Summary
Petal spots are widespread in Angiosperms and are often implicated in pollinator attraction. Clarkia gracilis petals each have a single red-purple spot that contrasts against a pink background. The position and presence of spots in C. gracilis are determined by the epistatic interaction of alleles at two, as-yet-unidentified loci.
We used HPLC to identify the different pigments produced in the petals, and qualitative and quantitative RT-PCR to assay for spatio-temporal patterns of expression of different anthocyanin pathway genes.
We found that spots contain different pigments from the remainder of the petal, being composed of cyanidin/peonidin-based, instead of malvidin-based anthocyanins. Expression assays of anthocyanin pathway genes show that Dfr2 has a spot-specific expression pattern and acts as a switch for spot production. Co-segregation analyses implicate the gene products of the P and I loci as trans-regulators of this switch. Spot pigments appear earlier in development due to early expression of Dfr2 and F3′h1. Pigments in the background appear later, due to later expression of Dfr1 and F3′5′h1.
The evolution of this spot production mechanism appears to have been facilitated by duplication of the Dfr gene and to have required substantial reworking of the anthocyanin pathway regulatory network.
Keywords: Clarkia, Dfr, flower color, petal spot, pigment pattern
Introduction
A major objective of evolutionary developmental biology (‘EvoDevo’) is to elucidate the genetic changes that result in the evolution of novel characters. One unresolved issue is whether novel characters can arise through changes in the activity of genes that are already active in developmental programs or if they require recruitment of additional genes into those programs (Keys et al., 1999). In addition, while the evolution of novel morphologies is sometimes associated with gene duplication (Xiao et al., 2008, Parker et al., 2009), it is unclear how frequently duplication contributes to the emergence of novel traits. Because of the apparent simplicity and ecological relevance of wing spotting patterns, the developmental control of evolutionarily novel spots in animals, particularly butterflies, has been used to address these issues (Nijhout 1980, Carroll et al, 1994, Keys et al. 1999, Beldade et al. 2002, Reed & Serfas, 2004). These investigations have tentatively indicated that new wing spots have arisen by the modification of existing developmental programs.
In plants, petals with large spots— discrete depositions of visible pigments that contrast with the background coloration of the flower—occur in the families Liliaceae, Orchidaceae, Asteraceae, Papaveraceae, Fabaceae, and many others, indicating that they have evolved independently numerous times. Moreover, field studies have frequently demonstrated that spots are important in mediating interactions with pollinators (Jones, 1996; Johnson & Midgley, 1997; Van Kleunen et al., 2007; Goulson et al., 2009). Nevertheless, the genetic and developmental control of petal spots, and more generally, of petal color patterning, have seldom been investigated. Studies in model species such as petunia and snapdragon have clarified the genetic control of some patterns, namely vein-associated pigmentation (Schwinn et al., 2006; Shang et al., 2011) or variation in pigment intensity in different regions of the corolla (Jackson et al., 1992; Schwinn et al., 2006; Albert et al., 2011). However, our general understanding of how floral pigment patterns develop and are regulated at the molecular level, specifically in relation to petals spots, remains poor. In this report, we describe experiments that begin to elucidate the biochemical, genetic and developmental basis of petal spots in the California wildflower, Clarkia gracilis.
Clarkia gracilis (Onagraceae) is native to northern California, Oregon and Washington. It is the only polyploid in section Rhodanthos. Based on cytological analyses, (Abdel-Hameed & Snow, 1972) suggested that this allotetraploid derived from hybridization between two diploid species, one closely related to C. amoena ssp. huntiana and one related to C. lassenensis and C. arcuata. However, the parentage of C. gracilis has not been tested adequately by modern molecular systematic methods.
Clarkia gracilis is composed of four different subspecies that have somewhat overlapping ranges: C. g. ssp. albicaulis, C. g. ssp. gracilis, C. g. ssp. sonomensis, and C. g. ssp. tracyi. The subspecies differ in their floral morphologies, particularly with respect to the presence and position of petal spots. Subspecies albicaulis and tracyi have a single spot at the base of each petal, ssp. sonomensis has a single spot in the center of each petal, and ssp. gracilis lacks petal spots (Fig. 1). In all cases, mature petal spots are dark reddish-purple concentrations of pigment on a pale pink background. However, there is some variation within some subspecies: C. g. sonomensis and C.g. albicaulis can be found in either spotted or unspotted morphs.
