 1989, Lee et al., 1991). The resultant protein product, polyprotein 1a/b, is then proteolytically cleaved by virus-encoded proteases into 16 nonstructural proteins, termed nsp 1–16, many of which have enzymatic activities, such as papain-like proteases (nsp3), 3C-like protease (nsp5), RNA-dependent RNA polymerase (RdRp, nsp12), helicase (nsp13), exoribonuclease and methyltransferase (nsp14), endoribonuclease (nsp15), 2-O-ribosyl-transferase (nsp16) (Baker et al., 1989, Bost et al., 2001, Denison et al., 1992, Denison et al., 1998, Harcourt et al., 2004, Snijder et al., 2003, Thiel et al., 2001, Ziebuhr, 2005). These nsps are believed to form replication/transcription complexes along with putative cellular factors that catalyze the synthesis of genomic RNA (replication) and subgenomic RNAs (transcription). Coronaviral replication/transcription is thought to take place in double-membranous vesicles localized in the ERGIC (Endoplasmic Reticulum-to Golgi Intermediate Complex) compartment of the infected cells (Gosert et al., 2002, Knoops et al., 2008, Snijder et al., 2006). These vesicles are originated from rough endoplasmic reticulum (RER) that is modified by the viral nsps (Gosert et al., 2002, Knoops et al., 2008, Snijder et al., 2006).
nsp15 is conserved among coronaviruses (Bhardwaj et al., 2004). Although the overall deduced amino acid sequence identity among all coronaviral nsp15s sequenced so far is about 30%, the sequence identities within certain functional domains can reach to around 48% (Cao and Zhang, unpublished results). Importantly, the 3D structure of several coronaviral nsp15s is virtually identical as revealed by crystallography (Bhardwaj et al., 2008, Joseph et al., 2007, Ricagno et al., 2006, Xu et al., 2006), suggesting conservation of biological functions of nsp15s. Indeed, comparative sequence analysis has predicted that coronaviral nsp15 may have endoribonuclease activity similar to Xenopus endoU (XendoU) that cleaves U on single-stranded RNA molecule and requires Mn2+ as a cofactor (Bhardwaj et al., 2004, Gioia et al., 2005, Laneve et al., 2003). The nidoviral endoU (NendoU) activity was first demonstrated biochemically in vitro for SARS-CoV nsp15 expressed from bacteria (Bhardwaj et al., 2004), and subsequently confirmed in bacteria-expressed nsp15s from MHV (Kang et al., 2007, Xu et al., 2006), human CoV-229E (Ivanov et al., 2004), turkey CoV (Cao et al., 2008), and in nsp11 of equine arteritis virus, a cousin of coronavirus within the order of Nidovirales (Nedialkova et al., 2009). Unlike XendoU which acts as a monomer, NendoU appears to maintain its activity as a hexamer in vitro (Guarino et al., 2005, Joseph et al., 2007, Ricagno et al., 2006, Xu et al., 2006). CoV nsp15 has a sequence ranging from 327 to 375 amino acids with an estimated molecular weight of 38–42 kDa. Biochemical and crystal data have shown that the N-terminal domain of nsp15 is involved in protein–protein interaction while the C-terminal domain contains enzymatic activity (Guarino et al., 2005). The key amino acids at the enzymatically active site have been identified by mutagenic studies and have been shown highly conserved among all CoV nsp15s (Bhardwaj et al., 2004, Ivanov et al., 2004). Furthermore, reverse genetics experiments using infectious cDNA clones have demonstrated that amino acid mutations within the enzymatically active site of MHV nsp15 reduced viral genome replication and mRNA transcription by 22–56%, although trans-complementation with ectopically expressed-wild-type MHV nsp15 could rescue viral replication to some extent (Kang et al., 2007). Similarly, in human CoV-229E, a single amino acid mutation in nsp15 resulted in disruption of viral genome synthesis (Ivanov et al., 2004). These data support the notion that nsp15 is an important component of the replication/transcription complexes that are essential for CoV RNA synthesis. However, little is known about the biological properties of CoV nsp15s in vivo.
In the present study, we determined the intracellular biochemical and biological properties of CoV nsp15s in the context of virus infection and following ectopic expression as enhanced green fluorescence (EGFP) fusion. Our results showed that, in MHV-infected cells, nsp15 localized not only in the perinuclear region where the viral replication complex is usually located, but also in other areas throughout the cytoplasm. When ectopically expressed, the EGFP-nsp15 of MHV predominantly formed speckles while that of SARS-CoV and TGEV distributed smoothly throughout the cell without forming speckles. Systematic deletion mapping and domain swapping experiments uncovered specific sequence elements that are responsible either for inducing or suppressing speckle formation. Furthermore, a novel protein–protein interaction domain was identified at the C-terminus of MHV nsp15. The biological functions of the 3 coronaviral nsp15s were further assessed in MHV-infected cells. It was found that the augmentative or inhibitory effects of the expressed EGFP-nsp15s on MHV replication depended on virus species and the specific domains of the nsp15 being expressed. These findings thus underscore the differential biochemical and biological functions among the nsp15s of MHV, TGEV and SARS-CoV in host cells.
2. Materials and methods
2.1. Virus and cells
Mouse fibroblast 17Cl-1, mouse astrocytoma DBT cells, and 293T cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing 10% fetal bovine serum (FBS) (Invitrogen), 100 U/ml penicillin (Invitrogen), and 100 μg/ml streptomycin (Invitrogen) at 37 °C in a humidified incubator containing 5% CO2. Murine coronavirus mouse hepatitis virus (MHV) strain A59 (MHV-A59) and a recombinant MHV-A59 expressing the firefly luciferase (MHV-2aFLS) (kindly provided by Dr. Peter Rottier, Utrecht University, The Netherlands) were propagated in 17Cl-1 cells and the virus titer was determined in DBT cells using plaque assay.
2.2. Plasmid construction
Standard molecular methods were used to clone the coding sequence of nsp15 from MHV-A59 genomic RNA and all deletion mutants as N-terminal enhanced green fluorescence protein (EGFP) fusion proteins into expression vector pTriEx-4 (Novagen). The name, sequence and direction of all primers used in RT-PCR, PCR, and constructions are described in Table 1 . Briefly, EGFP was amplified from plasmid pEGFP-N1 with primer pairs EGFPF and EGFPR. The PCR product was purified, digested with Nco I and BamH I, and cloned into the Nco I–BamH I sites of pTriEx-4 to generate pTriEGFP. To clone MHV-nsp15, total RNA was isolated from MHV-A59-infected 17Cl-1 cells with Trizol reagent (Invitrogen). Approximately 10 μg of total RNA was used for cDNA synthesis with a random primer in 20 μl reaction using SuperScript II system (Invitrogen). Two microliters of cDNA was directly used in a 50 μl PCR containing dNTPs (0.5 mM final concentration), primer pairs mNsp15F and mNsp15R (0.2 μM), 1 unit of GoTaq and 1× GoTaq PCR buffer (Promega). The PCR was carried out on PTC-200 Peltier Thermal Cycler (MJ Research) with the following parameters: initial denaturing at 95 °C for 3 min followed by 30 cycles of denaturing at 93 °C for 10 s, annealing at 55 °C for 30 s, and extension at 72 °C for 90 s, and a final extension at 72 °C for 5 min. The PCR product was cloned into pCR2-TOPO vector directly and transformed into TOP10 chemical competent cells (Invitrogen) for colony formation. Positive colonies were identified by PCR screening and cultured for plasmid preparation. Plasmids were isolated with QIAprep Spin Columns (Qiagen) and were sequenced at the DNA sequencing core facility (UAMS). The plasmid with correct sequence of MHV nsp15 was designated as MHVnsp15. MHVnsp15 was then served as a template for amplification of nsp15 by PCR using the primer pair MHVnsp15F1bamHI and MHVnsp15R1. The PCR products were purified and digested with BamH I and Not I to release the full-length MHV nsp15, which was cloned into the BamH I and Not I sites of pTriEGFP, resulting in plasmid MHVEGFPnsp15. The full-length nsp15 fragments of SARS-CoV and TGEV were amplified from template plasmids (kindly provided by Dr. Ralph Baric, University of North Carolina at Chapel Hill) and were cloned into the same BamH I and Not I sites of the pTriEGFP, resulting in SARSEGFPnsp15 and TGEVEGFPnsp15, respectively.
Table 1.
