Abstract
Background
Staphylococcus aureus is an important pathogen that causes biofilm-associated infection in humans. Autoinducer 2 (AI-2), a quorum-sensing (QS) signal for interspecies communication, has a wide range of regulatory functions in both Gram-positive and Gram-negative bacteria, but its exact role in biofilm formation in S. aureus remains unclear.
Results
Here we demonstrate that mutation of the AI-2 synthase gene luxS in S. aureus RN6390B results in increased biofilm formation compared with the wild-type (WT) strain under static, flowing and anaerobic conditions and in a mouse model. Addition of the chemically synthesized AI-2 precursor in the luxS mutation strain (ΔluxS) restored the WT phenotype. Real-time RT-PCR analysis showed that AI-2 activated the transcription of icaR, a repressor of the ica operon, and subsequently a decreased level of icaA transcription, which was presumably the main reason why luxS mutation influences biofilm formation. Furthermore, we compared the roles of the agr-mediated QS system and the LuxS/AI-2 QS system in the regulation of biofilm formation using the ΔluxS strain, RN6911 and the Δagr ΔluxS strain. Our data indicate a cumulative effect of the two QS systems on the regulation of biofilm formation in S. aureus.
Conclusion
These findings demonstrate that AI-2 can decrease biofilm formation in S. aureus via an icaR-activation pathway. This study may provide clues for therapy in S. aureus biofilm-associated infection.
Background
Staphylococcus aureus is an opportunistic pathogen that can adhere to many tissues and implants in humans to form biofilms causing refractory chronic infections [1,2]. Many therapies have been proposed but the potential efficacy is limited [3]. Given this situation, intensive research into the molecular mechanism of biofilm formation in S. aureus could facilitate the development of novel therapeutic devices.
Biofilms are complex communities of microorganisms encased in slime that can attach to surfaces [4]. Protein, polysaccharide, and extracellular DNA are supposed to be important components of Staphylococcal biofilms [5-7]. Biofilm formation is established using at least two properties: the adherence of cells to a surface and accumulation to form multi-layered cell clusters [8,9]. The latter process is closely related to polysaccharide intercellular adhesion (PIA), a polysaccharide composed of β-1,6-linked N-acetylglucosamine residues in Staphylococci[10]. The intercellular adhesion (ica) locus is composed of four open reading frames (ORFs) icaA, icaD, icaB and icaC in an operon [11,12], and is responsible for generating PIA, which is required for biofilm formation in S. aureus. Moreover, decreased PIA level is considered to be the main factor leading to the destructive ability of biofilm formation in S. aureus RN6390B [13]. In recent years, many factors including glucose, glucosamine, oleic acid, urea, anaerobiosis and iron limitation have been identified as influencing the expression of PIA [12,14-18]. In addition, it has been demonstrated that IcaR represses ica expression by binding to the icaA promoter region [19]. Furthermore, QS has been recently shown to control the expression of the ica operon [20].
Quorum sensing is a widespread system used by bacteria for cell-to-cell communication, which regulates expression of multiple genes in a cell density-dependent manner [21,22]. The unique QS system shared by Gram-positive and Gram-negative bacteria is mediated by AI-2 [23], which is a signalling molecule synthesized by the luxS gene [24,25]. AI-2 originates from the auto-cyclization of precursor 4, 5-dihydroxy-2, 3-pentanedione (DPD) [26,27], and has been reported to regulate luminescence, motility and virulence [28-30]. Biofilm formation is known as the "bacterial social behaviour", in part owing to an organised mode of growth in a hostile environment. Many studies have described the role of AI-2 in biofilm formation. For example, synthetic AI-2 directly stimulates Escherichia coli biofilm formation and controls biofilm architecture by stimulating bacterial motility [31]. Subsequently, several studies also indicated that AI-2 indeed controls biofilm formation [32-34]. In contrast, some researchers reported that addition of AI-2 failed to restore biofilm phenotype of the parental strain [35-40], owing to the central metabolic effect of LuxS or difficulty in complementation of AI-2 [41]. There exists a conserved luxS gene in S. aureus, and it has been proved to be functional for generating AI-2 [42]. Previous work indicated that AI-2-mediated QS modulated capsular polysaccharide synthesis and virulence in S. aureus[43], deletion of the luxS gene led to increased biofilm formation in Staphylococcus epidermis[20], and biofilm enhancement due to luxS repression was manifested by an increase in PIA [44].
In this study, we provide evidence that S. aureus ΔluxS strain formed stronger biofilms than the WT strain RN6390B, and that the luxS mutation was complemented by adding chemically synthesized DPD, the exogenous precursor of AI-2. AI-2 activated the transcription of icaR, and subsequently led to decreased icaA transcription, as determined by real-time RT-PCR analysis. Furthermore, the differences in biofilm-forming ability of S. aureus RN6911, ΔluxS strain, and the ΔagrΔluxS strain were also investigated. Our data suggest that AI-2 could inhibit biofilm formation in S. aureus RN6390B through the IcaR-dependent regulation of the ica operon.
Methods
Bacterial strains, plasmids and DNA manipulations
The bacterial strains and plasmids used in this study are described in Table 1. E. coli cells were grown in Luria-Bertani (LB) medium (Oxoid) with appropriate antibiotics for cloning selection. S. aureus strain RN4220, a cloning intermediate, was used for propagation of plasmids prior to transformation into other S. aureus strains. S. aureus cells were grown at 37°C in tryptic soy broth containing 0.25% dextrose (TSBg) (Difco No. 211825). In the flow cell assay, biofilm bacteria were grown in tryptic soy broth without dextrose (TSB) (Difco No. 286220). Medium was supplemented when appropriate with ampicillin (150 μg/ml), kanamycin (50 μg/ml), erythromycin (2.5 μg/ml) and chloramphenicol (15 μg/ml).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| RN6390B |
Standard laboratory strain |
NARSAa |
| RN4220 |
8325-4 r- |
NARSA |
| ΔluxS |
RN6390B luxS::ermB |
This study |
| RN6911 |
RN6390B derivative; agr locus replaced with tetM cassette |
NARSA |
| ΔagrΔluxS |
RN6911 luxS::ermB, agr/luxS double mutant |
This study |
| ΔluxSpluxS |
Complemented strain of ΔluxS; Apr Cmr |
This study |
| RN6390BG |
RN6390B/pgfp |
This study |
| ΔluxSG |
ΔluxS/pgfp |
This study |
| RN6911G |
RN6911/pgfp |
This study |
| ΔagrΔluxSG |
ΔagrΔluxS/pgfp |
This study |
| NCTC8325 |
Standard Laboratory strain |
NARSA |
| NCTC8325ΔluxS |
NCTC8325 luxS::ermB |
60 |
|
E. coli strains |
|
|
| TOP10 |
Cloning |
Invitrogen |
| Plasmids |
|
|
| pEASY-Blunt |
Clone vector, Kanr Apr |
Transgen |
| pBTluxS |
Vector used for luxS mutagenesis, Apr Cmr Emr |
60 |
| pLI50 |
E. coli-S. aureus shuttle cloning vector, Apr Cmr |
Addgene |
| pLIluxS |
pLI50 with luxS and its promoter, Apr Cmr |
60 |
| pgfp | gfp expression with the promoter of S10 ribosomal gene, Apr, Cmr |
aNARSA, Network on Antimicrobial Resistance in Staphylococcus aureus.
Construction of bacterial strains
To construct the ΔluxS strain from S. aureus RN6390B and the Δagr ΔluxS strain from S. aureus RN6911, the purified pBTluxS plasmid was used for allele replacement by erythromycin-resistance gene insertional mutagenesis as described previously [45]. Briefly, the appropriate upstream and downstream fragments of luxS were amplified from the genome of RN6390B, and the erythromycin-resistance gene was amplified from pEC1 with the relevant primers. The three fragments were ligated with each other with the erythromycin-resistance gene in the middle, and then ligated with the temperature-sensitive shuttle vector pBT2. The resulting plasmid pBTluxS [43] was introduced by electroporation into S. aureus strain RN4220 for propagation, and then transformed into S. aureus RN6390B for luxS mutation and S. aureus RN6911 for agr luxS double-gene mutation. All primers used in this study are listed in Table 2.