Fig. 1.
Flowers of subspecies in the C. gracilis species complex. From left: C. g. ssp. tracyi, C. g. ssp. sonomensis, C. g. ssp. gracilis, and C. g. ssp. albicaulis.
The ecological significance of petal spots in C. gracilis has been studied in some detail. In mixed populations of central spotted and unspotted C. gracilis ssp. sonomensis, spotted plants have higher fitness, producing as much as 32% more seed than their unspotted counterparts in some growing seasons and sites (Jones, 1996). This difference might reflect pollinator preference, since petal spots were also shown to influence pollinator foraging behavior (Jones, 1996). In another species of Clarkia, C. xantiana, the maintenance of both spotted and unspotted morphs has been attributed to pollinator-based negative frequency-dependent selection (Eckhart et al., 2006).
Previous investigations have established that C. gracilis petal spots are under simple genetic control. Gottlieb & Ford (1988) identified two independent loci that interact epistatically to give rise to all of the morphs present in this species complex. One locus, P, has co-dominant alleles that determine the position of the spot within the petal: the PB allele causes the production of basal spots, and the PC allele causes the production of central spots. The other locus, I, affects the presence of basal spots: the IA allele (or I, in (Gottlieb & Ford, 1988)) suppresses basal spots, while the IP allele (i, in Gottlieb & Ford, 1988) allows basal spots to form. Neither allele at the I locus affects the formation of central spots. Gottlieb & Ford (1988) determined that C. gracilis ssp. gracilis generally has a PBPBIAIA genotype, whereas spotted plants of ssp. sonomensis often have the genotype, PCPCIPIP. However, a variable dominance relationship was also found between IA and IP suggesting that additional modifiers or alternative alleles are present in different backgrounds.
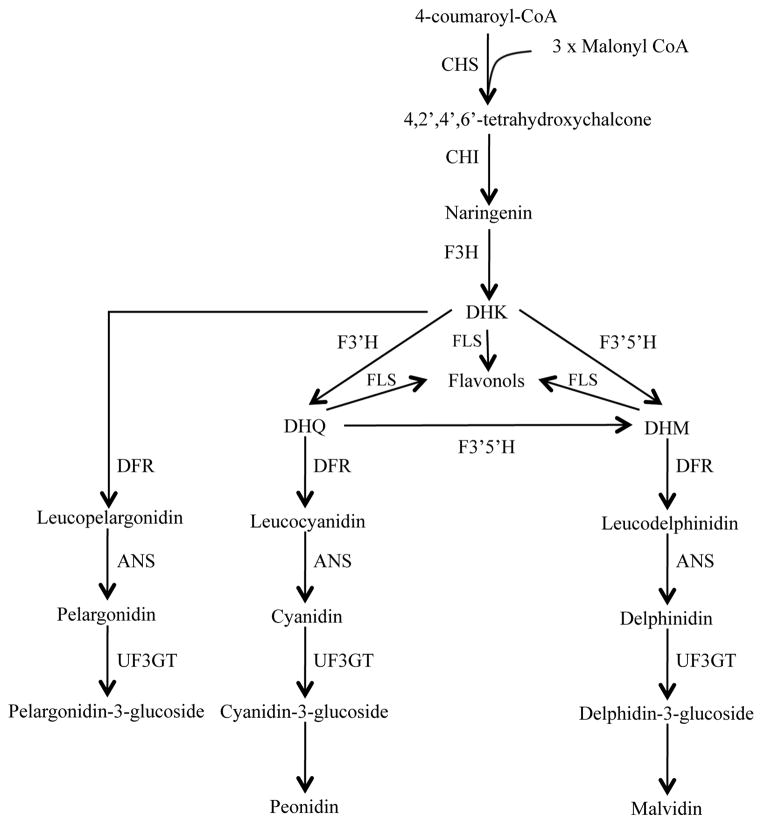
The identity of the pigments underlying this phenotype can provide important clues as to the genes that may be involved in spot formation. Species in the genus Clarkia produce anthocyanin pigments, mostly derivatives of malvidin and cyanidin, although not all species produce these pigments (Dorn & Bloom, 1984; Soltis, 1986). Production of pelargonidin and derivatives is rare in the genus, being known only in C. unguiculata (Robinson & Robinson, 1931), suggesting that the biochemical pathway branch leading to pelargonidin production is inactive in most species (Fig. 2). In C. gracilis flowers, the anthocyanidins malvidin, cyanidin and delphinidin (a precursor of malvidin) have been reported (Soltis, 1986).