Primer names and sequences used for PCR and plasmid construction.
| Plasmids | Forward primer (5′ → 3′) | Reverse primer (5′ → 3′) |
|---|---|---|
| pTriEGFP | EGFPF (actgccatggTGAGCAAGGGCGAGGA)a | EGFPR (gcatggatccTTGTACAGCTCGTCCAT) |
| MHVnsp15 | mNSP15F (CAACCACAGCGGGTTTTACT) | mNSP15R (GGCTTGCCATAATTCCAGAG) |
| MHVEGFPnsp15 | MHVnsp15F1bamHI (agtcggatccAAGTTTAGAAAATGTA) | MHVnsp15R1 (gactgcggccgcCTGCAAACGAGGATAGAA) |
| F50 | MHVnsp15F50 (agtcggatccAACTAATGTGGCTGTCGAA) | MHVnsp15R1 |
| F100 | MHVnsp15F100 (agtcggatccAGTCTGCAAATACACAGAT) | MHVnsp15R1 |
| F150 | MHVnsp15F150 (agtcggatccACCACAACGTGCCGATTT) | MHVnsp15R1 |
| F200 | MHVnsp15F200 (agtcggatccAAATCCGGGTGGTAATCGCGT) | MHVnsp15R1 |
| F250 | MHVnsp15F250 (agtcggatccAGCAAAATATAGTTTACA) | MHVnsp15R1 |
| F300 | MHVnsp15F300 (agtcggatccATCTAGCATTCATTCGTACTT) | MHVnsp15R1 |
| R50 | MHVnsp15F1bamHI | MHVnsp15R50 (gactgcggccgcGGGGAATGGCGTGTT) |
| R100 | MHVnsp15F1bamHI | MHVnsp15R100 (gactgcggccgcCTTATACGTCGAACTGCA) |
| R150 | MHVnsp15F1bamHI | MHVnsp15R150 (gactgcggccgcGCCTTTAATCATCGACA) |
| R200 | MHVnsp15F1bamHI | MHVnsp15R200 (gactgcggccgcACCTTGTGGGCTCCGGTA) |
| R250 | MHVnsp15F1bamHI | MHVnsp15R250 (gactgcggccgcAATGAACACATCATCAT) |
| R300 | MHVnsp15F1bamHI | MHVnsp15R300 (gactgcggccgcGTCGTATGTCACGAACTCTT) |
| MHVnsp15P101-150 | MHVnsp15F100 | MHVnsp15R150 |
| MHVnsp15P301-374 | MHVnsp15F300 | MHVnsp15R1 |
| mP301-324 | MHVnsp15F300 | MHVnsp15R324 (gactgcggccgcTTAATCAATAACAGTGCACA) |
| mP301-350 | MHVnsp15F300 | MHVnsp15R350 (gactgcggccgcTTAAACATTAACAACCTTA) |
| mP325-350 | MHVnsp15F325 (agtcggatccATTATTGTTAGATGATTT) | MHVnsp15R350 |
| mP350-374 | MHVnsp15F350 (agtcggatccAGATTTTAAAGATTTCCA) | MHVnsp15R1 |
| SARSEGFPnsp15 | SARSnsp15F1 (agtcggatccGAGTTTAGAAAATGTGGCTTA) | SARSnsp15R1 (gactgcggccgcTTGTAGTTTTGGGTAGAA) |
| SARSnsp15P101-156 | SARSnsp15F101 (agtcggatccAGTCTGCACAATGACTGA) | SARSnsp15R156 (gactgcggccgcTTATCCCTTTGAAGGTGTTA) |
| SARSnsp15P273-346 | SARSnsp15F273 (agtcggatccAAGCACAGTGAAAAATTA) | SARSnsp15R1 |
| SARSnsp15minicore | SARSnsp15F287 (agtcggatccATCATCAAAATGTGTGTGT) | SARSnsp15R323 (gactgcggccgcTTAGTCAATTGTAACCTTGA) |
| TGEVEGFPnsp15 | TGEVnsp15F1 (agtcggatccGAGTCTAGAAAATGTGGCTTT) | TGEVnsp15R1 (gactgcggccgcTTGGAGTTGTGGATAGAA) |
| TGEVnsp15P101-148 | TGEVnsp15F101 (agtcggatccAGTGTGTTCCTACACTGAT) | TGEVnsp15R148 (gactgcggccgcTTATAATTTAATGGCACTA) |
| TGEVnsp15P266-339 | TGEVnsp15F266 (agtcggatccAAGTACACTGAAAAGTT) | TGEVnsp15R1 |
| TGEVnsp15minicore | TGEVnsp15F280 (agtcggatccATCTTCTAAGAATGTGT) | TGEVnsp15R316 (gactgcggccgcTTAATCTACAATGACATCCA) |
| SARS/MHV-Domain I | SARSnsp15F1 | SMnsp15R1 (ATCTGTGTATTTGCAGACACCTATTGTAGATACAT) |
| SMnsp15F2 (ATGTATCTACAATAGGTGTCTGCAAATACACAGAT) | SMnsp15R2 (TTTGAAGGTGTTAGGCCTTTAATCATCGA) | |
| SMnsp15F3 (TCGATGATTAAAGGCCTAACACCTTCAAA) | SARSnsp15R1 | |
| TGEV/MHV-Domain I | TGEVnsp15F1 | TMnsp15R1 (ATCTGTGTATTTGCAGACTTGCTTAGTGAAATTT) |
| TMnsp15F2 (AAATTTCACTAAGCAAGTCTGCAAATACACAGAT) | TMnsp15R2 (ATCATTCAAAAGGCCGCCTTTAATCATCGA) | |
| TMnsp15F3 (TCGATGATTAAAGGCGGCCTTTTGAATGAT) | TGEVnsp15R1 | |
| SARS/MHV miniD | SARSnsp15F273 | SMR1 (TGCACACACTCTTACTACTACCTGTTTGCGCATCT) |
| SMF2 (AGATGCGCAAACAGGTAGTAGTAAGAGTGTGTGCA) | SMR2 (AATGAAATTTCAGCATAATCAACATTAACATTAA) | |
| SMF3 (TTAATGTTAATGTTGATTATGCTGAAATTTCATT) | SARSnsp15R1 | |
| TGEV/MHV miniD | TGEVnsp15F266 | TMR1 (ACACACTCTTACTACTTGGATCATCAGCATAT) |
| TMF2 (ATATGCTGATGATCCAAGTAGTAAGAGTGTGT) | TMR2 (ATCTCCATGCCTTACAATCAACATTAACATT) | |
| TMF3 (AATGTTAATGTTGATTGTAAGGCATGGAGAT) | TGEVnsp15R1 | |
| pTriGST | GSTNcoI (atgcccATGgCCCCTATACTAGGTTATT) | GSTNotI (atgcGCGGCCGCcTCGAGTCGACCCGGGAA) |
| pTriGSTnsp15C300 | MHVnsp15F300 | MHVnsp15R1 |
The noncoding sequence is indicated in low case letter and the restriction enzyme site is underlined.
For construction of nsp15 deletion mutants, the full-length nsp15 plasmids were used as templates for PCR amplification with a pair of primers specific for individual deletion mutants as described in Table 1. The PCR products for all deletion constructs were digested with BamH I and Not I and cloned into the BamH I–Not I sites of pTriEGFP.
For generating chimeric MHV/SARS or MHV/TGEV nsp15s, overlapping PCR was performed. The primer pairs used in PCR were as follow: SARSnsp15F1/SMnsp15R1, SMnsp15F2/SMnsp15R2, and SMnsp15F3/SARSnsp15R1 for swapping MHV nsp15 Domain I to SARS nsp15; SARSnsp15F273/SMR1, SMF2/SMR2, and SMF3/SARSnsp15R1 for swapping MHV mini domain to SARS nsp15; TGEVnsp15F1/TMnsp15R1, TMnsp15F2/TMnsp15R2, and TMnsp15F3/TGEVnsp15R1 for swapping MHV nsp15 Domain I to TGEV nsp15; TGEVnsp15F266/TMR1, TMF2/TMR2, and TMF3/TGEVnsp15R1 for swapping MHV mini domain to TGEV nsp15. As an example for swapping MHV nsp15 Domain I to SARS nsp15, three PCRs were first performed using primer pairs SARSnsp15F1/SMnsp15R1 (for SARS nsp15 template), SMnsp15F2/SMnsp15R2 (for MHV nsp15 template), and SMnsp15F3/SARSnsp15R1 (for SARS nsp15 template). The 3 PCR fragments were separated by agarose gel (1%) electrophoresis, excised from the gel and purified with Zymoclean Gel DNA Recovery Kit (Zymo Research). The 3 DNA fragments were then mixed in 50 μl PCR and the full-length DNA of the chimeric SARS/MHV Domain I was amplified by PCR using a pair of external primers (SARSnsp15F1/SARSnsp15R1). The chimera were digested with BamH I and Not I and were directionally cloned into pTriEGFP.