Table 2.
Oligonucleotide primers used in this study
| Primer | Sequence |
|---|---|
| rt-16S-f |
CGTGGAGGGTCATTGGA |
| rt-16S-r |
CGTTTACGGCGTGGACTA |
| rt-icaA-f |
TTTCGGGTGTCTTCACTCTAT |
| rt-icaA-r |
CGTAGTAATACTTCGTGTCCC |
| rt-icaR-f |
ATCTAATACGCCTGAGGA |
| rt-icaR-r |
TTCTTCCACTGCTCCAA |
| rt-clfB-f |
TTTGGGATAGGCAATCATCA |
| rt-clfB-r |
TCATTTGTTGAAGCTGGCTC |
| rt-fnbA-f |
ATGATCGTTGTTGGGATG |
| rt-fnbA-r |
GCAGTTTGTGGTGCTTGT |
| rt-fnbB-f |
ACAAGTAATGGTGGGTAC |
| rt-fnbB-r |
AATAAGGATAGTATGGGT |
| rt-map-f |
AAACTACCGGCAACTCAA |
| rt-map-r |
TGTTACACCGCGTTCATC |
| rt-efb-f |
TAACATTAGCGGCAATAG |
| rt-efb-r | CCATATTCGAATGTACCA |
To make the luxS-complemented strain, the pLIluxS plasmid, which contains the native promoter of luxS and its intact open reading frame, was constructed in our previous work [43]. We purified the pLIluxS plasmid and transformed it into the ΔluxS strain for complementation, thus constructing the ΔluxSpluxS strain. WT and ΔluxS strains were also transformed with the empty plasmid pLI50 constructing strains WTp and ΔluxSp, which were used as the control. These strains transformed with plasmid were cultured in medium with chloramphenicol (15 μg/ml). The AI-2 precursor molecule, DPD, of which the storage concentration is 3.9 mM dissolved in water, was purchased from Omm Scientific Inc., TX, USA.
Biofilm formation and analysis
Biofilm formation under static conditions was determined by the microtitre plate assay based on the method described previously [46]. Briefly, the overnight cultures were made at a 1:100 dilution using fresh TSBg. The diluted cell suspension was inoculated into flat-bottom 24-well polystyrene plates (Costar 3599, Corning Inc., Corning, NY), 1 ml for each well. The plates were incubated at 37°C for different time courses and the wells were rinsed gently with water five times to remove non-adherent cells. Subsequently, the plates were stained with 0.5% crystal violet for 15 m, and then rinsed again with water to remove unbound stain. After that, the plates were dried, and the optical density at 560 nm (OD560) was determined with an enzyme-linked immunosorbent assay reader in a 5 × 5 scan model. To investigate the effect of AI-2, the medium was supplemented with chemically synthesized DPD with a concentration range of 0.39 nM to 390 nM.
Biofilm formation was also examined in a flow cell (Stovall, Greensboro, USA), which was assembled and prepared according to the manufacturer's instructions. Flow cell experiments and laser scanning confocal microscope (CLSM) were performed as described previously [47]. Overnight cultures of different strains were adjusted to OD600 of 6.5 and made at a 1:100 dilution in fresh 2% TSB. Flow cells were inoculated with 4 ml of these culture dilutions and incubated at 37°C for 1 h, and then laminar flow (250 μl/m) was initiated. Biofilms of different strains were cultivated at 37°C in 2% TSB in three individual channels. The strains were transformed with the GFP plasmid for fluorescence detection, thus chloramphenicol was added to the flow cell medium to maintain plasmid selection. CLSM was performed on a Zeiss LSM710 system (Carl Zeiss, Jena, Germany) with a 20 × 0.8 n.a. apochromatic objective. Z-stacks were collected at 1 μm intervals. Confocal parameters set for WT biofilm detection were taken as standard settings. Selected confocal images stood for similar areas of interest and each confocal experiment was repeated four times. The confocal images were acquired from Zeiss ZEN 2010 software package (Carl Zeiss, Jena, Germany) and the three-dimensional biofilm images were rendered with Imaris 7.0 (Bitplane, Zurich, Switzerland). Biofilm biomass and average thickness were analysed with the COMSTAT program [48] and were indicated as the mean ± standard deviation calculated from three images obtained from a given biofilm.
Ethical statement
The use and care of mice in this study was performed strictly according to the Institutional Animal Care and Use Committee guideline of University of Science and Technology of China (USTCACUC1101053).
In vivo model of catheter-associated biofilm formation
Biofilm formation was assessed in vivo using a murine model of catheter-associated infection [49]. Briefly, male BALB/c mice (6- to 8-weeks old) were obtained from Shanghai Laboratory Animal Centre of Chinese Academy of Sciences (Shanghai, China). The mice were anaesthetised with 1% pentobarbital sodium (0.01 ml/g of body weight) and surgically dissected. Specifically, a 1-cm 18G FEP polymer catheter (Introcan, Melsungen, Germany) was implanted subcutaneously in the dorsal area of the mice. The wound was closed with surgical glue. After incubation of 24 h, 5 × 107 colony-forming units (CFU) of the test strains in a total volume of 100 μl were introduced directly into the lumen of the catheters. Mice were euthanised after 3 days of infection, and then the catheters were removed carefully and washed briefly with phosphate-buffered saline (PBS). Catheters were placed in 1 ml of sterile PBS and sonicated for 30 s to remove the adherent bacteria. The number of bacteria was determined by plating on tryptic soy agar (TSA).
Anaerobic conditions
Biofilm formation was also monitored under anaerobic conditions. The Forma Anaerobic System (Thermo, Waltham, USA) was used to provide strictly anaerobic conditions for bacterial growth and related operations. Overnight cultures were adjusted to OD600 of 6.5, and then the bacterial cultures were carried into the anaerobic system for 1:100 dilution and inoculated into 24-well plates. Resazurin, which is used as an indicator for anaerobic conditions, was added to final concentration of 0.0002% (w/v). The plates were incubated at 37°C for 4 h and OD560 was determined after crystal violet staining.
A standard anaerobic jar of 120 ml volume was used to monitor the biofilm formation of the WT strain and the mutants under anaerobic conditions. Medium and containers with thorough scavenging were prepared as follows. Water was boiled using a three-necked bottle to degas the water while nitrogen was bubbled into the bottle to keep the contents anaerobic. TSBg medium was prepared with this degassed water. Then each anaerobic jar was dispensed with 50 ml TSBg while nitrogen was gassed into the jar to drive out the oxygen. The rubber plug was quickly stuffed up following by an aluminium cap added, and then the jar containing TSBg was autoclaved at 121°C, 15 m. After preparation of the medium, biofilm formation under anaerobic conditions was examined and the operations were carried out in the anaerobic system.
Scanning electron microscopy (SEM)
Biofilm bacteria were grown on coverslips for five days, and then the coverslips were cut from the flow-cell settings and immediately fixed with 2.5% (vol/vol) glutaraldehyde in Dulbecco PBS (pH 7.2) overnight. According to the methods described previously [50], the coverslips were rinsed with PBS three times and dehydrated through an ethanol series (30%, 50%, 75%, 85% and 95%). Samples were dried and gold-palladium coated prior to SEM examination and micrographs were made with a XL-30 SEM at × 1500 to × 5000 magnification (FEI, Hillsboro, USA).