Fig. 2.
Simplified schematic representation of the anthocyanin biosynthetic pathway.
Abbreviations for enzymes: CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3 hydroxylase; F3′H, flavanoid 3′ hydroxylase; F3′5′H, flavonoid 3′-5′ hydroxylase; DFR, dihydroflavonol-4-reductase; ANS, anthocyanidin synthase; FLS, flavonol synthase; UF3GT, UDP-flavonoid-3-glucosyl-transferase. Dihydroflavonols: DHK, dihydrokaempferol; DHQ, dihydroquercetin; DHM, dihydromyricetin.
Our objectives in this investigation were to identify genes associated with spot formation in C. gracilis and characterize their pattern of regulation. Specifically, we sought to answer the following questions:
What pigments are produced in C. gracilis flowers? Do pigments in spots differ from the rest of the petal? Do pigments in central spots differ from those in basal spots?
Is transcriptional regulation of specific anthocyanin biosynthetic genes involved in petal spot formation? If so, does any gene correspond to the spot position P locus?
Materials and Methods
Plant growth and crosses
Seeds from C. gracilis (Piper) A. Nelson & J. F. Macbride were germinated in vermiculite at 15°C in the dark, and transferred to a 1:1 metromix:peatmoss mixture once seedlings emerged. Seedlings were then transferred to 5-inch pots and were grown in a glasshouse (20–24°C) under natural light. Plants were periodically randomized to avoid possible light-induced changes in flower color.
Several individuals from various populations were grown, and only those from monomorphic (with respect to spots) populations were used as the parents for crosses. Two independent crosses were carried out to separate the P and I loci. For the ‘P’ cross, a central-spotted C. g. sonomensis was crossed to a basal-spotted C. g. albicaulis. For the ‘I’ cross, an unspotted C. g. gracilis was crossed to a basal-spotted C. g. albicaulis. Voucher information is available in Supporting Information Table S1. Pollen recipient flowers were emasculated several days before full stigma receptivity to avoid self-pollination. Stigmas were pollinated by touching them with a dehiscing anther from the designated sire. F1 individuals were selfed to obtain F2 progeny.
Pigment extraction and High Performance Liquid Chromatography (HPLC) analyses
Anthocyanins were extracted from F2 individuals derived from the ‘P’ cross mentioned above as well as from C. gracilis individuals derived from several natural populations. Seven F2 individuals were examined: two central spotted, two basal spotted, and three double-spotted. Five to ten F1 individuals (all double spotted) were pooled into one sample. Individuals from natural populations included three C. g. ssp. albicaulis, and six C. g. ssp. sonomensis plants (three spotted and three unspotted, see Table S1 for voucher information). Each sample contained either the top, center, or base of 4–5 adult flowers per plant (Fig. S1).
Anthocyanidins were extracted as in Harborne (1984). Pigments were separated using HPLC and identified using the retention times of commercially available standards (delphinidin, pelargonidin, cyanidin, peonidin, malvidin, and petunidin, Polyphenols Laboratory, Sandnes, Norway) that were run using the same conditions as the unknown samples (previously described in Smith & Rausher, 2011).
DNA extraction, gene amplification and sequencing
Genomic DNA from C. gracilis ssp. for gene isolations and phylogenetic analyses was extracted from fresh plant tissue using the CTAB method (Doyle & Doyle, 1987). Genomic DNA from C. amoena ssp. huntiana, C. lassenensis, C. arcuata, C. franciscana, C. rubicunda, and C. breweri were kindly provided by Kenneth Sytsma (University of Wisconsin-Madison; voucher information in Table S1). We identified the two homeologs of C. gracilis genes (arbitrarily called A and B) by including sequences from other species in section Rhodanthos in phylogenetic analyses. This was facilitated by C. amoena and C. lassenensis (the putative parents of C. gracilis) not being sister taxa, allowing for the homeologs present in C. gracilis to be identified more easily.
