Defining clear hepatitis C virus (HCV) infection outcomes, including reinfection and viral intercalation after clearance of infection, requires ongoing, frequent follow-up, most importantly with longitudinal viral sequencing. Patients who have cleared HCV infection may demonstrate sustained viral clearance despite ongoing HCV exposure.
Keywords: hepatitis C virus, viral sequencing, reinfection, intercalation, young IDU
Abstract
Background. Detection of hepatitis C virus (HCV) reinfection and intercalation (ie, intermittent recurrent bouts of viremia with homologous virus interspersed with aviremic periods) requires extensive and frequent evaluation and viral sequencing.
Methods. HCV infection outcomes were studied prospectively in active injection drug users with recurrent HCV RNA–positive tests after serial negative results. HCV viremia and viral sequences (Core/E1) were assessed from monthly blood samples.
Results. Viral clearance, reinfection, and intercalating infection were all detected. Among 44 participants with apparently resolved HCV (26 incident HCV clearers and 18 enrolled with already resolved infection), 36 (82%) remained persistently HCV RNA negative, but 8 demonstrated intermittent recurrent viremia. Four of these (50%) had confirmed reinfection with a heterologous virus; 3 demonstrated viral intercalation, and 1 was not classifiable as either. Estimated incidence of first reinfection was 5.4 per 100 person-years (95% confidence interval, 2.0–14.5). Six (75%) participants, including 3 of 4 with reinfection, demonstrated sustained viral clearance for a median of 26 months since last HCV RNA test.
Conclusions. These results show that frequent monitoring and viral sequencing are required to correctly assess HCV outcomes and estimate incidence of reinfection (which was previously overestimated). Sustained clearance may take many months and occur after episodes of reinfection and viral intercalation. Three of 4 subjects who had confirmed reinfection showed evidence of long-term clearance. Viral intercalation occurs with significant frequency. Further studies of these events, especially immunological, are needed to inform HCV clinical care and vaccine development.
Hepatitis C virus (HCV) infection is spontaneously cleared in 15%–40% of seropositive individuals, depending on factors including sex, race, and genetics [1–3]. Reinfection after clearance of infection has been documented in several case studies [4–8], and prospective cohorts of active injection drug users (IDUs) have provided data showing frequency of these events [1, 9–16]. Significantly, some of these studies have provided compelling evidence that people who have spontaneously resolved HCV exhibit signs of protection against chronic HCV after reinfection, including frequent reclearance of the virus, lower concentration of HCV RNA during reinfection, and shorter durations of infection prior to reclearance [9, 11]. With respect to the mechanisms of such protection, Osburn et al [9] analyzed immune responses following repeated cycles of HCV infection and clearance, and identified increased breadth of both adaptive and humoral responses in reinfected compared with chronically infected individuals. Both new T-cell responses and a higher prevalence of broadly reactive antibodies provide strong evidence for protective immunity preventing progression to chronic infection following repeated HCV exposure, leading many to deduce that understanding and inducing these responses may provide a key vaccination strategy [17].
We have conducted further analyses and follow-up of clearance, reinfection, and reclearance events among 44 young IDUs who resolved HCV, many described formerly [1]. Previously, based on 7 participants with recurrent intermittent viremic events and 28.5 person-years (py) of follow-up, estimated incidence of first reinfection (24.6 per 100 py [95% confidence interval {CI}, 11.7–51.6]) was equivalent to the incidence of primary infection (26.7 per 100 py [95% CI, 21.5–31.6]) [1]. We have continued prospective observation of these cases, and others who resolved infection. We also conducted sequence analysis of HCV RNA from stored blood samples of all those demonstrating intermittent viremia to phylogenetically distinguish reinfection from recurrent viremia with the same virus. Here, we describe results and implications of the follow-up, which confirmed reinfection in fewer cases than previously reported, and show that “intercalating” events—intermittent recurrent bouts of viremia with the same primary infecting virus, interspersed with aviremic periods—occurred frequently.
METHODS
Study Population
Participants are from an ongoing study of young IDUs in San Francisco; details of enrollment, follow-up, and HCV infection detection have been previously published [1]. In brief, young (<30 years) active IDUs (past 30 days) are tested for HCV RNA and anti-HCV. The HCV RNA–negative patients are followed quarterly and those with newly detected HCV infection, monthly. Referrals for follow-up care are provided to all participants. The study protocol was reviewed and approved by the University of California, San Francisco, institutional review board.
Follow-up of HCV Viral Clearers, and HCV Infection Outcomes
Outcomes were assessed in subjects who had follow-up for at least 180 days after first HCV detection. HCV viral clearers were defined as (1) patients who were followed after a confirmed acute or incident infection and subsequently had 2 consecutive HCV RNA negative results at monthly intervals, and (2) patients who presented as anti-HCV positive but HCV RNA negative, indicating resolution of a previous infection at an earlier (undefined) time point. Reinfection was defined by the presence of new viremia, identified as a genetically unique HCV by Core-E1 phylogenetic analysis, in HCV viral clearers. Among participants who presented with evidence of previously resolved infection, analyses were limited to subsequent infections where HCV viremia was detectable and sequenced. HCV viral reclearance was defined as undetectable HCV RNA for at least 2 monthly visits following a confirmed reinfection. HCV viral intercalation was defined as recurrent periods of detectable HCV viremia with closely related Core-E1 sequences with alternating intervals of undetectable viremia [18]. Persistent reinfection was defined as continuous viremia with the confirmed reinfecting virus.