To construct glutathione-S-transferase (GST) fusion with nsp15 C-terminal, GST was cloned into pTriEx-4 with primers GSTNcoI and GSTNotI to create pTriGST vector. Primers MHVNsp15F300 and Nsp15R1 were then used to amplify MHV nsp15 C-terminal region (aa300–374) and cloned into pTriGST via BamH I and Not I sites to generate pTriGSTnsp15C300 for expression of GSTnsp15C300 fusion protein.
2.3. DNA transfection
One day before transfection, cells were subcultured in 96-well plate at 50% confluence in the absence of antibiotics. On the day of transfection, plasmids were prepared in Xfect reagent (Clontech) following supplier's protocol and transfected into the cells. Briefly, for each well, 100 ng of DNA was diluted to 25 μl with Opti-MEM I reduced serum-free medium (Invitrogen) and mixed with 25 μl of Opti-MEM I reduced serum-free medium containing 0.03 μl of Xfect reagent. The mixture was incubated for 10 min at room temperature. Following incubation, the DNA/Xfect mixture was added to the cells that had its culture medium removed. At 4 h post transfection, the medium was changed to 100 μl of DMEM containing 10% FBS for protein expression and image capture. Fluorescence was observed under a fluorescence microscope (Olympus IX70) at different times post transfection and images were captured with an attached digital camera (MagnaFire).
2.4. Immunofluorescence assay (IFA)
Cells in 6-well or 96-well plates were infected with MHV-A59 at a multiplicity of infection (m.o.i.) of 5. At various times post infection (p.i.), cells were fixed in 4% paraformaldehyde for 5 min at 4 °C and permeabilized with 0.3% Triton x-100 for 3 min. Fixed cells were then blocked in 5% bovine serum albumin (BSA) for 10 min, and incubated with a primary rabbit anti-nsp15 antibody D23 (1/400 dilution) (kindly provided by Dr. Susan Baker, Loyola University Stritch School of Medicine) at 37 °C for 30 min and then with a secondary goat anti-rabbit IgG conjugated with fluorescein isothiocyanate (FITC) (1/400 dilution, Sigma) for an additional 30 min. For double staining, cells were incubated with D23 (1/400) and a monoclonal antibody J3.3 against MHV nucleocapsid protein (1/100) (kindly provided by Dr. John Fleming, University of Wisconsin Medical Center in Madison) followed by incubation with goat anti-rabbit IgG-FITC (1/400) and goat anti-mouse IgG conjugated with tetramethylrhodamine-isothiocyanate (TRITC) (1/600, Sigma). To view co-localization of GST-nsp15C300 and EGFP-nsp15C300, plasmids pTriGSTnsp15C300 and F300 were co-transfected into 293T cells at 100 ng:100 ng per well in a 96-well plate using Xfect (Clontech). At 2 days post transfection, cells were fixed with 4% paraformaldehyde and IFA was performed as described above using anti-GST mAb and goat anti-mouse IgG-TRITC. Fluorescence was observed under a microscope (Olympus IX70) and the images were captured with an attached digital camera (MagnaFire).
2.5. Immunoprecipitation and Western blot analysis
To determine the interaction between the C-terminal region of nsp15, pTriGSTnsp15C300 and F300 (pTriEGFPnsp15C300) were co-transfected into 293T cells at 2.5 μg:2.5 μg per well in a 6-well plate using Xfect. Plasmids pTriGST and F300 were co-transfected to serve as a negative control. At 2 days post transfection, cells were washed with cold PBS twice and lysed in 250 μl radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 10 mM Tris, pH 7.2, 0.1% SDS, 1.0% Triton X-100, 1% deoxycholate, 5 mM EDTA) containing protease inhibitor cocktail (Roche) on ice for 30 min. After centrifugation at 16,000 × g for 5 min at 4 °C, supernatant was transferred to a 1.5 ml tube and was incubated with 10 μl of anti-GST mAb (2 μg) for 1 h at 4 °C with gentle shaking. The antibody–GST complex was then captured with protein A/G agarose (Roche) following incubation for 1 h at 4 °C with gentle shaking. After 5 washes with RIPA buffer containing 300 mM NaCl, the final pellet was suspended in 20 μl of sodium dodecyl sulfate (SDS) sample buffer and boiled for 5 min at 95 °C. After spinning for 1 min, 15 μl of the supernatant was loaded on 12% SDS-polyacrylamide gel electrophoresis (PAGE) and the proteins were transferred to nitrocellulose membrane for Western blotting. The membrane was blotted with a primary rabbit anti-GFP antibody (Invitrogen) and a secondary goat anti-rabbit IgG (H + L) antibody conjugated with horse radish peroxidase (HRP) (Sigma). The EGFP-containing protein band was then visualized with the enhanced chemiluminescence (ECL) reagent (Pierce).
2.6. Luciferase assay
The steady-Glo luciferase assay system (TM051) was carried out to determine the firefly luciferase activity in MHV-2aFLS-infected cells according to the manufacturer's protocol (Promega). Briefly, DBT cells were cultured in 96-well white plate at approximately10,000 cells/well/100 μl overnight. Culture medium was then removed and cells were transfected with plasmid DNAs containing various EGFP-nsp15 constructs. At 9 h post transfection, cells were infected with 50 μl of MHV 2aFLS at a multiplicity of infection (MOI) of 5 at 37 °C. At 14 h post infection (p.i.), the plate was transferred to 4 °C for 5 min and then kept at room temperature. One hundred microliter Steady-Glo luciferase reagent was then added to each well of the 96-well white plate followed by shaking gently for 5 min at room temperature. Luciferase activity was read with an automatic plate reader (Synergy 2) using Gen5 software (BioTek), and was expressed as percent relative to the negative control that expresses EGFP alone without the EGFP-nsp15 fusion protein.
3. Results
3.1. Features and subcellular localization of MHV nsp15 expressed in infected cells
To gain an understanding on the biological roles of coronaviral nsp15 in viral replication and host cell functions, we began with characterizing the subcellular localization of nsp15 in virus-infected cells. Mouse fibroblast 17Cl-1 cells were infected with MHV-A59 at an m.o.i. of 5. At various times p.i., cells were fixed, and the expression and intracellular localization of nsp15 were detected with immunofluorescence staining using an antibody specific to MHV nsp15 (D23). In general, fluorescence could be detected beginning at 3 h p.i. and the fluorescence intensity and the number of fluorescence-positive cells increased from 3 to 12 h p.i. (data not shown). Interestingly, while all fluorescence-positive cells exhibited cytoplasmic distribution, the pattern and subcellular localization appeared diverse among different cells (Fig. 1A). For example, in some cells the fluorescence was concentrated to one of the perinuclear regions or both sides of the spindle cell while in other cells it was relatively widely distributed (panels a and b); some cells had relatively smooth fluorescence staining whereas some other cells exhibited granular or punctate staining (panels c and d); sometimes the fluorescence formed a ring around the nucleus (panels e and f). No fluorescence was detected in mock-infected cells (panels g and h), indicating that the fluorescence detected in virus-infected cells was specific. However, the overall staining was relatively weak as compared to the staining with an antibody specific to the viral N protein (data not shown). Thus, to rule out the possibility that the detected fluorescence resulted from overexposure of nonspecific staining with the D23 antibody, infected cells were doubly stained for nsp15 and N protein. Indeed, all nsp15-positive cells (Fig. 1B, green) had viral N protein (Fig. 1B, red) although the amount of the two proteins did not always correlate in a given cell, or the intracellular localization of the two proteins did not always overlap (Fig. 1B, merge). In no instance, however, did the nsp15-positive cells have no red fluorescence staining (for N protein) (also see white arrows in Fig. 1B). These results demonstrate that the D23 antibody specifically recognized MHV nsp15 in infected cells.
Fig. 1.