We used sequences available in Genbank to design degenerate primers to isolate partial sequences corresponding to the anthocyanin biosynthetic genes (F3h, F3′h, F3′5′h, Dfr and Ans). Primers and conditions used for each gene/species are available upon request. PCR bands corresponding to predicted amplicon sizes were transformed into E. coli cells. At least 10 colonies per ligation were sequenced, and additional clones were sequenced as needed. Sequencing reactions used the BigDye Terminatorv. 3.1 system following the manufacturer’s protocol (Applied Biosystems) and electrophoresis was performed at the University of Wisconsin-Madison Biotechnology Center.
Sequence and Phylogenetic analyses
Sequencherv. 4.7 (Gene Codes Corp.) was used to edit, assemble and align sequence fragments with further manual alignment in MacClade v. 4.05 (Maddison & Maddison,2000). To confirm gene identity, sequences were submitted to the Basic Local Alignment Search Tool at the National Center for Biotechnology Information. All phylogenetic analyses were performed in PAUP* v. 4.0b10 using maximum likelihood (ML, Swofford, 2002). Potential PCR recombinants were excluded from the analyses. For all datasets, the GTR model with estimated base frequencies was assumed. Support values were obtained by performing 1000 replicates of ML bootstrapping. Only coding sequences were used in analyses (alignments of included sequences provided in Notes S1.
mRNA expression analyses
Total RNA was extracted from petals using RNeasy Plant Mini kit (Qiagen), followed by a DNAse step (MBI Fermentas) and cDNA synthesis using ImpromII Reverse Transcription System (Promega), or Superscript III First Strand Synthesis (Invitrogen), all according to respective manufacturer’s protocol.
For qualitative gene expression experiments, F2 plants of three different phenotypes, central, basal, and double spotted, from the P locus cross, and of two different phenotypes (basal spotted or unspotted) from the I locus cross, were used. Each RNA sample consisted of either the top, base, or center of three to four flower buds from each plant and three plants for each phenotype. Buds were collected at a stage of development when spot pigments were first appearing (or in equivalently sized petals of unspotted plants), but before background color was visible (Fig. S2).
For quantitative gene expression experiments, plants of three different phenotypes (central-, basal-, and non-spotted), derived from the P and I crosses, were used. Buds representing four different stages of development were collected, with three replicates of each stage. ‘Stage’ was based on the emergence and quantity of pigments, because petal length proved to be an unreliable marker for age, as flower size and maturation times varied greatly, within and between individuals. Change in floral pigmentation has been used previously in Clarkia spp. to determine floral developmental stage (Pichersky et al., 1994). In C. gracilis, color develops in the following order: no color, central spot appears (if present), basal spot appears (if present), background color appears (Fig. S3).
Quantitative assays of gene expression were carried out using Clontech’s SYBR Advantage qPCR premix and run in a Stratagene Mx3000P instrument. Dissociation curves and sequences of qPCR products were obtained to verify amplification of only the desired product. Absolute quantification of mRNA transcript levels was determined using a standard curve, followed by normalization to Actin. qPCR runs were performed in duplicate. Primers used for RT-PCR and qPCR (Table S2) flanked introns, with the exception of F3h and Ans. PCR products were sequenced to confirm that the desired copy was being amplified.
Genotyping F2 individuals from P and I crosses at the Dfr2-A and Dfr2-B loci
Thirty-two and 31 F2 plants from the P cross were genotyped for Dfr2-A and Dfr2-B, respectively. Homeolog specific primers were designed to amplify a portion of exon 2 and intron 2 (Dfr2-A), and a portion of intron 2 (Dfr2-B). Fifty plants from the F2 generation from the I cross were genotyped at Dfr2-A and Dfr2-B by amplifying intron 3 and partial sequences of exon 3 and 4. The same Dfr2 primers used for qPCR analyses were used. All alleles were identified using fragment length polymorphisms, with the exception of alleles of Dfr2-B for the P cross, which were genotyped by the presence of a SNP using restriction enzyme digestion (Tables S3 and S4). Fragment length runs were completed in an ABI 3730XL DNA Analyzer at the Duke Genome Sequencing & Analysis Core Resource, visualized using the GeneMapper software (Applied Biosystems) and scored by eye. Fisher’s exact test was used to test for possible co-segregation between spot presence and Dfr2-A/B alleles.