Sample collection dates were used to determine midpoint estimates for analyses of event times and durations (viremic and aviremic periods, etc). Subjects were excluded if they met any of the following criteria: (1) had gaps in follow-up >365 days, or (2) had no data on genotype, serotype, or Core-E1 sequencing of the initial HCV infection. Data were censored on 31 August 2011.
Procedures
Laboratory Tests
HCV RNA Testing
Plasma samples were tested for HCV RNA using a nucleic acid amplification test, the discriminatory HCV transcription-mediated amplification (dHCV TMA) assay component of the Procleix human immunodeficiency virus type 1 (HIV-1)/HCV assay (Gen-Probe Inc, San Diego, CA; Novartis Vaccines and Diagnostics, Emeryville, CA). Repeatedly reactive samples were considered confirmed positive, and further tested to determine quantitative HCV RNA levels using either the HCV Monitor assay (Roche Amplicor Monitor HCV 2.0, Roche Molecular Systems) performed on the Cobas Amplicor semiautomatic instrument, or the Bayer HCV RNA branched DNA assay (Versant HCV 3.0; Bayer Diagnostics, Tarrytown, NY). Samples <50 IU/mL were judged HCV RNA undetectable. Samples reactive by dHCV TMA testing but with undetectable quantitative results were judged positive and quantified as half the value of the lower limit of detection. Samples negative on both qualitative and quantitative tests were judged HCV RNA negative.
HCV Sequencing and Phylogenetic Analysis
HCV Core-E1 sequences were obtained from samples collected closest to the first and last visits where HCV viremia was detected. In cases where no or insufficient specimen was available, specimens from the most proximate visit were tested. Procedures for RNA extraction, sequencing, amplification, and interpretation have been described in detail by Osburn et al [9]. Viral sequences were identified as unique when the Core-E1 divergence between 2 sequences was ≥0.05. Genotypes were determined by comparing Core-E1 sequences to HCV genotype reference sequences using the Los Alamos National Laboratories HCV Phylogenetic Placement Service with pairwise distance analysis [19]. Sequencing results were compared with genotyping results obtained using a commercial genotyping assay (LiPA Line Probe Assay, Bayer Diagnostics, Tarrytown, NY) or serotyping assay (Murex Biotech Limited, Dartford, UK) [20] as previously described [1].
Other Tests
Testing for alanine aminotransferase (ALT) levels, HIV, and hepatitis B surface antigen, core, and surface antibody were conducted in a local commercial laboratory. Interleukin 28B (IL-28B) genotyping was performed using the ABI TaqMan allelic discrimination kit and the ABI7900HT Sequence Detection System (Applied Biosystems), or the Illumina 610 array for genome-wide association studies.
Injection Drug Use Risk Exposures
Risk behavior data, including number of days injected, borrowing previously used needles, and/or drug preparation equipment, were collected quarterly on exposures in the preceding 30 days.
Statistical Analyses
HCV infection and reinfection dates were estimated using midpoints between date of detection of HCV RNA (viremia) and the preceding HCV RNA–negative dates. HCV clearance date was estimated as the midpoint between last RNA positive and first RNA negative tests indicating viral resolution (of at least 2 serial negative HCV RNA results). Time to first reinfection (second HCV infection) was calculated as the number of days elapsed from estimated date of clearance to the reinfection date. Duration of viremia was calculated as the number of days between estimated date of reinfection or recurrent viremia date and date of viral resolution. Time between viremic events was calculated as the number of days between estimated date of viral resolution and subsequent viremic event. Peak quantitative HCV RNA level and ALT measurements were compared (1) in participants with primary acute HCV infection relative to those with recurrent and intercalating infection; (2) during each viremic episode (defined by 1 or more visits with detectable viremia); and (3) between those with reinfection and intercalation. We assessed associations between injection risk exposure and reinfection and intercalation outcomes using generalized estimating equation regression models with an exchangeable correlation structure to account for repeated measures. HCV viremia (log10 IU/mL) and ALT (IU/L) values were examined by Kruskal-Wallis test and a nonparametric equality of median test. Incidence of first reinfection was estimated using person-time methods.