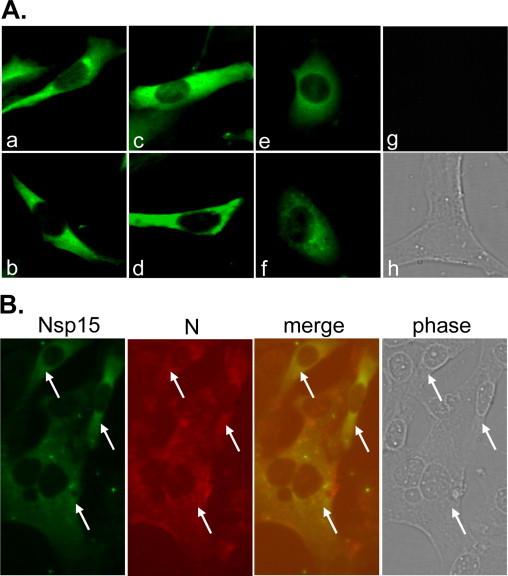
Nsp15 expression in MHV-A59 infected cells. (A) 17Cl-1 cells were infected with MHV-A59 at an m.o.i. of 5. At 7 h p.i., nsp15 protein expression was detected with immunofluorescence staining using rabbit anti-nsp15 antibody D23 and goat anti-rabbit IgG-FITC. Panels a–f show the various subcellular localizations of nsp15 in infected cells. Mock-infected cells were used as a negative control (g for fluorescence staining and h for phase contrast). (B) Dual immunofluorescence staining. Infected cells were stained with D23 and monoclonal antibody J3.3 to MHV N protein and detected with anti-rabbit IgG-FITC (Nsp15, green) and anti-mouse IgG-TRITC (N, red). The two colors are then super-imposed (merge) and the phase contrast image (phase) shows both infected and uninfected cells in the same field. The white arrows highlight that all nsp15-expressing cells (green) are virus-infected cells (red). Note that the exposure time for panels A and B was different. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
3.2. Features and subcellular localization of MHV nsp15 that was ectopically expressed as a green fluorescence fusion protein
To extend our observation in virus-infected cells and to further characterize the properties of MHV nsp15, we cloned the nsp15-coding sequence from MHV-A59 into the expression vector pTriEx-4 as a fusion protein with enhanced green fluorescence protein (EGFP) such that the expression and intracellular localization of nsp15 can be continuously monitored in living cells (Fig. 2A). Thus, 17Cl-1 cells were transfected with MHVEGFPnsp15 fusion construct or the same vector expressing EGFP alone as a control. Green fluorescence was observed as early as 6 h post transfection. We found that the green fluorescence distributed smoothly throughout the cells transfected with the EGFP vector alone while unevenly in the cytoplasm expressing MHV EGFP-nsp15 fusion protein (Fig. 2B, left 2 panels). To further confirm that the observed green fluorescence in MHVEGFPnsp15-transfected cells represents EGFP-nsp15 fusion protein rather than EGFP alone (although the fusion site of the construct was verified by sequencing), we used the D23 antibody to detect nsp15 in immunofluorescence assay. Results showed that nsp15 was detected only in MHVEGFPnsp15- but not in pTriEGFP-transfected cells (Fig. 2B, middle panels) and that the distribution of nsp15 (red) and EGFP (green) completely overlapped (Fig. 2B, top right panel). Thus, the EGFP-fusion system may be useful for monitoring the intracellular distribution of nsp15 in real time. Further, there was no uniform pattern of intracellular distribution for EGPF-nsp15; the majority of fluorescence was seen as speckles in the cytoplasm (Fig. 2C). These speckles were found in one or multiple locations of each cell. Some were close to the nucleus or formed a ring-like appearance in the perinuclear region while others were distantly away from the nucleus. While some of the fluorescence distribution patterns were similar to those found in virus-infected cells, the fluorescence speckles were markedly extensive in transfected cells than in infected cells (compare Fig. 2C with Fig. 1A). As there is no uniformed subcellular localization of the speckles, experiments for colocalization with cellular markers were not performed subsequently. It is noted that speckle formation did not appear to be associated with cytotoxicity as cells overexpressing EGFP-nsp15 continued to divide 3 days after transfection as determined by the XTT cytotoxicity assay (data not shown).
Fig. 2.
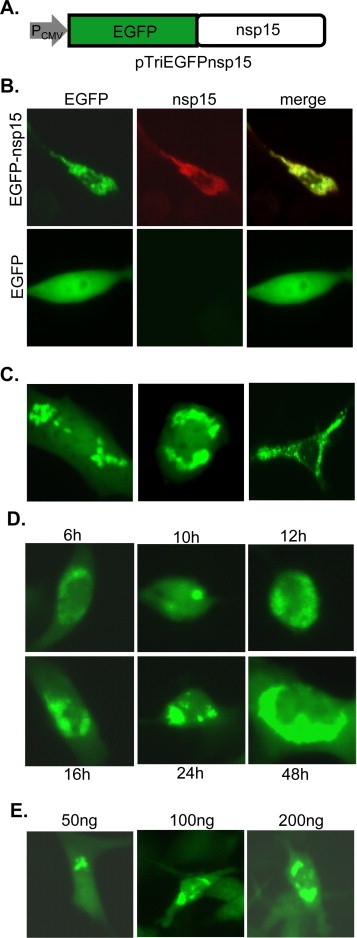
Ectopic expression of MHV nsp15 as EGFP fusion protein in transfected cells. (A) Diagram of expression plasmid MHVEGFPnsp15 showing the MHV nsp15 coding sequence fused at the N-terminus to EGFP. (B) Expression of MHV EGFP-nsp15 or EGFP alone following plasmid transfection. Left panels indicate direct detection of EGFP (EGFP) while middle panels show the detection of nsp15 following immunofluorescence staining with anti-nsp15 antibody D23 and anti-rabbit IgG-TRITC (red). Color-merged images are shown on the right (merge). (C) Examples of detailed speckle formation at various subcellular localizations following the expression of MHV EGFP-nsp15. (D) Time course experiment showing speckle formation from 6 to 48 h post transfection with MHVEGFPnsp15. (E) Speckle formation at 24 h post transfection with MHVEGFPnsp15 DNA at various concentrations (50–200 ng). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
To determine whether the observed fluorescence speckles were dependent on the amount of EGFP-nsp15 expressed, we examined fluorescence distribution from the earliest time point (6 h) post transfection, at which time EGFP-nsp15 was expressed at a minimal level. We found that the fluorescence was concentrated in one or a few distinct spots within the cytoplasm and not distributed throughout the cytoplasm as seen in virus-infected cells, although the size of the fluorescent spots increased with time (Fig. 2D). In addition, when cells were transfected with a lower amount of MHVEGFPnsp15, similar results were obtained (Fig. 2E). These findings suggest that the fluorescent speckles are specific to EGFP-nsp15 independent of the amount (concentration) of the expressed protein.
3.3. Comparative analysis of nsp15 from MHV, SARS-CoV and TGEV
The unusual fluorescence clusters seen in MHV EGFP-nsp15-expressed cells raised an important question as to whether this property is unique to MHV nsp15 or is common among coronaviral nsp15s. To address this question, we subcloned the coding sequence of nsp15 of SARS-CoV and TGEV into pMHVEGFPnsp15 to replace the MHV nsp15 counterpart, such that the expressed protein has an identical sequence context to MHV nsp15 as an EGFP fusion protein. Expression of the corresponding fusion proteins and their intracellular distribution following plasmid DNA transfection were then monitored by observing the fluorescence in living cells. As shown in Fig. 3 , both SARS-CoV and TGEV nsp15s did not form any fluorescence speckle in either 17Cl-1 or DBT cells. Their intracellular distribution was similar to that of EGFP alone but was distinct from that of MHV EGFP-nsp15. We confirmed by sequencing the constructs that both SARS-CoV and TGEV nsp15s had a correct in-frame fusion with EGFP and that there was no stop codon introduced into the nsp15 coding sequence following PCR amplification and cloning. Thus, the ability to form speckles in transfected cells appears unique for MHV nsp15 independent of cell types.
Fig. 3.
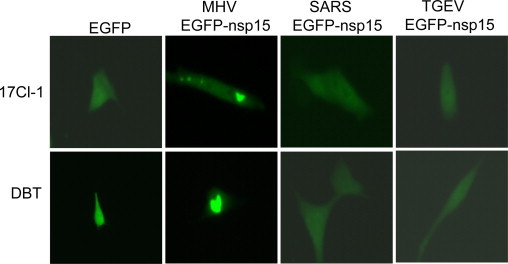
Intracellular distribution of ectopically expressed nsp15 of SARS-CoV and TGEV as EGFP fusion proteins. 17CL-1 or DBT cells were transfected with plasmids expressing EGFP alone, MHV EGFP-nsp15, SARS-CoV EGFP-nsp15, and TGEV EGFP-nsp15. Fluorescence images were captured at 24 h post transfection.