Results
Crosses confirm the two-locus model and provide segregating families for further analysis
Although the genetics of petal spot inheritance had already been determined, we established populations of individuals with known genotypes to facilitate genetic and molecular analyses and to confirm previous genetic results. We developed two separate segregating F2 populations for the two independent loci, thus removing the epistatic interaction that could mask the genotype at the P locus.
Segregation patterns (Table 1) confirmed those previously reported (Gottlieb & Ford, 1988; Jones, 1996). In the P-cross (see Materials and Methods), 100% of F1 progeny were double-spotted. Segregation in the F2 population did not deviate significantly from the expected 1:2:1 ratio of basal spotted:double spotted:central spotted plants (chi-square test, P = 0.74) consistent with a model of a single locus with co-dominant alleles. According to this model PBPB plants have basal spots, PCPC plants have central spots, and PBPC plants have both a central and a basal spot.
Table 1.
Segregation for presence versus absence and position of petal spot in two separate crosses, in F1 and F2 generations of C. gracilis plants
| Spot Position | |||||
|---|---|---|---|---|---|
|
| |||||
| n | Central | Basal | Double | Absent | |
| P cross | |||||
| F1 | 23 | - | 23 | - | |
| F2 | 82 | 18 | 23 | 41 | - |
| I cross | |||||
| F1 | 7 | - | 7 | - | - |
| F2 | 101 | - | 77 | - | 24 |
P cross refers to central-spotted C. g. sonomensis basal-spotted C. g. albicaulis cross, while I cross refers to unspotted C. g. gracilis basal-spotted C. g. albicaulis cross.
In the I-cross (see Materials and Methods), all F1 individuals had basal spots, suggesting that the suppressing IA allele was recessive to the spot-permitting allele IP. In the F2 population, we saw variation in spot intensity, from a strongly defined spot to an ill-defined ‘smudge’, to unspotted petals. Because it was difficult to distinguish spots from smudges, plants showing any color at the base were coded as ‘basal spotted’ for statistical analyses. As shown in Table 1, using this scoring, a 3:1 pattern of spotted:unspotted plants could not be rejected (chi-square test, P= 0.77), suggesting that the IP allele isolated is generally (but variably) dominant to the IA allele. Genotypes for P and I and expected phenotypes under this genetic model are presented in Fig. S4.
It should be noted that, while in our crosses the IP allele appears to be dominant, previous work has shown that alleles at the I locus can switch from dominant to recessive in different backgrounds, possibly due to other modifiers (Gottlieb & Ford, 1988). Further genetic studies are necessary to fully understand the phenotypic expression of this allele in different C. gracilis populations and subspecies.