RESULTS
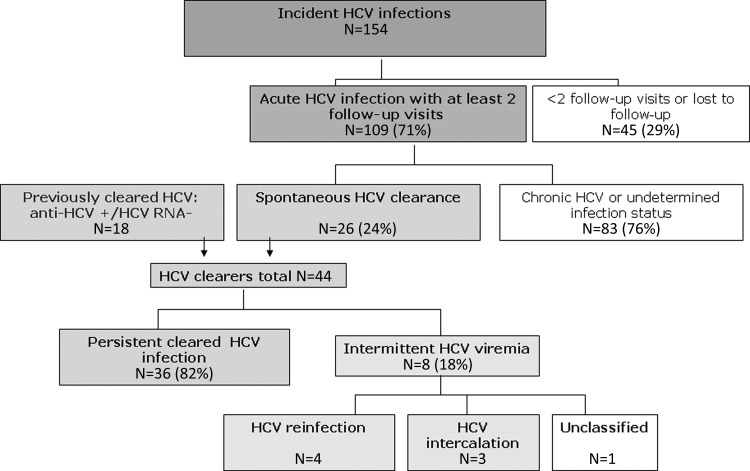
Between January 2000 and August 2011, 18 individuals with previously cleared HCV infection and 109 individuals with incident acute infections were followed (Figure 1). Prospectively, 26 of 109 (24%) cleared HCV infection, resulting in 44 clearers followed. Thirty-six of the 44 (82%) clearers remained persistently HCV RNA negative, and 8 demonstrated intermittent recurrent viremia (Table 1). Viral sequencing demonstrated that 4 of the 8 (50%) cases with intermittent viremia had confirmed reinfection with a heterologous virus (Figure 2A); 4 of these also demonstrated viral intercalation (ie, intermittent detection of the same virus) during some infection episodes. Three individuals (38%) were not reinfected but had multiple intercalating viremia bouts (Figure 2B), and one could not be classified (Figure 1C). Reinfection cases were younger (median age, 23 years [interquartile range {IQR}, 20–26 years) than intercalating cases (median age, 27 years [IQR, 26–28 years]). There were no significant differences between reinfection and intercalation cases with respect to frequency of injecting or sharing/borrowing needles or drug preparation equipment (data not shown). Based on 4 confirmed reinfections observed over 74.2 py, incidence of first reinfection after resolved primary HCV infection was 5.4 per 100 py (95% CI, 2.0–14.4).
Figure 1.
Prospective follow-up of hepatitis C virus infection outcomes after spontaneous resolution, in young injection drug users in the UFO Study. Abbreviation: HCV, hepatitis C virus.
Table 1.
Characteristics, Exposures, and Risk Behaviors Among 127 Injection Drug Users With Acute Hepatitis C Virus Infection
| Baseline Characteristic | Chronic HCV No. (%) | Cleared or Intermittent HCV Infection (n = 43),a No. (%)b |
|||
|---|---|---|---|---|---|
| Total | Persistent Cleared | Reinfection | Intercalation | ||
| Overall | 83 | 44c | 36 (83.7) | 4 (9.3) | 3 (7.0) |
| Age | |||||
| ≤22 y | 40 (48.2) | 15 (34.1) | 13 (86.7) | 1 (13.3) | 0 (0) |
| >22 y | 43 (51.8) | 29 (65.9) | 23 (79.38) | 3 (10.3) | 3 (10.3) |
| Race/ethnicity | |||||
| White | 62 (74.7) | 36 (81.8) | 30 (85.7) | 3 (8.6) | 2 (5.7) |
| Nonwhite | 21 (25.3) | 8 (18.2) | 6 (75.0) | 1 (12.5) | 1 (12.5) |
| Sex | |||||
| Male | 57 (68.7) | 20 (45.5) | 14 (73.7) | 3 (15.8) | 2 (10.5) |
| Female | 26 (31.3) | 24 (54.5) | 22 (91.7) | 1 (4.2) | 1 (4.2) |
| HCV genotype, primary infection | |||||
| 1 | 41 (49.4) | 17 (63.0) | 13 (76.5) | 2 (11.8) | 2 (11.8) |
| 2 | 8 (9.6) | 3 (11.1) | 1 (33.3) | 1 (33.3) | 1 (33.3) |
| 3 | 29 (34.9) | 4 (14.8) | 2 (66.7) | 1 (33.3) | 0 (0 (0) |
| 4 | 1 (1.2) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Not available | 4 (4.8) | 3 (11.1) | 3 (100) | 0 (0) | 0 (0) |
| Peak HCV viremia during primary infection, log IU/mL, median (IQR) | 6.3 (5.4–6.9) | 5.8 (2.7–7.2) | 6.0 (3.0–7.2) | 0.8 (0.8–0.8)d | 7.4 (7.4–7.4) |
| Peak ALT during primary infection, U/L, median (IQR) | 138 (75–250) | 39 (20–195) | 66 (33–219) | 20 (20–20) | 97 (15–179) |
Data are presented as No. (%) unless otherwise specified.
Abbreviations: ALT, alanine aminotransferase; HCV, hepatitis C virus; IQR, interquartile range.
a Reinfection status for 1 case could not be determined and is excluded from 3 right-hand columns.
b Percentages are estimated from the number of cleared infections by strata.
c N = 44 including n = 26 incident HCV clearers with 2 or more visits after HCV infection and n = 18 HCV resolved at baseline.
d Reinfection vs cleared infection: P = .10.
Figure 2.