3.4. Identification of MHV nsp15 domains responsible for speckle formation
Because speckle formation appears unique for MHV nsp15, we asked whether some specific sequences or domains of MHV nsp15 are responsible for speckle formation. Thus, we made a series of sequential N-terminal or C-terminal deletion mutants of MHV nsp15 and fused each to EGFP as a fusion protein (Fig. 4A). Following transfection of these plasmids into 17Cl-1 cells, the expression and localization of mutant EGFP-nsp15s were then observed. It was found that some of the deletion mutants formed speckles while others did not (Fig. 4B). Collectively, two regions were found to form speckles independently, i.e., Domain I between amino acid (aa) 101 and aa150 and Domain III between aa301 and aa374. Interestingly, when the Domain III was extended to include an upstream domain (II) between aa151 and aa250, speckles disappeared (Fig. 4B, constructs F150 and F200), suggesting that Domain II had an inhibitory effect on speckle formation induced by Domain III. However, the same Domain II could not inhibit the speckle formation mediated by Domain I as evidenced in the constructs R300, R250 and R200 (Fig. 4). These results revealed a complex intra-molecular interaction of MHV nsp15 (Fig. 4C).
Fig. 4.
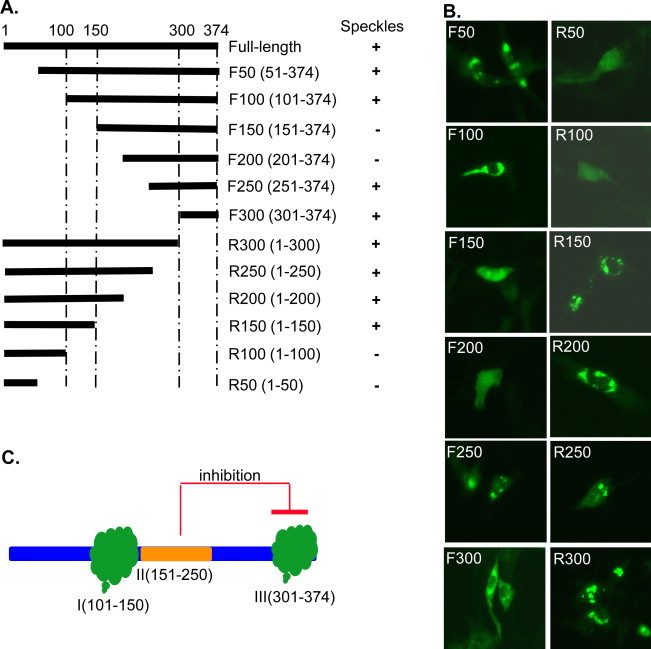
Mapping of MHV nsp15 domains for speckle formation. Schematic diagram, name and amino acid position of MHV nsp15 deletion mutants used in the experiments in reference to the full-length nsp15. These deleted fragments were fused with EGFP as fusion proteins as in Fig. 2A. (B) Expression and distribution of EGFP-nsp15 deletion mutants. A summary of the ability of individual fusion proteins to form speckles is shown on the right to the corresponding construct in panel (A). (C) Diagram illustrating the domain mapping results, highlighting 3 potentially separate domains (Domain I: aa101–150, Domain II: aa151–250, and Domain III: aa301–374) that regulate speckle formation.
Because the C-terminal region (Domain III) of nsp15 is highly conserved and contains enzymatically active sites, this domain was further dissected to identify minimal sequences required for speckle formation. Expression of the fusion proteins in 17Cl-1 cells revealed that all fusion proteins containing a region between aa325 and aa350 of MHV nsp15 formed speckles in transfected cells (Fig. 5 , panels b, c, and d) whereas those regions that lack this 25 amino acid sequence did not (Fig. 5, panels a and e), thus narrowing Domain III down to roughly 25 amino acids between aa325 and aa350 that is responsible for speckle formation.
Fig. 5.
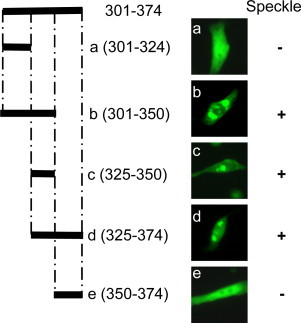
Determination of mini domains in the C-terminal region of MHV nsp15. Diagram of deletion constructs in the C-terminal region of MHV nsp15 with names and amino acid positions is shown on the left, while the expression and intracellular localization of the individual EGFP fusion proteins are shown in the middle. A summary of mini-domain mapping results with respect to the ability to form speckle is indicated on the right.
3.5. Evidence for inter-molecular interaction involving Domain III of MHV nsp15
The speckle formation involving Domain III of MHV nsp15 described above suggests that Domain III may be involved in protein–protein interaction. To test this hypothesis and to provide biochemical evidence, the inter-molecular interaction was assessed with two approaches. The 74aa sequence (Domain III) at the C-terminus of MHV nsp15 was fused with EGFP or GST and was co-expressed in the same cells (Fig. 6A). In the first experiment, the intracellular localization of the two fusion proteins was detected with immunofluorescence staining using antibodies against GST (rhodamine-red). It was found that the two fusion proteins essentially co-localized in the cells (Fig. 6B), suggesting potential interaction between them. In the second experiment, cell lysates were isolated at 48 h post transfection and were subject to immunoprecipitation with an antibody specific to GST. The immunoprecipitates were separated by SDS-PAGE, and were subsequently detected by Western blotting with an antibody specific to EGFP. Results showed that the C-terminal 74aa of nsp15 (EGFP-nsp15C300) was co-precipitated with GST-nsp15C300 but not with GST by the anti-GST antibody (Fig. 6C). These results thus provide direct evidence, demonstrating that the C-terminal domain of MHV nsp15 involves homotypic interactions in ectopically expressed cells.
Fig. 6.
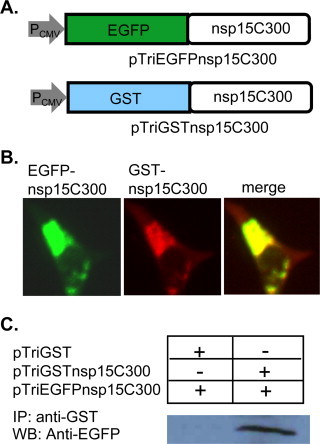
Protein-protein interaction involving the C-terminal domains of MHV nsp15. (A) Diagram of expression plasmids pTriEGFPnsp15C300 and pTriGSTnsp15C300 showing the MHV nsp15 C-terminal 74aa (from aa300 forward) coding sequence fused at the N-terminus to EGFP or GST, respectively. (B) Detection of co-localization (right panel, merge) of EGFP-nsp15C300 and GST-nsp15C300 by immunofluorescence staining with antibodies specific to GFP (left panel, green) and GST (middle panel, red) at 48 h post transfection. (C) Detection of the homotypic interaction involving the C-terminal domain of MHV nsp15 by co-immunoprecipitation and Western blot analysis. Cells were co-transfected with pTriEGFPnsp15C300 and pTriGSTnsp15C300 or pTriGST. Cell lysates were then precipitated with an antibody specific to GST. The immunocomplex was then detected by Western blot with an antibody specific to GFP. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
3.6. Comparative analysis of nsp15 domains from MHV, SARS-CoV, and TGEV
The striking difference in speckle formation among the 3 coronaviral nsp15s prompted us to further compare their amino acid sequences with an emphasis on Domains I and III (Fig. 7A). We found that the overall amino acid sequence identity among the 3 nsp15s was low (≈30%). Pair-wise comparison showed the identity ranging from 41 to 46% (46% between MHV and SARS-CoV, 41% between MHV and TGEV, and 43% between SARS-CoV and TGEV). However, sequence within Domain I is less conserved (41% between MHV and SARS-CoV, 38% between MHV and TGEV, 32% between SARS-CoV and TGEV). In contrast, Domain III is relatively highly conserved among the 3 viruses (59% between MHV and SARS-CoV, 55% between MHV and TGEV, and 57% between SARS-CoV and TGEV) (Fig. 7A).
Fig. 7.