HPLC analyses show that spots contain different pigments from the rest of the petal
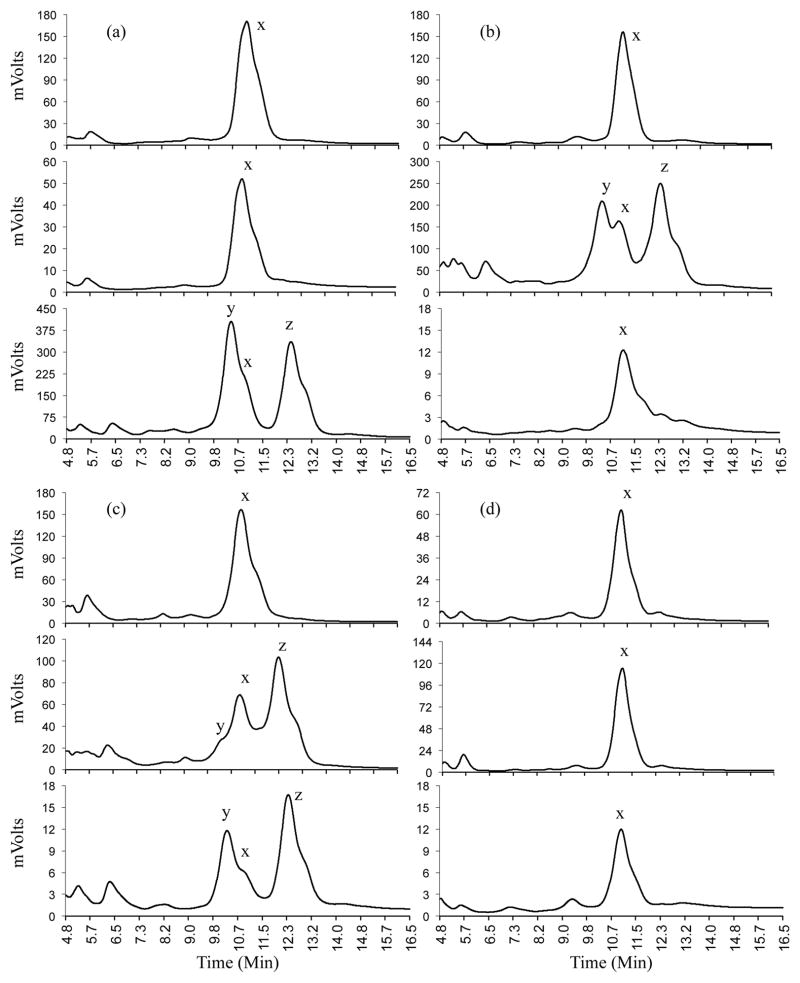
While the pigment composition of entire C. gracilis petals had been previously determined through chromatography (Soltis, 1986), it was unclear whether the spots contained the same pigments as the background color. Using HPLC, we analyzed anthocyanidins present in mature petals from several C. gracilis individuals, spanning all spot phenotypes (see Materials and Methods). In all cases, irrespective of subspecies or spot phenotype, the distribution of anthocyanidins in the petal followed the same pattern. Petals of C. gracilis were found to include the anthocyanidins malvidin, cyanidin, and peonidin (Fig. 3, Table 2). These results contrast with a published report where delphinidin was found instead of peonidin (Soltis, 1986). The discrepancy between this study and Soltis (1986) may be due to the different methodologies used (HPLC versus thin-layer chromatography).
Fig. 3.
Anthocyanidins found in C. gracilis flowers. HPLC traces showing pigments in different sections of petals. Each panel (a–d) shows traces for sections of the petal (top, center, and base, in that order) for one of the four phenotypic classes: Basal-spotted (a); central-spotted (b); double-spotted (c), and; unspotted (d). Peaks corresponding to malvidin, cyanidin, and peonidin (indicated x, y, and z, respectively) were identified by comparison to known standards.
Table 2.
Pigment composition in C. gracilis petals with different spot phenotypes
| Phenotype
|
|||||
|---|---|---|---|---|---|
| Basal spot | Central spot | Central & Basal spot | No spot | ||
|
| |||||
| Petal section | Top | 0.171 (±0.050), 5 | 0.132 (±0.026), 3 | 0.205 (±0.102), 3 | 0.137 (±0.033), 2 |
| Center | 0.227 (±0.078), 2 | 3.524 (±1.599), 5 | 2.717 (±0.179), 3 | 0.143 (±0.026), 2 | |
| Base | 4.55 (±1.020), 5 | 0.494*, 1 | 5.316 (±1.091), 3 | 0.355 (±0.115), 2 | |
|
|
|||||
Numbers refer to the ratio of cyanidin+peonidin: malvidin (mean ± standard deviations), followed by number of individuals sampled. Amount of each pigment was inferred from the average peak height between 10.4–10.5 min. for cyanidin, 11.1–11.2 min. for malvidin, and 12.3–12.4 min. for peonidin.
All but one central-spotted individuals had spots that extended close to the petal base (on the abaxial, but not the adaxial surface), impeding dissection of spot-free tissue. Therefore, only one individual was used.
Cyanidin and peonidin were present in all petal sections that included spots and were never detected in petal sections that lacked spots (Fig. 3, Table 2). Likewise, petal sections that lacked spots, only having background color, contained only malvidin. The central regions of central-spotted flowers sometimes contained small amounts of malvidin, consistent with the presence of some background color in the central sector. In contrast, the basal regions of basal-spotted flowers contained very little or no malvidin, consistent with the almost complete lack of background color near the petal base (Fig. S1).