RNA isolation and real-time RT-PCR
All the bacteria used for RNA isolation to investigate the expression of genes that affect biofilm formation were those that grew statically in the 24-well plate. Bacteria in the wells of biofilm formation at different time courses (4 h, 8 h, 12 h) were collected and re-suspended in TE (Tris-EDTA) buffer (pH 8.0) containing 10 g/l lysozyme and 40 mg/l lysostaphin. After incubation at 37°C for 8 m, S. aureus cells were prepared for total RNA extraction using the Trizol method (Invitrogen), and the residual DNA was removed with RNase-free DNase I (TaKaRa). The concentration of RNA was adjusted to 100 ng/μl, and the samples were stored at −70°C. cDNA templates were synthesized from 50 ng RNA with PrimeScript™ 1st strand cDNA Synthesis Kit (TaKaRa) and gene-specific primers at 42°C for 15 m, 85°C for 5 s. Real-time PCR was performed with the cDNA and SYBR Premix Ex Taq (TaKaRa) using a StepOne Real-Time PCR System (Applied Biosystems). The quantity of cDNA measured by real-time PCR was normalised to the abundance of 16S cDNA. Real-time RT-PCR was repeated three times in triplicate parallel experiments.
Statistical analysis
The paired t test was used for statistical comparisons between groups. The level of statistical significance was set at a P value of ≤ 0.05.
Results
AI-2 inhibits biofilm formation in a concentration-dependent manner under static conditions
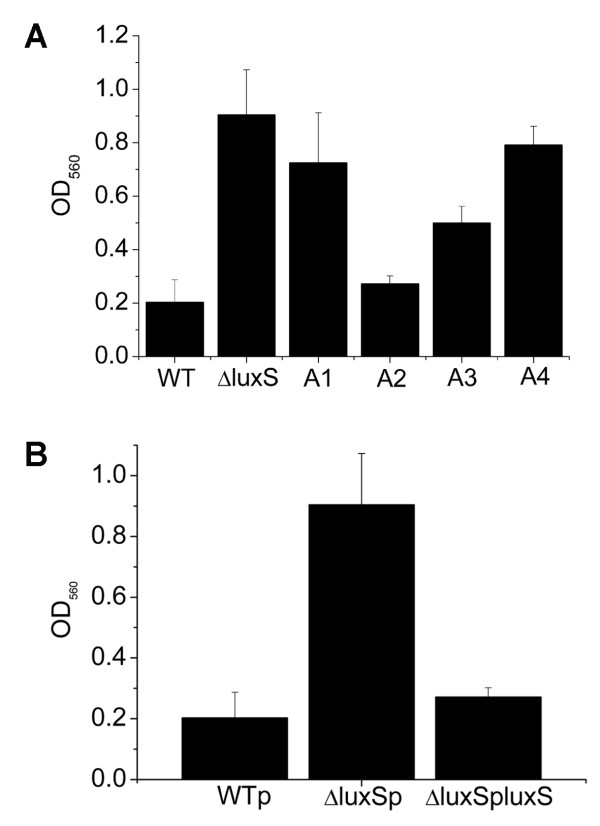
Previous studies showed that biofilm formation was influenced by the LuxS/AI-2 system both in Gram-positive and Gram-negative bacteria [32,34]. The genome of S. aureus encodes a typical luxS gene, which plays a role in the regulation of capsular polysaccharide synthesis and virulence [43]. In this study, to investigate whether LuxS/AI-2 system regulates biofilm formation in S. aureus, we monitored the biofilm formation of S. aureus WT strain RN6390B and the isogenic derivative ΔluxS strain using a microtitre plate assay. As shown in Figure 1A, the WT strain formed almost no biofilm after 4 h incubation at 37°C. However, the ΔluxS strain formed strong biofilms as measured by quantitative spectrophotometric analysis based on OD560 after crystal violet staining (Figure 1A). This discrepancy could be complemented by introducing a plasmid that contains the luxS gene (Figure 1B).
Figure 1.
Biofilm formation under static conditions and chemical complementation by DPD of different concentrations. Biofilm growth of S. aureus WT (RN6390B), ΔluxS and ΔluxS complemented with different concentrations of chemically synthesized DPD in 24-well plates for 4 h under aerobic conditions (A1: 0.39 nM, A2: 3.9 nM, A3: 39 nM, A4: 390 nM). The cells that adhered to the plate after staining with crystal violet were measured by OD560 .
The effects of LuxS could be attributed to its central metabolic function or the AI-2-mediated QS regulation, which has been reported to influence biofilm formation in some strains [32-34]. To determine if AI-2, as a QS signal, regulates biofilm formation in S. aureus, the chemically synthesized pre-AI-2 molecule DPD at concentrations from 0.39 nM to 390 nM was used to complement the ΔluxS strain. The resulting data suggested that exogenous AI-2 could decrease biofilm formation of the ΔluxS strain and the effective concentration for complementation was from 3.9 nM to 39 nM DPD (Figure 1A). As expected, these concentrations were within the range that has been reported [51]. The phenomenon that the higher concentration of AI-2 does not take effect on biofilm formation is very interesting, which has also been found in other species [51]. In the previous work [52,53], they indicated that AI-2 activity was associated with cyclic derivatives of this molecule that can be generated spontaneously. Therefore, it is possible that the concentration of effective molecules is different as the DPD concentration changes. These findings indicate that AI-2 could complement the effect of luxS mutation on biofilm formation and act in a concentration-dependent manner in S. aureus.
AI-2 inhibits biofilm formation in flow cell
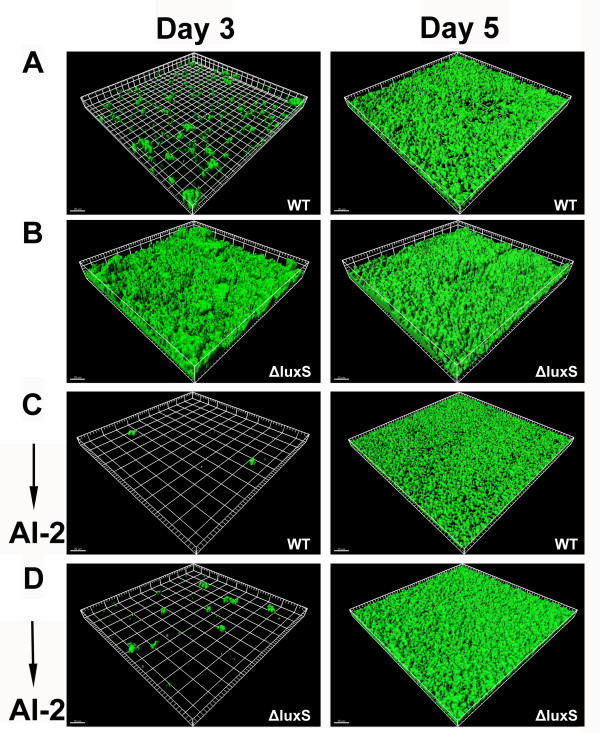
To further compare the different biofilm formation ability owing to luxS deletion, biofilm formation of WT and the ΔluxS strains was assessed using a flow-cell assay. After 3 days of incubation, biofilms produced by WT strain were undetectable as monitored by CLSM. In contrast, the ΔluxS strain began to form intact and rough biofilms. At the 5th day, the WT strain produced biofilms similar to that formed by the ΔluxS strain 2 days before; meanwhile, the ΔluxS strain formed thicker and stronger biofilms (Figure 2A and B). Analysis of the biofilms by COMSTAT is shown in Table 3. The ΔluxS strain exhibited significantly increased total biomass and average thickness of biofilms relative to those of the WT strain.
Figure 2.
Biofilm formation in flow cell and chemical complementation by DPD. Biofilms of WT (RN6390BG) and ΔluxS (ΔluxSG) were grown in a flow cell in 2% TSB with chloramphenicol (15 μg/ml). Biofilm integrity and GFP fluorescence were monitored at the 3rd day and the 5th day by CLSM. For chemical complementation, 3.9 nM DPD was added to the TSB medium at the beginning of the experiment. CLSM images are representative of two separate experiments and each grid square represents 20 μm (A) WT. (B) ΔluxS. (C) WT supplemented with DPD. (D) ΔluxS supplemented with DPD.