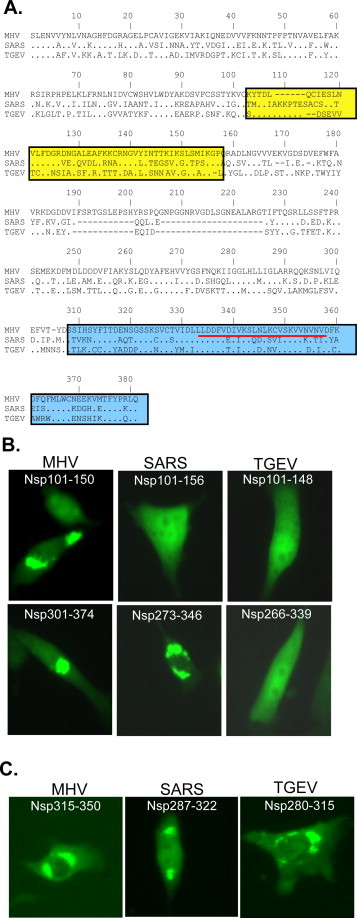
Comparison of domains among the 3 coronaviral nsp15s. (A) Amino acid sequence alignment of nsp15s of MHV, SARS-CoV and TGEV, with Domains I and III boxed, and the 25aa core sequence within Domain III is underlined. (B) Expression of Domains I and III of the three EGFP-nsp15 fusion proteins and their intracellular distribution. (C) Expression of the three EGFP-mini domain (core sequence) fusion proteins in 17CL-1 cells and their intracellular distribution.
Since Domains I and III of MHV nsp15 independently formed speckles (Fig. 4), we wanted to further confirm whether the corresponding domains in SARS-CoV and TGEV lack the ability to form speckles. To test this possibility, we cloned the corresponding Domain I and Domain III from SARS-CoV and TGEV nsp15 based on the sequence alignment shown in Fig. 7A. As with the corresponding domains of MHV nsp15, these domains were individually fused to EGFP as fusion proteins. Expression of the fusion proteins in transfected cells showed that both Domains I and III of TGEV nsp15 and Domain I of SARS-CoV nsp15 did not form any speckles (Fig. 7B). Surprisingly, Domain III (aa273–346) of SARS-CoV nsp15 alone formed speckles (Fig. 7B), suggesting that upstream sequence likely inhibited the speckle formation mediated by Domain III when expressed in full-length (compare with Fig. 3). This striking result prompted us to further delineate Domain III of SARS-CoV and TGEV nsp15s. Thus, the corresponding core sequence (35aa) that was essential for speckle formation in MHV nsp15 was cloned from SARS-CoV and TGEV and expressed as EGFP fusion proteins (Fig. 7A). Results showed that all 3 core sequence constructs formed speckles (Fig. 7C), suggesting that sequences surrounding this core sequence within Domain III had an inhibitory effect for TGEV nsp15, but not for MHV and SARS-CoV nsp15s.
3.7. Domain swapping confirmed the presence of suppressive sequences within Domain III of TGEV nsp15
To provide direct experimental evidence for potential suppressive sequence, we performed experiments by swapping domains among MHV, SARS-CoV and TGEV. In the first set of experiments, Domain I of SARS-CoV and TGEV nsp15s was swapped with the corresponding domain of MHV nsp15 (Fig. 8A). Results showed that the chimeric nsp15s (SARS/MHV-Domain I and TGEV/MHV-Domain I) bearing MHV nsp15 Domain I formed speckles when expressed as EGFP fusion proteins in 17Cl-1 cells (Fig. 8B), confirming that Domain I of MHV nsp15 alone is sufficient to confer the ability to form speckles. In the second set of experiments, the core sequence of Domain III in SARS-CoV nsp15 (aa287–322) and TGEV nsp15 (280–315) was swapped with the counterpart of MHV nsp15 (aa315–350) (Fig. 9A). As expected, swapping this region of SARS-CoV nsp15 with the corresponding region of MHV nsp15 did not alter the expression and intracellular distribution pattern, confirming that surrounding sequence in Domain III of SARS-CoV nsp15 did not have any inhibitory effect on speckle formation by the core sequence (Fig. 9B, left panel). However, when the same MHV nsp15 region was expressed in the sequence context of TGEV nsp15, no speckle was formed (Fig. 9B, right panel). This result thus established that the surrounding sequence in Domain III of TGEV nsp15 had a suppressive effect on speckle formation by the core sequence (aa280–315).
Fig. 8.
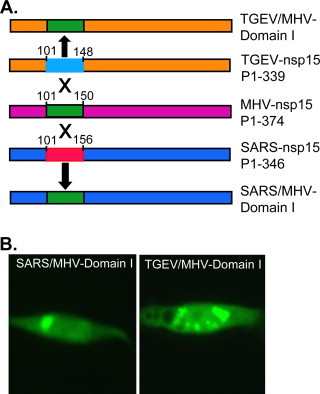
Swapping of Domain I between MHV and other CoV nsp15s. (A) Diagram showing the replacement of Domain I of SARS-CoV and TGEV nsp15 with the corresponding domain of MHV nsp15, and the resultant chimeric constructs. Only the nsp15 region of the EGFP fusion construct is shown. (B) Expression and detection of speckles for the chimeric constructs.
Fig. 9.
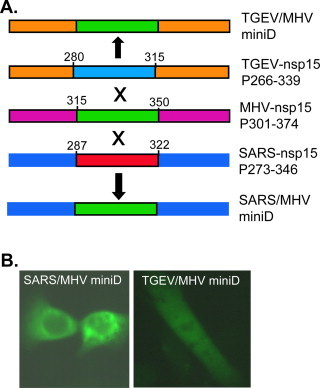
Swapping of mini domains within Domain III between MHV and other CoV nsp15s. (A) Diagram showing the replacement of the mini domain in Domain III of SARS-CoV and TGEV nsp15 with the corresponding mini domain of MHV nsp15, and the resultant chimeric constructs. Only the nsp15 region of the EGFP fusion construct is shown. (B) Expression and detection of speckle for the chimeric constructs.
3.8. Effect of ectopically expressed EGFP-nsp15 fusion proteins on MHV replication
The above experiments established that MHV nsp15 has different subcellular properties when expressed as EGFP fusion protein as compared with SARS-CoV and TGEV nsp15s. To further assess whether ectopically expressed EGFP-nsp15s have any biological function on MHV replication and whether their functions are interchangeable, DBT cells in 96-well plate were transfected with plasmids expressing EGFP fusion proteins of the full-length nsp15 or its deletion construct containing either Domain I (DI) or Domain III (DIII). Cells transfected with the plasmid expressing EGFP alone were used as a negative control. At 9 h post transfection, when fluorescence of EGFP or EGFP fusion proteins was seen in all transfected wells, the cells were infected with the luciferase-expressing recombinant MHV-2aFLS. Virus replication and gene expression were then determined at 14 h post infection by luciferase assay. Fig. 10 shows results of one representative experiment. Compared with the EGFP negative control, expression of MHV EGFP-nsp15 drastically enhanced luciferase activity (P < 0.01) while expression of SARS EGFP-nsp15 reduced luciferase activity slightly but statistically significantly (P < 0.05). Expression of TGEV EGFP-nsp15 appeared to have little effect on luciferase activity (P > 0.05). Interestingly, expression of Domain I alone of all three coronaviral nsp15s significantly inhibited luciferase activity (P < 0.05), suggesting a dominant-negative effect of the deletion mutants on MHV replication. In contrast, expression of Domain III of SARS-CoV and TGEV nsp15 had no effect on luciferase activity, whereas expression of Domain III of MHV nsp15 had a significant inhibitory effect on luciferase activity. These data suggest that ectopically expressed MHV EGFP-nsp15s are biologically functional. The data also suggest that some domains (e.g. Domain I) can be functionally interchangeable among the three viral nsp15s while some other domains (e.g. Domain III) are functionally distinct.
Fig. 10.
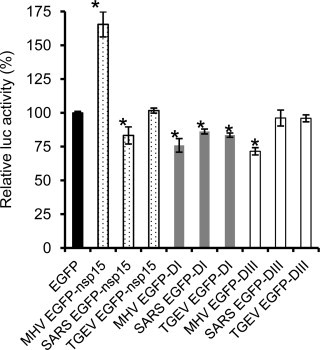
Effect of ectopically expressed EGFP-nsp15s on MHV replication. DBT cells were transfected with 100 ng each plasmid DNA containing either the full-length or deletion mutant (DI and DIII) nsp15s fused to EGFP as indicated at the bottom of the x-axis. Cells transfected with the vector plasmid expressing EGFP alone were used as a negative control. At 9 h post transfection, the cells were infected with MHV-2aFLS at m.o.i. of 5. Luciferase activity was determined at 14 h p.i., and was expressed as percent relative to the negative control, which is set as 100%. The results were analyzed for statistical significance between the testing and control groups using the Student's t-test. P values of <0.01 or <0.05 were considered statistically significant, which is indicated with an asterisk.