Event timelines for reinfection cases (A), intercalation cases (B), and an unclassified case (C) with multiple genotypes. A, Hepatitis C virus (HCV) viremia (IU/mL), alanine aminotransferase (ALT), genotype (GT), HCV RNA, and anti-HCV results showing acute HCV infection, reinfection, and reclearance among young injection drug users (IDUs) who have previously cleared HCV infection. Visit dates are shown in the lower boxes. Gray boxes show new acute HCV infections and black boxes show reinfections. Participants whose first infection serology was consistent with that of previously resolved infection (anti-HCV positive and HCV RNA negative) are shown in scored boxes. ^ in the GT box indicates that GT was determined by serotyping. B, HCV viremia (IU/mL), ALT, GT, HCV RNA, and anti-HCV results showing acute HCV infection and intercalation among young IDUs. Visit dates are shown in the lower boxes. Light gray boxes show new acute HCV infections and intercalations. Participants whose first infection serology was consistent with that of previously resolved infection (anti-HCV positive and HCV RNA. negative) are shown with scored boxes. ^ in the GT box indicates that GT was determined by serotyping. C, HCV viremia (IU/mL), ALT, GT, HCV RNA, and anti-HCV results in a young IDU whose infection could not be classified as either reinfection or intercalation. Results show acute HCV infection and either coinfection or superinfection. Visit dates are shown in the lower boxes. Lighter gray boxes show new acute HCV infection and darker gray box shows infection with a different genotype. Abbreviations: ALT, alanine aminotransferase; GT, genotype; HCV, hepatitis C virus; IL28B, interleukin 28B.
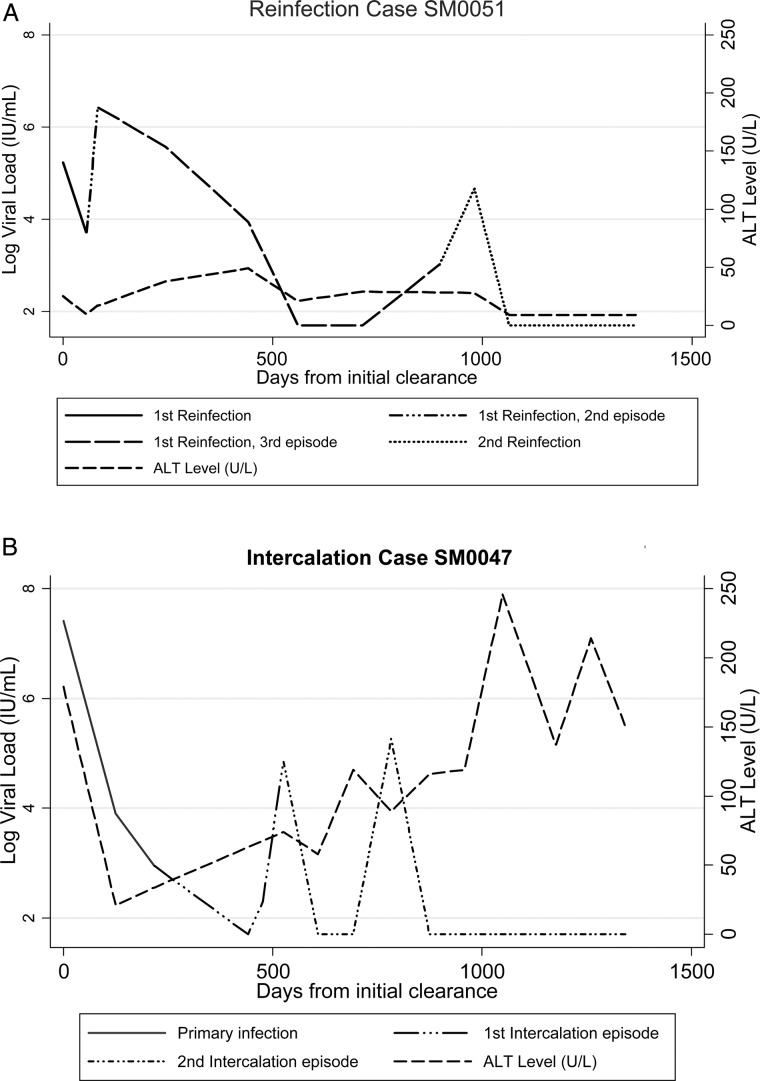
Figure 2A–C shows event timelines for each of the 8 participants, including individuals' IL-28B genotype, HCV genotype, time between and duration of viremic events, and HCV viremia levels (IU/mL). Median time between detected intercalating viremic events was 119 days (range, 56–364 days). Six (75%) participants had sufficient follow-up to determine infection outcome and demonstrated sustained viral clearance for a median of 26 months (range, 4–58 months). Comparisons of viremia and ALT levels between reinfection and intercalation cases are limited by the small sample sizes, but some qualitative differences are noted. Figure 3 shows representative graphs of viremia and ALT in a reinfection and an intercalating case. ALT levels tended to be higher in intercalating cases, with some decline, whereas in reinfection cases ALT declined and remained low.
Figure 3.
Representative graphs of viremia and alanine aminotransferase in (A) reinfection and (B) intercalation cases. Abbreviation: ALT, alanine aminotransferase.