Table 3.
Biofilm formation of WT and ΔluxS strains
| Strains | Biofilm biomass (μm3/μm2) | Average thickness (μm) | ||
|---|---|---|---|---|
| |
Day 3 |
Day 5 |
Day 3 |
Day 5 |
| WT |
3.01 ± 0.2 |
11.71 ± 1.25 |
3.81 ± 0.35 |
11.51 ± 0.92 |
| ΔluxS |
20.16 ± 1.59* |
25.67 ± 1.16* |
20.79 ± 1.47* |
26.18 ± 0.43* |
| WT + AI-2 |
0.11 ± 0.01 |
10.44 ± 0.51 |
0.12 ± 0.01 |
9.45 ± 0.5 |
| ΔluxS + AI-2 | 0.49 ± 0.018 | 14.31 ± 0.59 | 0.59 ± 0.06 | 13.53 ± 0.5 |
* Significantly different results compared with WT (P < 0.01).
In the flow-cell assay, 3.9 nM DPD was added to the culture medium at the beginning of the experiment. As expected, examination with CLSM showed that the ΔluxS strain complemented with 3.9 nM DPD did not produce biofilms after 3 days of growth in the flow cell, and formed biofilms similar to that of the WT strain at the 5th day (Figure 2C and D). As shown in Table 3, they both formed ~10-μm thick biofilms until the 5th day. These results suggest that AI-2 supplementation decreases biofilm formation under flow conditions.
Inactivation of luxS results in increased biofilm formation in vivo
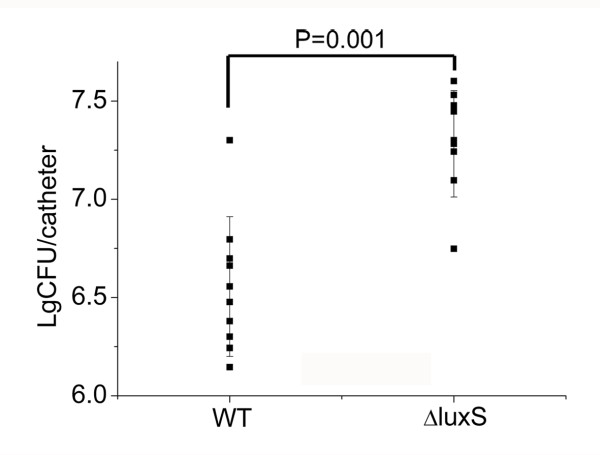
To further verify the effect of AI-2 on biofilm formation in vivo, a murine model of catheter-associated biofilm formation was used. In this assay, mice were separately infected with 5 × 107 CFU/ml of the WT strain and the ΔluxS strain. After incubation for 3 days, the catheters were taken out and the number of bacteria was counted. As shown in Figure 3, the ΔluxS strain exhibited significantly increased capacity to form biofilms compared to the WT strain (P = 0.001) in vivo. These results suggest that LuxS/AI-2 system is involved in the regulation of biofilm formation in vivo, which is consistent with the conclusion in vitro.
Figure 3.
Biofilm formation of S. aureus in vivo. Biofilm formation was assessed using a murine catheter-associated model of WT (NCTC8325) and ΔluxS (NCTC8325ΔluxS). Overnight culture of 5 × 107 CFU was injected into the catheters, which were implanted subcutaneously in the dorsal area of the mice. Results shown are the number of bacteria counted from the catheters after incubation for 3 days. Each point stands for one independent mouse. P value refers to a comparison between WT and ΔluxS and means statistically significant differences (P = 0.001) by Student's t test.
AI-2 represses the transcription of icaA via the activation of icaR
PIA is considered to be a major factor determining biofilm formation in some bacteria [10,54,55]. To test if AI-2-mediated biofilm reduction is due to a change in PIA expression, the transcription of icaA was examined using real-time RT-PCR with RNA isolated from biofilm bacteria at different time points. Transcription of icaA reached its peak at 4 h of biofilm formation and the maximum difference between the WT strain and the ΔluxS strain was also highlighted at this time (data not shown). Thus, RNA was isolated from 4 h biofilm bacteria of the WT strain, the ΔluxS strain, and the ΔluxS strain complemented with 3.9 nM DPD. Expression of icaA was examined using real-time RT-PCR. The resulting data showed that expression of icaA was elevated in the ΔluxS strain, and it could be complemented by 3.9 nM DPD (Figure 4A). As expected, corresponding to the biofilm formation in Figure 1, thicker biofilms were presented owing to the luxS mutation while the bacteria within the biofilms also displayed elevated icaA transcription. Moreover, we examined the expression of several main adhesion molecules. As shown in Additional file 1: Figure S1, there were no obvious differences between the WT, ΔluxS and ΔluxS transformed with the pLIluxS plasmid for complementation (ΔluxSpluxS). Here, the WT and ΔluxS strains were also transformed with an empty PLI50 plasmid constructing the WTp strain and ΔluxSp strain, which were used as the control. Besides, we added sodium-metapeiodate into the well-developed biofilms and found that biofilms dispersed after 2 h incubation at 37°C. Taken together, our results suggest that PIA is the main factor controlled by AI-2 in the regulation of biofilm formation in S. aureus.
Figure 4.
Transcriptional regulation of icaA and icaR by AI-2. Real-time RT-PCR of icaA and icaR transcription was measured. The bacteria used for RNA extraction were those that were incubated at 4 h for biofilm formation under aerobic conditions. Error bars indicate the variation between triplicate samples within the real-time RT-PCR. The relative cDNA abundance of the WT sample was assigned a value of 1. (A) Relative transcript levels of icaA of WT (RN6390B), ΔluxS and ΔluxS complemented with 3.9 nM DPD under aerobic conditions. (B) Relative transcript levels of icaR of WT (RN6390B), ΔluxS and ΔluxS complemented with 3.9 nM DPD under aerobic conditions.
It was reported that IcaR is a negative regulator of the icaA locus [19], and that icaR could be regulated by Rbf, SarA and SigB [56,57]. However, few studies indicate that the signalling molecule AI-2 could be an activator of icaR. We therefore investigated whether repression of icaA by AI-2 was mediated by IcaR by examining the icaR transcription in the biofilm bacteria of the WT strain, the ΔluxS strain and the ΔluxS strain complemented with 3.9 nM DPD. We found that the ΔluxS strain displayed decreased transcription of icaR compared to WT, and DPD supplementation could complement the effect of luxS mutation (Figure 4B). These data indicate that the repression of icaADBC transcription by AI-2 is through the activation of icaR. These results allow us to conclude that AI-2 activates icaR, which results in decreased icaADBC transcription and subsequently decreased biofilm formation.