4. Discussion
In the present study, the expression and in vivo biochemical and biological properties of MHV nsp15 were assessed in virus-infected and ectopically expressed cells, and were compared with the counterpart of SARS-CoV and TGEV. This study thus represents the first step toward understanding the in vivo biological role of coronaviral nsp15, as most of the previous studies have been limited to in vitro structural and functional analysis of bacteria-expressed nsp15. Our results show that, in virus-infected cells, a relatively higher amount of nsp15 is present near the perinuclear region than in the peripheral regions (Fig. 1). This observation is consistent with its putative role in viral replication and suggests that nsp15 is likely associated with other viral nsps and viral RNA in the replication/transcription complexes found in the perinuclear region during viral infection (Bost et al., 2001, Shi et al., 1999). However, we also found a substantial amount of nsp15 present outside the putative viral replication/transcription complexes and in some cases throughout the cytoplasm (Fig. 1). This may suggest that in addition to its putative role in viral replication/transcription, coronaviral nsp15 may have additional biological functions in host cells, as intracellular expression and localization are often associated with the biological properties of a protein. In support of this idea is a recent report, which shows that SARS-CoV nsp15 has an ability to inhibit mitochondrial anti-viral signal (MAVS) protein-mediated apoptosis when ectopically co-expressed in human 293T cells (Lei et al., 2009).
The observation that ectopically expressed MHV EGFP-nsp15 exhibited significantly more extensive speckles in the cytoplasm as compared to that in virus-infected cells (compare Fig. 1 with Fig. 2) prompted us to ask whether the speckles have resulted from artificial over-expression due to EGFP fusion. To address this question, we removed the EGFP coding sequence, which leaves an AUG initiation codon in place to fuse with nsp15 coding region in the expression plasmid (Fig. 2A). However, no nsp15 was detected by immunofluorescence assay or Western blot with the anti-nsp15 antibody (D23), suggesting that nsp15 was either expressed at a level below detection or was rapidly degraded after expression (data not shown). Thus, the effect of EGFP on speckle formation of the fusion protein cannot be conclusively ruled out. To circumvent this problem, we then replaced EGFP with GST. Expression of the GST-nsp15 fusion protein was detected with immunofluorescence staining using an anti-GST monoclonal antibody. Our results showed that similar speckle formation was observed for GST-nsp15 fusion protein (Fig. 6B and further data not shown), thus arguing that speckle formation is likely not due to EGFP fusion. Furthermore, replacement of MHV nsp15 with the corresponding coding sequences for nsp15 of SARS-CoV and TGEV did not result in speckle formation (Fig. 3). Thus, the speckle formation appears unique to MHV EGFP-nsp15. To further assess whether the speckles resulted from protein over-expression from the plasmid, we transfected cells with reducing amounts of plasmid DNAs. While the overall expression level was dose-dependent at any given time point, the phenomenon of localized speckle formation remained unchanged (Fig. 2E). Furthermore, when the fluorescence was carefully observed at the earliest time points detectable following transfection, similar speckles were readily seen, albeit to a less intensity (Fig. 2D). Taken together, these findings suggest that the intracellular behavior or the in vivo biochemical properties of MHV nsp15 are different from those of SARS-CoV and TGEV.
Why is the speckle formation much more pronounced in ectopically expressed nsp15 than during MHV infection? One possible explanation is that during ectopic expression, other viral proteins are absent and the expressed EGFP-nsp15 does not undergo proteolytic processing. Thus, self-interaction among MHV EGFP-nsp15 is greatly accelerated, resulting in speckle formation as homotypic interaction and hexamer formation have been demonstrated in vitro for bacteria-expressed nsp15 (Guarino et al., 2005). In contrast, during virus infection, nsp15 is expressed as a polypeptide precursor and undergoes cascades of proteolytic processing. As a result, intermediate precursors likely exist and interactions with other viral nsps may interfere with its self-interaction. In this regard, it is important to note that although bacteria-expressed nsp15 forms hexamers and exhibits endonuclease activity in vitro (Cao et al., 2008, Guarino et al., 2005), these biochemical properties have yet to be demonstrated in virus-infected or ectopically expressed cells.
An important finding from this comparative analysis is the marked difference in fluorescence speckle formation among the EGFP-nsp15 fusion proteins of MHV, SARS-CoV and TGEV (Fig. 3). This was a bit surprising as these proteins were expressed in the same sequence context as EGFP fusion and the overall sequence homologies among them are similar (Fig. 7A). However, this result also suggests that the speckle formation may result from specific domain(s) rather than the entire coding sequence. Interestingly, our systematic deletion and swapping experiments have not only identified specific sequence elements within nsp15 being involved in either inducing or suppressing speckle formation but also uncovered complex intra-molecular or inter-domain interactions in vivo. For example, when only the 35aa core sequence in Domain III from the 3 viral nsp15s was individually expressed, all formed speckles (Fig. 7C); when the entire Domain III was expressed, only those from MHV and SARS-CoV formed speckles (Fig. 7B, lower panels). Likewise, when Domains I and III of MHV nsp15 were separately expressed, either domain formed speckles (Fig. 7B). However, when Domains II and III of MHV nsp15 were expressed as a single peptide, speckle was no longer formed (Fig. 4B); but again, when Domains I and II were expressed as a single peptide, it still formed speckles (Fig. 4B). Thus, formation of speckles or absence thereof is indicative of their inter- and intra-molecular interactions. Crystal structure of SARS-CoV and MHV nsp15s has revealed that three nsp15 molecules formed a trimer via interaction between their N-terminal domains and two trimers interacted back-to-back to form a hexamer while their C-terminal domains exposed outside for interaction with RNA molecule (Bhardwaj et al., 2006, Guarino et al., 2005). In solution, nsp15 purified from Escherichia coli formed monomer, trimer, and hexamer, with hexamer being the enzymatically active form (Guarino et al., 2005, Xu et al., 2006). In our current study, we also found that the N-terminal domain of MHV-nsp15 forms speckles, suggesting that N-terminal domain may be involved in protein–protein interaction. However, protein-interaction involving the C-terminal domain of nsp15 has not been reported previously. This idea was further tested in this study. Our results demonstrate that the C-terminal domain of nsp15 could mediate direct protein–protein interactions (Fig. 6). To the best of our knowledge, this is the first demonstration that the C-terminal domain of MHV-nsp15 is involved in protein–protein interaction.
The differential intracellular features of the three viral nsp15s suggest a potential difference in their biological functions. This hypothesis was further tested for their ability to affect MHV replication as nsp15 has been shown to be critical in coronavirus replication (Ivanov et al., 2004, Kang et al., 2007). Indeed, our results demonstrate that ectopically expressed EGFP-nsp15 fusion proteins have the ability to modulate MHV replication and gene expression (Fig. 10). Thus, the EGFP-nsp15 fusion proteins are likely functional in vivo even though some of the fusion proteins substantially formed speckles. This may suggest that either the speckle formation does not affect its biological function or the remaining EGFP-nsp15 that does not form speckles is sufficient to exert its function. The functional compatibility appears both virus-strain- and domain-dependent. In the case of the full-length nsp15, expression of MHV EGFP-nsp15 augmented MHV replication, while TGEV EGFP-nsp15 did not. In contrast, expression of SARS-CoV EGFP-nsp15 inhibited MHV replication instead (Fig. 10). Similarly, when Domain III alone was expressed, only MHV nsp15 had an effect on MHV replication. These data indicate that the interaction between MHV replication complexes and the expressed EGFP-nsp15 is virus-strain-dependent, further underscoring the differential biological functions among the nsp15s of the three viruses. On the other hand, when Domain I was expressed, all three viral nsp15s had an inhibitory effect on MHV replication, while only MHV nsp15 Domain III did so (Fig. 10). These results indicate that the effect of coronaviral nsp15s on MHV replication is also domain-dependent. Taken together, these findings support the central thesis of this study that different coronaviral (MHV, SARS-CoV, and TGEV) nsp15s have differential structural and functional properties that may involve inter- and intra-molecular interactions during virus infection or in the absence of virus infection in host cells.