Reinfection Cases
Four individuals had 2 confirmed infections (first detected infection plus 1 reinfection), and 3 of these also had multiple intercalating HCV events during their infection episodes (Figure 2A). All reported injecting exposure (over 42 follow-up visits, median number of days injected per month, 28 [IQR, 0–30]). One female participant (MN0552) with 7 years of follow-up, first infected 2 years after enrollment (genotype 1 virus), demonstrated viral intercalation, then cleared, and after 6 months was reinfected with a genotype 3a virus. She cleared the virus quickly and has remained HCV RNA negative despite ongoing risk exposure. Patient VT0134, HCV naive at enrollment, returned almost 10 months later with serology showing a resolved genotype 1 infection. Reinfection (genotype 2a) was detected >2 years later. Patient MN0560 was found to be infected with a genotype 3a virus 6 months after enrollment (as a clearer), demonstrated viral intercalation, and then cleared viremia and was reinfected with a genotype 1a virus, which was intermittently detectable but not quantifiable. He has been HCV RNA negative for >1 year. Patient SM0051 was enrolled as a clearer and was followed for 4 years before detection of a new infection (genotype 2b). He demonstrated intercalating viremia over a 14-month period. Following more than a year (399 days) of negative HCV RNA tests, he was reinfected briefly with a type 1a virus, had quantifiably lower viremia during these subsequent infections, and subsequently became negative.
Intercalation Cases
Three individuals demonstrated alternating positive and negative viremic events with the same principal infecting virus; median time between detectable viremic events was 42 days (range, 28–186; Figure 2B). One participant had almost 6 years of follow-up (SM0047), with 5 detectable viremic events over that time. He has shown sustained viral clearance for 2.5 years, despite some ongoing injection exposure. Viremia first occurred 4 months prior to detection of anti-HCV and lasted an estimated 284 days; the second episode of viremia lasted 144 days, which occurred after an aviremic interval of 186 days. Each subsequent viremic bout was shorter; the last observed one lasting 56 days. The highest quantified level of viremia occurred during the first episode (25 447 183 IU/mL), and subsequent levels were much lower and finally not quantifiable. The 2 other participants with HCV viral intercalation were first enrolled into follow-up with evidence of cleared infection: a male with 2 documented viremic (genotype 1a) bouts after enrollment, who has remained aviremic for >2 years (and reported no injection exposure for that period); and a female who had 3 detectable bouts of viremia with genotype 1a virus and ongoing exposure.
Unclassified Case
TL0161's first infection was first detected with a very low-level (365 IU/mL) viremia, which could not be genotyped (Figure 2C). After 271 days, viremia (genotype 3a) was detected again, and quantified much higher (2 103 959 IU/mL). Without data in the infecting virus from the first viremic event, we have conservatively assumed these first 2 events were from the same infecting virus. A second infection (genotype 1a) was detected subsequently; however, there was no period of aviremia in between the genotype 3a and 1a infections. This patient may have had HCV coinfection or superinfection. He has remained persistently HCV RNA negative for >5 years despite ongoing injection risk.
DISCUSSION
This study provides a detailed assessment of HCV infection outcomes in young IDUs who have spontaneously cleared at least 1 HCV infection and, during follow-up with frequent serial testing intervals, demonstrated multiple episodes of HCV viremia, including both viral intercalation and reinfections. Because all had ongoing risk exposure during follow-up, accurate classification of the multiple bouts of alternating detectable and undetectable viremia was only possible with extensive HCV sequence analyses. Only half of these cases were confirmed as reinfections. Misclassification of reinfection as a result of lack of viral sequencing and limited follow-up can lead to significant overestimation of reinfection rates. Incidence of reinfection in this group, 5.4%, is substantively lower than our previously published estimate of 24.6%, due to these factors [1]. Significant variability in estimated reinfection rates may also result from differences in definitions of reinfection, testing intervals [21, 22], host factors (eg, age), and exposure, and measures of viral divergence [21–23]. Finally, anti-HCV was persistent in all HCV clearers, unlike in some studies [24, 25], potentially due to ongoing exposures. Our findings demonstrate the importance of frequent testing intervals and viral sequencing to accurately document infection events, and reduce misclassification of resolved, persistent, and reinfection outcomes after acute infection [21, 26] for both clinical and research assessments.
HCV viral intercalation is not well studied, and although there are published reports recording such events, few are in injecting populations [18, 27–33] or involve long follow-up with frequent sampling. As a result, the frequency and prognostic significance of these events is also unknown. Mosley et al [18] have provided the most extensive description of HCV intercalation in transfusion recipients with known infection dates, frequent sampling, and extensive follow-up (some for up to 10 years). Numerous patients demonstrated viremic intercalations, including 48% (12/25) of patients who were ultimately classified as spontaneous resolvers, and 16% (11/69) of patients whose last HCV RNA test was positive. The high frequency of detection of viral intercalation in this, and in Mosley et al's study, may also be due to sensitive nucleic acid amplification testing for HCV RNA, potentially enabling detection of more low-level viremia events than by quantitative assays with higher limits of RNA detection [34, 35].