AI-2 inhibits biofilm formation and represses the transcription of icaA under anaerobic conditions
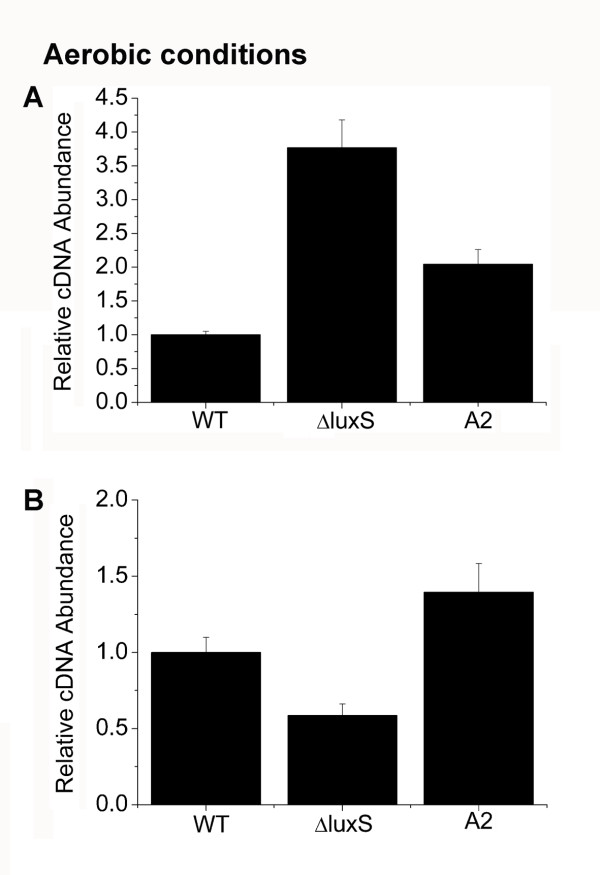
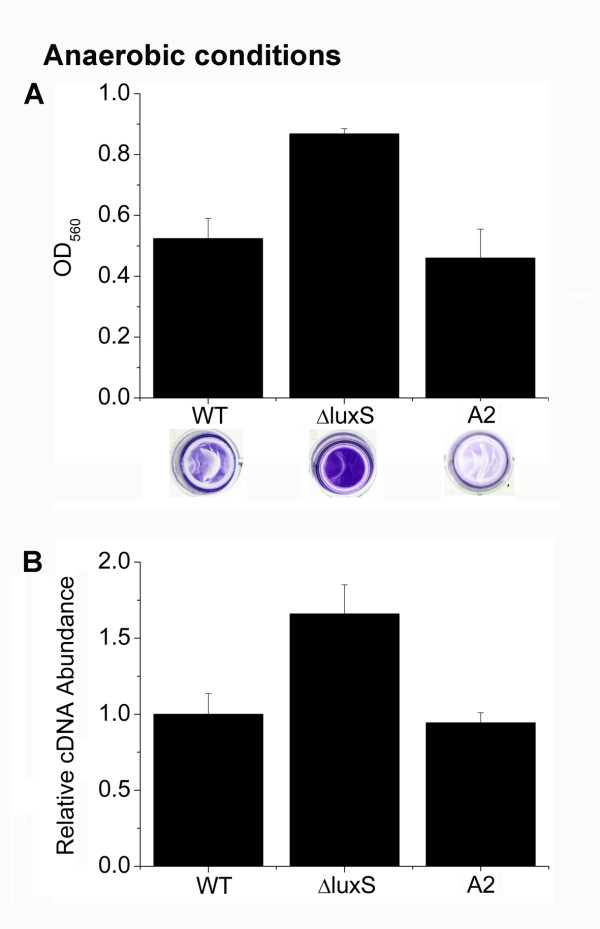
Hypoxia or anaerobic conditions is a common hostile environment that the biofilm bacteria suffer in vivo[3,58,59]. To determine whether or not AI-2 could also affect biofilm formation under anaerobic conditions, the microtitre plate assay was used to examine the biofilm growth. After incubation of the plate for 4 h under anaerobic conditions, we found that the ΔluxS strain displayed increased biofilm formation compared to the WT strain, and AI-2 supplementation restored the WT phenotype (Figure 5A). Consistently, AI-2 repressed the transcription of icaA under anaerobic conditions (Figure 5B).
Figure 5.
Analysis of biofilm formation and the icaA transcription under anaerobic conditions. (A) Biofilm formation of WT (RN6390B), ΔluxS and ΔluxS complemented with 3.9 nM DPD under anaerobic conditions. (B) Relative transcript levels of icaA of WT (RN6390B), ΔluxS and ΔluxS complemented with 3.9 nM DPD under anaerobic conditions.
The LuxS/AI-2 QS system and the agr-mediated QS system have a cumulative effect on the regulation of biofilm formation
It was reported that the agr QS system mediates biofilm dispersal in S. aureus[60]. To determine whether the LuxS/AI-2 QS system and the agr-mediated QS system have a cumulative or complementary effect on the regulation of biofilm formation, we constructed a Δagr ΔluxS strain and compared the biofilm formation among the WT strain and the mutants using different assays, including the microtitre plate assay, flow cell, anaerobic jar and SEM. Consistently, we found that the Δagr ΔluxS strain displayed the strongest capacity for biofilm formation among all the stains we investigated.
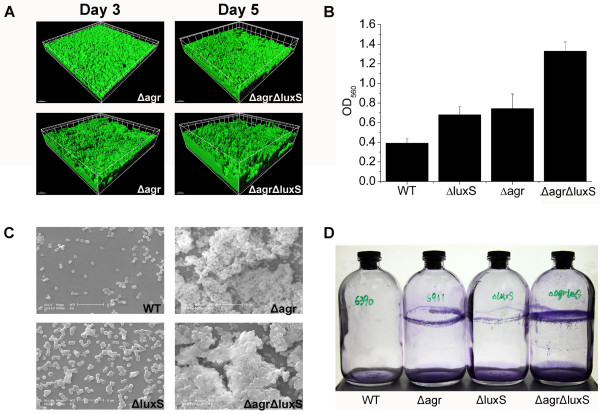
In the flow-cell assay, as shown in Figure 6A, the Δagr ΔluxS strain formed stronger biofilms than RN6911, as shown by CLSM, indicating that mutation of luxS indeed influences biofilm formation and that the two systems seem to play a cumulative effect. Moreover, similar results were obtained in the microtitre plate assay and the anaerobic jar assay under anaerobic conditions (Figure 6B and D).
Figure 6.
Additive effect played by the LuxS/AI-2 QS system and the agr-mediated QS system. (A) The ΔagrΔluxSG and RN6911G grew biofilms in the flow cell, and the representative images were measured by CLSM at the 3rd and 5th day of biofilm formation. Strains are indicated in the figure. (B) Overnight cultures of WT (RN6390B), Δagr (RN6911), ΔluxS and Δagr ΔluxS were inoculated in 24-well plate and formed biofilms under anaerobic conditions. (C) WT, Δagr, ΔluxS and Δagr ΔluxS formed 5 days biofilms in a flow cell on the upper surface of the coverslips, which were cut and examined by scanning electron microscopy. (D) The anaerobic jar was used for monitoring the biofilm formation of the WT, Δagr, ΔluxS and Δagr ΔluxS, OD560 was measured after crystal violet staining.
To accurately describe the distinct biofilm formation resulting from luxS deletion, SEM was used for evaluating the structure and surface appearance of the mature biofilm. Therefore, the coverslips of the flow-cell chamber on which 5 days biofilms of WT and the ΔluxS strain grew were cut out. SEM analysis showed that the ΔluxS strain produced a compact biofilm structure with increased coverage than that of the WT strain (Figure 6C). On closer inspection, we found that the ΔluxS strain displayed stronger intercellular adhesion and this was also reflected in the Δagr ΔluxS strain. The Δagr ΔluxS strain showed stronger intercellular adhesion ability than RN6911 (Figure 6C), indicating a possible result of elevated expression of PIA. Interestingly, microscopic analysis of the biofilm structure revealed that the agr mutation led to biofilms that adopted a "ridged" appearance with many channels, rather than the relatively smooth, confluent layer normally detected in the WT and ΔluxS strains, presumably because the thicker biofilms with a dense compact structure restrict the growth of bacteria inside. Based on these results, we speculate that the LuxS/AI-2 QS system and the agr-mediated QS system play a cumulative effect on the regulation of biofilm formation in S. aureus. It has been reported that induction of the agr system in established S. aureus biofilms detaches cells in an ica-independent manner and they also demonstrate that the dispersal mechanism requires extracellular protease activity [60]. Therefore, it seems that the influences of the LuxS/AI-2 QS system and the agr-mediated QS system on biofilm formation are through different pathways in S. aureus.