5. Conclusion
We have shown that MHV nsp15 expressed in virus-infected cells distributed unevenly throughout the cytoplasm but predominantly in the perinuclear region. When ectopically expressed as an EGFP fusion protein, nsp15 of MHV but not SARS-CoV or TGEV formed speckles in the cytoplasm without specific subcellular localization. Further deletion mapping experiments identified two domains of MHV nsp15 (I: aa101–150 and III: aa301–374) being independently responsible for the speckle formation while Domain II (aa151–250) had an inhibitory effect on Domain III- but not Domain I-mediated speckle formation. Expression of a small sequence (≈35aa) within Domain III of all three viral nsp15s alone was sufficient to form speckles but addition of surrounding sequences modulated the speckle formation. Furthermore, a novel protein-interaction sequence at the C-terminal domain of MHV nsp15 was identified. Additionally, it was found that the effects of EGFP-nsp15s on MHV replication were both virus species- and nsp15 domain-dependent. Thus, these results demonstrate the differential biochemical and biological functions among the nsp15s of MHV, TGEV and SARS-CoV in host cells.
Acknowledgements
We thank Susan Baker (Loyola University Stritch School of Medicine, Maywood, IL) for kindly providing the anti-nsp15 antiserum, John Fleming (University of Wisconsin Medical Center in Madison, WI) for the anti-N monoclonal antibody, Ralph Baric (University of North Carolina at Chapel Hill, NC) for the SARS-CoV and TGEV nsp15 constructs, and Peter Rottier (Utrecht University, the Netherlands) for the recombinant MHV-2aFLS. This work was supported in part by Public Health Service grant AI061204 from the National Institutes of Health.
References
- Baker S.C., Shieh C.K., Soe L.H., Chang M.F., Vannier D.M., Lai M.M. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. Journal of Virology. 1989;63(9):3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Guarino L., Kao C.C. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. Journal of Virology. 2004;78(22):12218–12224. doi: 10.1128/JVI.78.22.12218-12224.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Palaninathan S., Alcantara J.M., Yi L.L., Guarino L., Sacchettini J.C., Kao C.C. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. Journal of Biological Chemistry. 2008;283(6):3655–3664. doi: 10.1074/jbc.M708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj K., Sun J., Holzenburg A., Guarino L.A., Kao C.C. RNA recognition and cleavage by the SARS coronavirus endoribonuclease. Journal of Molecular Biology. 2006;361(2):243–256. doi: 10.1016/j.jmb.2006.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bost A.G., Prentice E., Denison M.R. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology. 2001;285(1):21–29. doi: 10.1006/viro.2001.0932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredenbeek P.J., Pachuk C.J., Noten A.F., Charite J., Luytjes W., Weiss S.R., Spaan W.J. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Research. 1990;18(7):1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Boursnell M.E., Binns M.M., Bilimoria B., Blok V.C., Brown T.D., Inglis S.C. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO Journal. 1987;6(12):3779–3785. doi: 10.1002/j.1460-2075.1987.tb02713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I., Digard P., Inglis S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell. 1989;57(4):537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., Wu C.C., Lin T.L. Turkey coronavirus non-structure protein NSP15 – an endoribonuclease. Intervirology. 2008;51(5):342–351. doi: 10.1159/000175837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denison M.R., Sims A.C., Gibson C.A., Lu X.T. Processing of the MHV-A59 gene 1 polyprotein by the 3C-like proteinase. Advances in Experimental Medicine and Biology. 1998;440:121–127. doi: 10.1007/978-1-4615-5331-1_16. [DOI] [PubMed] [Google Scholar]
- Denison M.R., Zoltick P.W., Hughes S.A., Giangreco B., Olson A.L., Perlman S., Leibowitz J.L., Weiss S.R. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology. 1992;189(1):274–284. doi: 10.1016/0042-6822(92)90703-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioia U., Laneve P., Dlakic M., Arceci M., Bozzoni I., Caffarelli E. Functional characterization of XendoU, the endoribonuclease involved in small nucleolar RNA biosynthesis. Journal of Biological Chemistry. 2005;280(19):18996–19002. doi: 10.1074/jbc.M501160200. [DOI] [PubMed] [Google Scholar]
- Gosert R., Kanjanahaluethai A., Egger D., Bienz K., Baker S.C. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. Journal of Virology. 2002;76(8):3697–3708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarino L.A., Bhardwaj K., Dong W., Sun J., Holzenburg A., Kao C. Mutational analysis of the SARS virus Nsp15 endoribonuclease: identification of residues affecting hexamer formation. Journal of Molecular Biology. 2005;353(5):1106–1117. doi: 10.1016/j.jmb.2005.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt B.H., Jukneliene D., Kanjanahaluethai A., Bechill J., Severson K.M., Smith C.M., Rota P.A., Baker S.C. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. Journal of Virology. 2004;78(24):13600–13612. doi: 10.1128/JVI.78.24.13600-13612.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov K.A., Hertzig T., Rozanov M., Bayer S., Thiel V., Gorbalenya A.E., Ziebuhr J. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(34):12694–12699. doi: 10.1073/pnas.0403127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J.S., Saikatendu K.S., Subramanian V., Neuman B.W., Buchmeier M.J., Stevens R.C., Kuhn P. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. Journal of Virology. 2007;81(12):6700–6708. doi: 10.1128/JVI.02817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H., Bhardwaj K., Li Y., Palaninathan S., Sacchettini J., Guarino L., Leibowitz J.L., Kao C.C. Biochemical and genetic analyses of murine hepatitis virus Nsp15 endoribonuclease. Journal of Virology. 2007;81(24):13587–13597. doi: 10.1128/JVI.00547-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K., Kikkert M., Worm S.H., Zevenhoven-Dobbe J.C., van der Meer Y., Koster A.J., Mommaas A.M., Snijder E.J. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biology. 2008;6(9):e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laneve P., Altieri F., Fiori M.E., Scaloni A., Bozzoni I., Caffarelli E. Purification, cloning, and characterization of XendoU, a novel endoribonuclease involved in processing of intron-encoded small nucleolar RNAs in Xenopus laevis. Journal of Biological Chemistry. 2003;278(15):13026–13032. doi: 10.1074/jbc.M211937200. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Shieh C.K., Gorbalenya A.E., Koonin E.V., La Monica N., Tuler J., Bagdzhadzhyan A., Lai M.M. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180(2):567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y., Moore C.B., Liesman R.M., O‘Connor B.P., Bergstralh D.T., Chen Z.J., Pickles R.J., Ting J.P. MAVS-mediated apoptosis and its inhibition by viral proteins. PLoS One. 2009;4(5):e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova D.D., Ulferts R., van den Born E., Lauber C., Gorbalenya A.E., Ziebuhr J., Snijder E.J. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. Journal of Virology. 2009;83(11):5671–5682. doi: 10.1128/JVI.00261-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S., Egloff M.P., Ulferts R., Coutard B., Nurizzo D., Campanacci V., Cambillau C., Ziebuhr J., Canard B. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(32):11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S.T., Schiller J.J., Kanjanahaluethai A., Baker S.C., Oh J.W., Lai M.M. Colocalization and membrane association of murine hepatitis virus gene 1 products and De novo-synthesized viral RNA in infected cells. Journal of Virology. 1999;73(7):5957–5969. doi: 10.1128/jvi.73.7.5957-5969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., Bredenbeek P.J., Dobbe J.C., Thiel V., Ziebuhr J., Poon L.L., Guan Y., Rozanov M., Spaan W.J., Gorbalenya A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. Journal of Molecular Biology. 2003;331(5):991–1004. doi: 10.1016/S0022-2836(03)00865-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder E.J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J.J., van der Meulen J., Koerten H.K., Mommaas A.M. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. Journal of Virology. 2006;80(12):5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel V., Herold J., Schelle B., Siddell S.G. Viral replicase gene products suffice for coronavirus discontinuous transcription. Journal of Virology. 2001;75(14):6676–6681. doi: 10.1128/JVI.75.14.6676-6681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Zhai Y., Sun F., Lou Z., Su D., Xu Y., Zhang R., Joachimiak A., Zhang X.C., Bartlam M., Rao Z. New antiviral target revealed by the hexameric structure of mouse hepatitis virus nonstructural protein nsp15. Journal of Virology. 2006;80(16):7909–7917. doi: 10.1128/JVI.00525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebuhr J. The coronavirus replicase. Current Topics in Microbiology and Immunology. 2005;287:57–94. doi: 10.1007/3-540-26765-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]