Several researchers have suggested that intermittent viremic episodes may be indicative of ongoing host immune efforts to resolve infection, and that those with intermittent negative HCV viremia in early infection may be more likely to resolve [11, 36]. Spada et al [29] studied 10 patients who cleared HCV infection and proposed that 1 negative RNA test during early (1–3 months after symptoms) acute infection may be a prognostic marker for clearance. Although Mosley et al [18] observed that intercalation does occur among people who subsequently develop chronic infection, the proportion with intercalation was lower in those with chronic infection compared with clearers (16% vs 48%). Similar to other reports, both reinfection and intercalation cases observed here demonstrated declining viral RNA levels during subsequent viremic episodes [9, 37]. Viral intercalation, including among reinfection cases, may reflect host immune system suppression of viral replication with transient viral control. Studies in both humans and chimpanzees have shown that cellular immune responses, including timing, magnitude, and breadth, are important determinants of infection outcome [33, 38, 39]. Further research assessing immune responses will further clarify these events and prognostic significance.
Both reinfection and intercalation cases reported ongoing but variable injection exposures. Despite the theoretical possibility that intercalation cases could be clearers who may be getting reinfected with the same virus from infected injecting partners, we do not think that this is the case. Our group has been following IDUs with injecting partners to assess transmission events, and we have reported on a high number and frequent turnover within injecting partnerships, as well as a reduced likelihood of engaging in receptive needle sharing among HCV-negative persons who perceive that their partners are HCV positive [40]. Injecting partnerships in San Francisco are very heterogeneous, and the majority of IDUs in our cohort report multiple and frequent changes in injecting partners, most lasting <6 months (data not shown). The frequency of intercalation seen in patients infected by blood transfusions [18] demonstrates that these intercalating events are not conditional on ongoing exposure, possibly reflecting lingering viremia fluctuating between detectable and nondetectable levels. Finally, although we assume that ongoing risk behavior results in exposure to HCV owing to the high prevalence among IDUs, misclassification may be occurring if participants are sharing with uninfected partners.
Our results, along with those of others, show that there can be significant heterogeneity in HCV infection course, and that defining clear dichotomous outcomes after infection and clearance is not always possible without ongoing, frequent follow-up, most importantly, with longitudinal viral sequencing [33]. A prior study testing for superinfecting strains as minority variants during viremia with the earlier strain, using strain-specific polymerase chain reaction, did not detect the later strain, indicating that the earlier quasispecies did not already contain the later strain [41]. Our observation that the majority of reinfection cases cleared infection even with ongoing risk exposure provides hope that vaccines can be developed that will limit chronic infection. Frequent sampling, larger sample sizes, and follow-up are needed to systematically study reinfection and intercalation and to accurately quantify incidence. This may be possible with pooled data from prospective studies of HCV infection [42, 43]. Such studies will inform both HCV clinical care and development of prophylactic vaccines.
Notes
Acknowledgments. The authors thank all the participants in the UFO Study, whose ongoing dedication makes this study possible and the findings relevant. We also recognize the important contributions the UFO Study receives from the San Francisco Department of Public Health, including the Communicable Disease Branch and Housing and Urban Health, the Tenderloin AIDS Resource Center, Street Outreach Services, the Homeless Youth Alliance, and the University of California, San Francisco (UCSF) Tenderloin Clinical Research Center. We also acknowledge helpful suggestions on the study from Dr Greg Dore.
Financial support. This work was supported by the National Institutes of Health (NIH; 2 R01 DA016017–03A1; 5 U19 AI40034–13 to K. P.; K01 DA023365 to J. A. H.; and U19AI088791 to W. O. and A. L. C.). Novartis Vaccines and Diagnostics provided support to this study for HCV EIA-3, RIBA-3, and HCV-TMA testing. Roche Molecular Systems, Inc, provided Amplicor HCV Monitor 2.0 assay kits for some of the HCV RNA testing conducted in this study. We also acknowledge support from the UCSF Clinical and Translational Science Institute (NIH UL1 RR024131) and the UCSF Liver Center (NIH P30 DK026743).