Discussion
Most previous studies of biofilm formation have been performed under one or two conditions to present this phenotype. However, biofilm is a kind of "smart community" that seems able to cope with different environments. Therefore, a single condition may lead to misunderstanding regarding the elaborate function of a specific gene. To provide sufficient and rigorous evidence, we demonstrate that the LuxS/AI-2 system is involved in the regulation of biofilm formation under different conditions. In contrast to the most commonly used microtitre plate assay, flow cell is increasingly used for detecting biofilm growth, of which the dynamic three-dimensional image could be monitored by CLSM dynamically. This setting simulates the environment of flowing surfaces in vivo, such as some interfaces between body fluids and implants. The result under this condition may offer more accurate information about flow surroundings in vivo. In addition, we also investigated biofilm formation under anaerobic conditions, which the biofilm bacteria undergo. The oxygen partial pressure of air-equilibrated medium is high in vitro, whereas hypoxic environments may prevail in body implants and human tissues distant from arterial blood flow [58,61]. Moreover, most locations in which the biofilm bacteria accumulate are usually niches of local low oxygen because of inflammatory cell aggregation [59,62].
The mouse model was used here to compare biofilm formation of the WT and the ΔluxS strains and our data were consistent with the in vitro data. Nevertheless, there are few studies regarding AI-2 complementation in the mouse model to date, and the specific mechanism of these signal molecules in vivo remains elusive. In general, we offer consistent results under different conditions to emphasise that the conclusion drawn is consistent and worthy of reference in most cases.
LuxS and AI-2 affect biofilm development, whereas the results may be different in the same strain due to various factors. Previous work has shown that AI-2 was produced in rich medium under aerobic and anaerobic conditions peaking during the transition to stationary phase, but cultures retained considerable AI-2 activity after entry into the stationary phase under anaerobic conditions. In addition, the S. aureus RN6390BΔluxS strain showed reduction in biofilm formation in TSB containing 1% glucose and 3% sodium chloride under anaerobic conditions [42]. However, in our study, analysis of biofilm growth in vitro and in vivo led to the conclusion that the ΔluxS strain exhibited increased biofilm formation compared to the WT strain. Importantly, the luxS mutation could be complemented by chemically synthesized DPD, indicating the effect of AI-2-mediated QS on biofilm formation in S. aureus. Hardie and Heurlier [41] summarised six main factors that influence the experimental results for doing research on the LuxS/AI-2 system: experimental design; genetic complementation; chemical complementation; conditioned supernatant complementation; and complementation with molecules linked to AI-2 production and that independent of luxS status. With detailed analysis, we found that the inconsistency of the results is in part owing to the different growth medium provided to the biofilm bacteria, especially the different concentrations of glucose and sodium chloride, which are both important factors enhancing biofilm formation [63].
In addition to the present evidence of AI-2-regulated biofilm formation in S. aureus, we found that the transcription of icaR is activated by AI-2, which is barely reported, although we have not yet identified the mechanism of the interaction between them. This is partly because the detailed mechanism of transport and action of AI-2 has only been described in several strains and different mechanisms are applied to different species, although AI-2 has been proven to act as a signalling molecule that could regulate series of gene expression. The first mechanism revealed was in Vibrio harveyi, which responds to AI-2 by initiating a phosphorylation cascade [64]. In Salmonella typhimurium[65] and E. coli[66,67], AI-2 seems to be taken up by an ABC transporter. However, the mechanism of AI-2 transport and functional performing in Staphylococci was still unknown. Therefore, the detailed mechanism through which AI-2 functions in S. aureus should be highlighted here, and the interaction between AI-2 and IcaR requires further study.
In addition to PIA, we do not observe any obvious increase of extracellular protein (Additional file 2: Figure S2) or bacterial autolysis in the ΔluxS strain (Additional file 3: Figure S3). Our results showed no significant differences in the transcriptional levels of several main adhesion molecules. Moreover, previous work indicated that S. aureus strains 8325-4 and RN4220 formed PIA-dependent biofilms [68]. We thus propose that AI-2 signalling represses the icaA expression, and subsequently leads to decreased biofilm formation in S. aureus.
More and more studies concerning multispecies biofilms gradually uncover the mechanisms of the interaction and communication of the different species inside the biofilms. One of the most popular approaches of the signalling regulation is directed towards the AI-2-controlled QS system for its extensive use of interspecies. The plaque biofilms on tooth surfaces consist of various oral bacteria including S. aureus and involve complex microbial interactions [69-71]. There is evidence that AI-2-mediated QS may play a significant role in oral biofilm formation [50]. As reported by others, airway infections by Pseudomonas aeruginosa afflicting patients with cystic fibrosis (CF) are among the most enigmatic of biofilm diseases [2]. S. aureus is also found to be a major pathogen associated with P. aeruginosa in CF lung infection [72]. Previous work has shown that PIA is expressed in lungs infected with S. aureus, whereas CP8 is not expressed because of limited oxygen [73]. Here, we provide evidence that AI-2 can regulate icaA expression under anaerobic conditions, suggesting a potential role of AI-2 in influencing S. aureus infection in lungs. However, few studies about biofilm formation cooperated by S. aureus and the other species are reported. Therefore, could S. aureus and the other species in their focus areas form multispecies biofilms? Could AI-2 play an important role in this process? It is interesting to discuss the actual complex-flora interaction in human and social behaviour of the bacteria. Therefore, revelation of the AI-2-regulated biofilm formation in S. aureus possesses instructive meaning for these related studies.
Conclusions
These findings demonstrate that AI-2 can decrease biofilm formation in S. aureus via an icaR-activation pathway. This study may provide clues for therapy in S. aureus biofilm-associated infection.
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
DY carried out the experiments and performed the data analyses. BS, ZL, and TX contributed to the design and coordination of the experiments. DY wrote the manuscript. BS, TX and ZL participated in editing the manuscript. All authors have read and approved the manuscript.
Supplementary Material
Relative transcript levels of several adhesions. The levels of transcription of these genes including map, fnbA, fnbB, clfB, efb were measured by real-time RT-PCR in S. aureus WTp, ΔluxSp and ΔluxS complemented with a plasmid containing luxS gene for genetic complementation (ΔluxSpluxS). As the control, WT and ΔluxS were transformed with empty plasmid PLI50, constructing WTp and ΔluxSp.
Extracellular protein loaded on SDS-PAGE. The levels of extracellular-protein expression of biofilm bacteria, which were incubated at 37°C for 4 h and 24 h, were measured.
Triton X-100-stimulated autolysis. The autolysis of WT, ΔluxS and ΔluxSpluxS induced in 0.05 M Tris–HCl buffer containing 0.05% (vol/vol) Triton X-100 were measured.
Contributor Information
Dan Yu, Email: yudanaz@mail.ustc.edu.cn.
Liping Zhao, Email: zhaolp@mail.ustc.edu.cn.
Ting Xue, Email: xueting@ustc.edu.cn.
Baolin Sun, Email: sunb@ustc.edu.cn.
Acknowledgments
We thank our colleagues X. Zhang, Y. Bao for their kind help with the experiments, and X. Wu, Z.B Liu for their technical assistance of the CLSM detection in the Experimental Centre of Life Science of University of Science and Technology of China. We thank the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for providing the bacterial strains.
This study was supported by the National Natural Science Foundation of China (30970118, 31021061).
References
- Harris LG, Richards RG. Staphylococci and implant surfaces: a review. Injury. 2006;37(Suppl 2):S3–S14. doi: 10.1016/j.injury.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu Rev Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- Cooper R, Okhiria O. Biofilms, wound infection and the issue of control. Wounds UK. 2006;2(3):48–56. [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–228. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci USA. 2007;104(19):8113–8118. doi: 10.1073/pnas.0610226104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74(2):470–476. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C, Gerke C, Perdreau-Remington F, Gotz F. Characterization of Tn917 insertion mutants of Staphylococcus epidermidis affected in biofilm formation. Infect Immun. 1996;64(1):277–282. doi: 10.1128/iai.64.1.277-282.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C, Gotz F. Further characterization of Staphylococcus epidermidis transposon mutants deficient in primary attachment or intercellular adhesion. Zentralbl Bakteriol. 1998;287(1–2):69–83. doi: 10.1016/s0934-8840(98)80149-7. [DOI] [PubMed] [Google Scholar]
- Mack D, Fischer W, Krokotsch A, Leopold K, Hartmann R, Egge H, Laufs R. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear beta-1,6-linked glucosaminoglycan: purification and structural analysis. J Bacteriol. 1996;178(1):175–183. doi: 10.1128/jb.178.1.175-183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Gotz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20(5):1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- Gerke C, Kraft A, Sussmuth R, Schweitzer O, Gotz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273(29):18586–18593. doi: 10.1074/jbc.273.29.18586. [DOI] [PubMed] [Google Scholar]
- Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67(10):5427–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack D, Siemssen N, Laufs R. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect Immun. 1992;60(5):2048–2057. doi: 10.1128/iai.60.5.2048-2057.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell IM, Crozier DN, Pawagi AB, Buivids IA. In vitro response of Staphylococcus aureus from cystic fibrosis patients to combinations of linoleic and oleic acids added to nutrient medium. J Clin Microbiol. 1983;18(2):408–415. doi: 10.1128/jcm.18.2.408-415.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjelm E, Lundell-Etherden I. Slime production by Staphylococcus saprophyticus. Infect Immun. 1991;59(1):445–448. doi: 10.1128/iai.59.1.445-448.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramton SE, Ulrich M, Gotz F, Doring G. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect Immun. 2001;69(6):4079–4085. doi: 10.1128/IAI.69.6.4079-4085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deighton M, Borland R. Regulation of slime production in Staphylococcus epidermidis by iron limitation. Infect Immun. 1993;61(10):4473–4479. doi: 10.1128/iai.61.10.4473-4479.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson KK, Pier DB, Goldmann DA, Pier GB. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J Bacteriol. 2004;186(8):2449–2456. doi: 10.1128/JB.186.8.2449-2456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Li H, Vuong C, Vadyvaloo V, Wang J, Yao Y, Otto M, Gao Q. Role of the luxS quorum-sensing system in biofilm formation and virulence of Staphylococcus epidermidis. Infect Immun. 2006;74(1):488–496. doi: 10.1128/IAI.74.1.488-496.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler BL. How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr Opin Microbiol. 1999;2(6):582–587. doi: 10.1016/S1369-5274(99)00025-9. [DOI] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- De Keersmaecker SC, Sonck K, Vanderleyden J. Let LuxS speak up in AI-2 signaling. Trends Microbiol. 2006;14(3):114–119. doi: 10.1016/j.tim.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96(4):1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauder S, Shokat K, Surette MG, Bassler BL. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol Microbiol. 2001;41(2):463–476. doi: 10.1046/j.1365-2958.2001.02532.x. [DOI] [PubMed] [Google Scholar]
- Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature. 2002;415(6871):545–549. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- Miller ST, Xavier KB, Campagna SR, Taga ME, Semmelhack MF, Bassler BL, Hughson FM. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol Cell. 2004;15(5):677–687. doi: 10.1016/j.molcel.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Lupp C, Ruby EG. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J Bacteriol. 2004;186(12):3873–3881. doi: 10.1128/JB.186.12.3873-3881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rader BA, Campagna SR, Semmelhack MF, Bassler BL, Guillemin K. The quorum-sensing molecule autoinducer 2 regulates motility and flagellar morphogenesis in Helicobacter pylori. J Bacteriol. 2007;189(17):6109–6117. doi: 10.1128/JB.00246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal T, Jesudhasan P, Pillai S, Wood TK, Jayaraman A. Temporal regulation of enterohemorrhagic Escherichia coli virulence mediated by autoinducer-2. Appl Microbiol Biotechnol. 2008;78(5):811–819. doi: 10.1007/s00253-008-1359-8. [DOI] [PubMed] [Google Scholar]
- Gonzalez Barrios AF, Zuo R, Hashimoto Y, Yang L, Bentley WE, Wood TK. Autoinducer 2 controls biofilm formation in Escherichia coli through a novel motility quorum-sensing regulator (MqsR, B3022) J Bacteriol. 2006;188(1):305–316. doi: 10.1128/JB.188.1.305-316.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Lamont RJ, Demuth DR. Autoinducer 2 is required for biofilm growth of Aggregatibacter (Actinobacillus) actinomycetemcomitans. Infect Immun. 2007;75(9):4211–4218. doi: 10.1128/IAI.00402-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal JE, Ludewick HP, Kunkel RM, Zahner D, Klugman KP. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect Immun. 2011;79(10):4050–4060. doi: 10.1128/IAI.05186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auger S, Krin E, Aymerich S, Gohar M. Autoinducer 2 affects biofilm formation by Bacillus cereus. Appl Environ Microbiol. 2006;72(1):937–941. doi: 10.1128/AEM.72.1.937-941.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock GW, Ghazally S, Walter J, Loach D, Brooks H, Cook G, Surette M, Simmers C, Bremer P, Dal Bello F. et al. Ecological behavior of Lactobacillus reuteri 100–23 is affected by mutation of the luxS gene. Appl Environ Microbiol. 2005;71(12):8419–8425. doi: 10.1128/AEM.71.12.8419-8425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challan Belval S, Gal L, Margiewes S, Garmyn D, Piveteau P, Guzzo J. Assessment of the roles of LuxS, S-ribosyl homocysteine, and autoinducer 2 in cell attachment during biofilm formation by Listeria monocytogenes EGD-e. Appl Environ Microbiol. 2006;72(4):2644–2650. doi: 10.1128/AEM.72.4.2644-2650.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeer S, De Keersmaecker SC, Verhoeven TL, Fadda AA, Marchal K, Vanderleyden J. Functional analysis of luxS in the probiotic strain Lactobacillus rhamnosus GG reveals a central metabolic role important for growth and biofilm formation. J Bacteriol. 2007;189(3):860–871. doi: 10.1128/JB.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Learman DR, Yi H, Brown SD, Martin SL, Geesey GG, Stevens AM, Hochella MF Jr. Involvement of Shewanella oneidensis MR-1 LuxS in biofilm development and sulfur metabolism. Appl Environ Microbiol. 2009;75(5):1301–1307. doi: 10.1128/AEM.01393-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker SCJ, Varszegi C, van Boxel N, Habel LW, Metzger K, Daniels R, Marchal K, De Vos D, Vanderleyden J. Chemical synthesis of (S)-4,5-dihydroxy-2,3-pentanedione, a bacterial signal molecule precursor, and validation of its activity in Salmonella typhimurium. J Biol Chem. 2005;280(20):19563–19568. doi: 10.1074/jbc.M412660200. [DOI] [PubMed] [Google Scholar]