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Page K, Hahn JA, Evans J, et al. Acute hepatitis C virus infection in young adult injection drug users: a prospective study of incident infection, resolution, and reinfection. J Infect Dis. 2009;200:1216–26. doi: 10.1086/605947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Micallef JM, Kaldor JM, Dore GJ. Spontaneous viral clearance following acute hepatitis C infection: a systematic review of longitudinal studies. J Viral Hepat. 2006;13:34–41. doi: 10.1111/j.1365-2893.2005.00651.x. [DOI] [PubMed] [Google Scholar]
- 3.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proust B, Dubois F, Bacq Y, et al. Two successive hepatitis C virus infections in an intravenous drug user. J Clin Microbiol. 2000;38:3125–7. doi: 10.1128/jcm.38.8.3125-3127.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asselah T, Vidaud D, Doloy A, et al. Second infection with a different hepatitis C virus genotype in a intravenous drug user during interferon therapy. Gut. 2003;52:900–2. doi: 10.1136/gut.52.6.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai ME, Mazzoleni AP, Argiolu F, et al. Hepatitis C virus in multiple episodes of acute hepatitis in polytransfused thalassaemic children. Lancet. 1994;343:388–90. doi: 10.1016/s0140-6736(94)91224-6. [DOI] [PubMed] [Google Scholar]
- 7.den Hollander JG, Rijnders BJ, van Doornum GJ, van der Ende ME. Sexually transmitted reinfection with a new hepatitis C genotype during pegylated interferon and ribavirin therapy. AIDS. 2005;19:639–40. doi: 10.1097/01.aids.0000163947.24059.68. [DOI] [PubMed] [Google Scholar]
- 8.Cotte L, Chevallier Queyron P, Schlienger I, et al. Sexually transmitted HCV infection and reinfection in HIV-infected homosexual men. Gastroenterol Clin Biol. 2009;33:977–80. doi: 10.1016/j.gcb.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Osburn WO, Fisher BE, Dowd KA, et al. Spontaneous control of primary hepatitis C virus infection and immunity against persistent reinfection. Gastroenterology. 2009;138:315–24. doi: 10.1053/j.gastro.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerlach JT, Diepolder HM, Jung MC, et al. Recurrence of hepatitis C virus after loss of virus-specific CD4(+) T-cell response in acute hepatitis C. Gastroenterology. 1999;117:933–41. doi: 10.1016/s0016-5085(99)70353-7. [DOI] [PubMed] [Google Scholar]
- 11.Mehta SH, Cox A, Hoover DR, et al. Protection against persistence of hepatitis C. Lancet. 2002;359:1478–83. doi: 10.1016/S0140-6736(02)08435-0. [DOI] [PubMed] [Google Scholar]
- 12.Micallef JM, Macdonald V, Jauncey M, et al. High incidence of hepatitis C virus reinfection within a cohort of injecting drug users. J Viral Hepat. 2007;14:413–8. doi: 10.1111/j.1365-2893.2006.00812.x. [DOI] [PubMed] [Google Scholar]
- 13.Currie SL, Ryan JC, Tracy D, et al. A prospective study to examine persistent HCV reinfection in injection drug users who have previously cleared the virus. Drug Alcohol Depend. 2008;93:148–54. doi: 10.1016/j.drugalcdep.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Aitken CK, Lewis J, Tracy SL, et al. High incidence of hepatitis C virus reinfection in a cohort of injecting drug users. Hepatology. 2008;48:1746–52. doi: 10.1002/hep.22534. [DOI] [PubMed] [Google Scholar]
- 15.van de Laar TJ, Molenkamp R, van den Berg C, et al. Frequent HCV reinfection and superinfection in a cohort of injecting drug users in Amsterdam. J Hepatol. 2009;51:667–74. doi: 10.1016/j.jhep.2009.05.027. [DOI] [PubMed] [Google Scholar]
- 16.Grebely J, Conway B, Raffa JD, Lai C, Krajden M, Tyndall MW. Hepatitis C virus reinfection in injection drug users. Hepatology. 2006;44:1139–45. doi: 10.1002/hep.21376. [DOI] [PubMed] [Google Scholar]
- 17.Halliday J, Klenerman P, Barnes E. Vaccination for hepatitis C virus: closing in on an evasive target. Expert Rev Vaccines. 2011;10:659–72. doi: 10.1586/erv.11.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosley JW, Operskalski EA, Tobler LH, et al. The course of hepatitis C viraemia in transfusion recipients prior to availability of antiviral therapy. J Viral Hepat. 2008;15:120–8. doi: 10.1111/j.1365-2893.2007.00900.x. [DOI] [PubMed] [Google Scholar]
- 19.Los Alamos National Laboratory. HCV phylogenetic placement service. Available at: http://hcv.lanl.gov/content/sequence/phyloplace/PhyloPlace.html. Accessed 10 August 2011. [Google Scholar]
- 20.Dixit V, Quan S, Martin P, et al. Evaluation of a novel serotyping system for hepatitis C virus: strong correlation with standard genotyping methodologies. J Clin Microbiol. 1995;33:2978–83. doi: 10.1128/jcm.33.11.2978-2983.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vickerman P, Grebely J, Dore GJ, et al. The more you look, the more you find: effects of hepatitis C virus testing interval on reinfection incidence and clearance and implications for future vaccine study design. J Infect Dis. 2012;205:1342–50. doi: 10.1093/infdis/jis213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grebely J, Prins M, Hellard M, et al. Hepatitis C virus clearance, reinfection, and persistence, with insights from studies of injecting drug users: towards a vaccine. Lancet Infect Dis. 2012;12:408–14. doi: 10.1016/S1473-3099(12)70010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corson S, Greenhalgh D, Palmateer N, Weir A, Hutchinson S. Risk of hepatitis C virus re-infection following spontaneous viral clearance in injecting drug users: a systematic review. Int J Drug Policy. 2011;22:102–8. doi: 10.1016/j.drugpo.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Beld M, Penning M, van Putten M, et al. Quantitative antibody responses to structural (Core) and nonstructural (NS3, NS4, and NS5) hepatitis C virus proteins among seroconverting injecting drug users: impact of epitope variation and relationship to detection of HCV RNA in blood. Hepatology. 1999;29:1288–98. doi: 10.1002/hep.510290442. [DOI] [PubMed] [Google Scholar]