- Li L, Xu Z, Zhou Y, Li T, Sun L, Chen H, Zhou R. Analysis on Actinobacillus pleuropneumoniae LuxS regulated genes reveals pleiotropic roles of LuxS/AI-2 on biofilm formation, adhesion ability and iron metabolism. Microb Pathog. 2011;50(6):293–302. doi: 10.1016/j.micpath.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Hardie KR, Heurlier K. Establishing bacterial communities by 'word of mouth': LuxS and autoinducer 2 in biofilm development. Nat Rev Microbiol. 2008;6(8):635–643. doi: 10.1038/nrmicro1916. [DOI] [PubMed] [Google Scholar]
- Doherty N, Holden MT, Qazi SN, Williams P, Winzer K. Functional analysis of luxS in Staphylococcus aureus reveals a role in metabolism but not quorum sensing. J Bacteriol. 2006;188(8):2885–2897. doi: 10.1128/JB.188.8.2885-2897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Xue T, Shang F, Sun H, Sun B. Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect Immun. 2010;78(8):3506–3515. doi: 10.1128/IAI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehl R, Al-Bataineh S, Gordon O, Luginbuehl R, Otto M, Textor M, Landmann R. Furanone at subinhibitory concentrations enhances staphylococcal biofilm formation by luxS repression. Antimicrob Agents Chemother. 2009;53(10):4159–4166. doi: 10.1128/AAC.01704-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner R. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol Lett. 1997;151(1):1–8. doi: 10.1016/S0378-1097(97)00116-X. [DOI] [PubMed] [Google Scholar]
- Beenken KE, Blevins JS, Smeltzer MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun. 2003;71(7):4206–4211. doi: 10.1128/IAI.71.7.4206-4211.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood JM, Bartels DJ, Volper EM, Greenberg EP. Quorum sensing in Staphylococcus aureus biofilms. J Bacteriol. 2004;186(6):1838–1850. doi: 10.1128/JB.186.6.1838-1850.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersboll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146(Pt 10):2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186(14):4665–4684. doi: 10.1128/JB.186.14.4665-4684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida A, Ansai T, Takehara T, Kuramitsu HK. LuxS-based signaling affects Streptococcus mutans biofilm formation. Appl Environ Microbiol. 2005;71(5):2372–2380. doi: 10.1128/AEM.71.5.2372-2380.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickard AH, Palmer RJ, Blehert DS, Campagna SR, Semmelhack MF, Egland PG, Bassler BL, Kolenbrander PE. Autoinducer 2: a concentration-dependent signal for mutualistic bacterial biofilm growth. Mol Microbiol. 2006;60(6):1446–1456. doi: 10.1111/j.1365-2958.2006.05202.x. [DOI] [PubMed] [Google Scholar]
- Feather MS. Amine-assisted sugar dehydration reactions. Prog Food Nutr Sci. 1981;5:37–45. [Google Scholar]
- Nedvidek W, Ledl F, Fischer P. Detection of 5-hydroxymethyl-1-2-methyl-3(2H)-furanone and of α-dicarbonyl compounds in reaction mixtures of hexoses and pentoses with different amines. Z Lebensm UntersForsch. 1992;194:222–228. doi: 10.1007/BF01198411. [DOI] [Google Scholar]
- Gotz F. Staphylococcus and biofilms. Mol Microbiol. 2002;43(6):1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- Mack D, Haeder M, Siemssen N, Laufs R. Association of biofilm production of coagulase-negative staphylococci with expression of a specific polysaccharide intercellular adhesin. J Infect Dis. 1996;174(4):881–884. doi: 10.1093/infdis/174.4.881. [DOI] [PubMed] [Google Scholar]
- Cue D, Lei MG, Luong TT, Kuechenmeister L, Dunman PM, O'Donnell S, Rowe S, O'Gara JP, Lee CY. Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J Bacteriol. 2009;191(20):6363–6373. doi: 10.1128/JB.00913-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerca N, Brooks JL, Jefferson KK. Regulation of the intercellular adhesin locus regulator (icaR) by SarA, sigmaB, and IcaR in Staphylococcus aureus. J Bacteriol. 2008;190(19):6530–6533. doi: 10.1128/JB.00482-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman G, Garbutt IT, Demnitz U. Ability of a Staphylococcus aureus isolate from a chronic osteomyelitic lesion to survive in the absence of air. Eur J Clin Microbiol. 1983;2(6):595–597. doi: 10.1007/BF02016574. [DOI] [PubMed] [Google Scholar]
- Simmen HP, Blaser J. Analysis of pH and pO2 in abscesses, peritoneal fluid, and drainage fluid in the presence or absence of bacterial infection during and after abdominal surgery. Am J Surg. 1993;166(1):24–27. doi: 10.1016/S0002-9610(05)80576-8. [DOI] [PubMed] [Google Scholar]
- Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4(4):e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst JF, Tielker D. Responses to hypoxia in fungal pathogens. Cell Microbiol. 2009;11(2):183–190. doi: 10.1111/j.1462-5822.2008.01259.x. [DOI] [PubMed] [Google Scholar]
- McGovern NN, Cowburn AS, Porter L, Walmsley SR, Summers C, Thompson AA, Anwar S, Willcocks LC, Whyte MK, Condliffe AM. et al. Hypoxia selectively inhibits respiratory burst activity and killing of Staphylococcus aureus in human neutrophils. J Immunol. 2011;186(1):453–463. doi: 10.4049/jimmunol.1002213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretro T, Hermansen L, Holck AL, Sidhu MS, Rudi K, Langsrud S. Biofilm formation and the presence of the intercellular adhesion locus ica among staphylococci from food and food processing environments. Appl Environ Microbiol. 2003;69(9):5648–5655. doi: 10.1128/AEM.69.9.5648-5655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler BL, Wright M, Silverman MR. Sequence and function of LuxO, a negative regulator of luminescence in Vibrio harveyi. Mol Microbiol. 1994;12(3):403–412. doi: 10.1111/j.1365-2958.1994.tb01029.x. [DOI] [PubMed] [Google Scholar]
- Taga ME, Miller ST, Bassler BL. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol Microbiol. 2003;50(4):1411–1427. doi: 10.1046/j.1365-2958.2003.03781.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Hashimoto Y, Tsao CY, Valdes JJ, Bentley WE. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J Bacteriol. 2005;187(6):2066–2076. doi: 10.1128/JB.187.6.2066-2076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J Bacteriol. 2005;187(1):238–248. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill E, Pozzi C, Houston P, Smyth D, Humphreys H, Robinson DA, O'Gara JP. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol. 2007;45(5):1379–1388. doi: 10.1128/JCM.02280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ Jr. Communication among oral bacteria. Microbiol Mol Biol Rev. 2002;66(3):486–505. doi: 10.1128/MMBR.66.3.486-505.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didilescu AC, Skaug N, Marica C, Didilescu C. Respiratory pathogens in dental plaque of hospitalized patients with chronic lung diseases. Clin Oral Investig. 2005;9(3):141–147. doi: 10.1007/s00784-005-0315-6. [DOI] [PubMed] [Google Scholar]
- Sumi Y, Miura H, Michiwaki Y, Nagaosa S, Nagaya M. Colonization of dental plaque by respiratory pathogens in dependent elderly. Arch Gerontol Geriatr. 2007;44(2):119–124. doi: 10.1016/j.archger.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Govan JR. Infection control in cystic fibrosis: methicillin-resistant Staphylococcus aureus, Pseudomonas aeruginosa and the Burkholderia cepacia complex. J R Soc Med. 2000;93(Suppl 38):40–45. [PMC free article] [PubMed] [Google Scholar]
- McKenney D, Pouliot KL, Wang Y, Murthy V, Ulrich M, Doring G, Lee JC, Goldmann DA, Pier GB. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science. 1999;284(5419):1523–1527. doi: 10.1126/science.284.5419.1523. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative transcript levels of several adhesions. The levels of transcription of these genes including map, fnbA, fnbB, clfB, efb were measured by real-time RT-PCR in S. aureus WTp, ΔluxSp and ΔluxS complemented with a plasmid containing luxS gene for genetic complementation (ΔluxSpluxS). As the control, WT and ΔluxS were transformed with empty plasmid PLI50, constructing WTp and ΔluxSp.
Extracellular protein loaded on SDS-PAGE. The levels of extracellular-protein expression of biofilm bacteria, which were incubated at 37°C for 4 h and 24 h, were measured.
Triton X-100-stimulated autolysis. The autolysis of WT, ΔluxS and ΔluxSpluxS induced in 0.05 M Tris–HCl buffer containing 0.05% (vol/vol) Triton X-100 were measured.