- 25.Lefrere JJ, Girot R, Lefrere F, et al. Complete or partial seroreversion in immunocompetent individuals after self-limited HCV infection: consequences for transfusion. Transfusion. 2004;44:343–8. doi: 10.1111/j.1537-2995.2004.00656.x. [DOI] [PubMed] [Google Scholar]
- 26.Dore GJ, Micallef J. Low incidence of HCV reinfection: exposure, testing frequency, or protective immunity? Hepatology. 2007;45:1330. doi: 10.1002/hep.21577. [DOI] [PubMed] [Google Scholar]
- 27.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Larghi A, Zuin M, Crosignani A, et al. Outcome of an outbreak of acute hepatitis C among healthy volunteers participating in pharmacokinetics studies. Hepatology. 2002;36:993–1000. doi: 10.1053/jhep.2002.36129. [DOI] [PubMed] [Google Scholar]
- 29.Spada E, Mele A, Berton A, et al. Multispecific T cell response and negative HCV RNA tests during acute HCV infection are early prognostic factors of spontaneous clearance. Gut. 2004;53:1673–81. doi: 10.1136/gut.2003.037788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang CT, Tobler LH, Haesche C, Busch MP, Phelps B, Leparc G. Fluctuation of HCV viral load before seroconversion in a healthy volunteer blood donor. Transfusion. 2003;43:541–4. doi: 10.1046/j.1537-2995.2003.00355.x. [DOI] [PubMed] [Google Scholar]
- 31.Beld M, Penning M, McMorrow M, Gorgels J, van den Hoek A, Goudsmit J. Different hepatitis C virus (HCV) RNA load profiles following seroconversion among injecting drug users without correlation with HCV genotype and serum alanine aminotransferase levels. J Clin Microbiol. 1998;36:872–7. doi: 10.1128/jcm.36.4.872-877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santantonio T, Sinisi E, Guastadisegni A, et al. Natural course of acute hepatitis C: a long-term prospective study. Dig Liver Dis. 2003;35:104–13. doi: 10.1016/s1590-8658(03)00007-0. [DOI] [PubMed] [Google Scholar]
- 33.Cox AL, Mosbruger T, Lauer GM, Pardoll D, Thomas DL, Ray SC. Comprehensive analyses of CD8+ T cell responses during longitudinal study of acute human hepatitis C. Hepatology. 2005;42:104–12. doi: 10.1002/hep.20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fytili P, Tiemann C, Wang C, et al. Frequency of very low HCV viremia detected by a highly sensitive HCV-RNA assay. J Clin Virol. 2007;39:308–11. doi: 10.1016/j.jcv.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 35.Desombere I, Van Vlierberghe H, Couvent S, Clinckspoor F, Leroux-Roels G. Comparison of qualitative (COBAS AMPLICOR HCV 2.0 versus VERSANT HCV RNA) and quantitative (COBAS AMPLICOR HCV monitor 2.0 versus VERSANT HCV RNA 3.0) assays for hepatitis C virus (HCV) RNA detection and quantification: impact on diagnosis and treatment of HCV infections. J Clin Microbiol. 2005;43:2590–7. doi: 10.1128/JCM.43.6.2590-2597.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farci P, Alter HJ, Govindarajan S, et al. Lack of protective immunity against reinfection with hepatitis C virus. Science. 1992;258:135–40. doi: 10.1126/science.1279801. [DOI] [PubMed] [Google Scholar]
- 37.Gerlach JT, Diepolder HM, Zachoval R, et al. Acute hepatitis C: high rate of both spontaneous and treatment-induced viral clearance. Gastroenterology. 2003;125:80–8. doi: 10.1016/s0016-5085(03)00668-1. [DOI] [PubMed] [Google Scholar]
- 38.Bukh J, Forns X, Emerson SU, Purcell RH. Studies of hepatitis C virus in chimpanzees and their importance for vaccine development. Intervirology. 2001;44:132–42. doi: 10.1159/000050040. [DOI] [PubMed] [Google Scholar]
- 39.Post J, Ratnarajah S, Lloyd AR. Immunological determinants of the outcomes from primary hepatitis C infection. Cell Mol Life Sci. 2009;66:733–56. doi: 10.1007/s00018-008-8270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hahn JA, Evans J, Davidson PJ, Lum PJ, Page K. Hepatitis C virus risk behaviors within the partnerships of young injecting drug users. Addiction. 2010;105:1254–64. doi: 10.1111/j.1360-0443.2010.02949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herring BL, Page-Shafer K, Tobler LH, Delwart EL. Frequent hepatitis C virus superinfection in injection drug users. J Infect Dis. 2004;190:1396–403. doi: 10.1086/424491. [DOI] [PubMed] [Google Scholar]
- 42.Cox AL, Page K, Bruneau J, et al. Rare birds in North America: acute hepatitis C cohorts. Gastroenterology. 2009;136:26–31. doi: 10.1053/j.gastro.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grebely J, Morris M, Rice T, et al. Cohort profile: the International Collaboration of Incident HIV and Hepatitis C in Injecting Cohorts (InC3) study. Int J Epidemiol. doi: 10.1093/ije/dys167. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]