Given these findings, we conclude that, regardless of spot position, anthocyanins in C. gracilis petal spots are derived from cyanidin and peonidin, two similar anthocyanidins that differ in that peonidin has a methyl substitution on the B-ring. In contrast, the background anthocyanins present in the petal appears to be malvidin-based, an anthocyanidin that has two methyl substitutions on the B-ring, and derives from a different branch of the anthocyanin pathway (Fig. 2).
Clarkia gracilis carries multiple copies of anthocyanin biosynthetic genes
In the anthocyanin pathway the branch-point between cyanidin/peonidin and malvidin occurs after the F3H-mediated synthesis of DHK (Fig. 2). This points to the regulation or action of F3′h and/or F3′5′h as possible determinants of spot formation. Another possible candidate for the synthesis of different pigments is Dfr. Numerous studies show that single or very few amino acid changes in DFR can lead to changes in substrate specificity that redirect flux down a different branch of the pathway (Johnson et al., 2001; Fischer et al., 2003; Shimada et al., 2005; DesMarais and Rausher, 2008). Consequently, we focused on documenting whether differences in the patterns of regulation of F3′h, F3′5′h and Dfr are associated with the different petal spot phenotypes of C. gracilis. In addition, we also examined F3h and Ans, which are involved in earlier and later steps of pigment biosynthesis, respectively, to serve as controls and provide a more comprehensive understanding of patterns of expression of anthocyanin biosynthetic genes.
We conducted PCR amplification of partial gene sequences using multiple degenerate and specific primers, designed to conserved regions of the genes encoding the targeted enzymes, and cloned numerous PCR products in an attempt to identify all homologous gene copies in C. gracilis and, when possible, in related diploid species. We analyzed sequences for the presence of stop codons to assess gene functionality and used phylogenetic analyses to confirm gene identity (Fig. S5). However, it should be noted that we obtained only partial sequences for most genes from most species.
Two copies of F3h, F3′h, and F3′5′h and three copies of Dfr were found in diploid species in Sect. Rhodanthos, while a single copy of Ans was found. For F3′h and F3′5′h, only one copy appeared to be functional in the diploids: one copy of each gene had a premature stop codon or a frameshift mutation. For F3h, one copy from C. amoena and one from C. arcuata appeared to be divergent (as evidenced by longer branches, see Fig. S5). There were no indications from the sequences to suggest that any Dfr genes lack function (Fig. S5). We focused our efforts on isolating the putative functional or non-divergent copies of these genes from C. gracilis, and recovered one copy (two homeologs) corresponding to the putative functional F3h and F3′h, and all three copies (and 2 homeologs for each copy) of Dfr. We isolated both homeologs of the functional copy of F3′5′h, and one homeolog of the nonfunctional F3′5′h gene, which was confirmed to have an early stop codon. We isolated a single sequence corresponding to Ans, but is unclear whether only one homeolog exists, or whether both homeologs are present but do not differ in sequence within the short fragment obtained.
Spatial expression of Dfr2 shows a tight correlation to spot location in C. gracilis petals
If spot formation depends on location-specific expression of any of the anthocyanin biosynthetic genes, it follows that such gene(s) would be expressed mainly in areas where spots are formed. To examine spatial expression patterns, we performed RT-PCR on RNA extracted from three different petal sections (top, middle and base) from young buds, from plants with central (PCPC), basal (PBPB), or double-spotted (PBPC) flowers. Copy-specific primers were used for all of the putatively functional genes, with homeolog-specific primers being used in C. gracilis when that proved possible.
As shown in Fig. 4, the gene encoding the most upstream enzyme examined in the anthocyanin pathway, F3h, and one of the two copies of the gene encoding the next enzyme, F3′h1-A, showed consistent, robust expression in all sections of all petals. F3′h1-B transcript was not found in floral tissue, despite multiple attempts using several primers, suggesting it is either not expressed or limited to other tissues. Genes encoding enzymes further down the pathway, such as F3′5′h, Dfr1, and Ans (hereafter, ‘downstream genes’) showed variable expression that did not correlate with genotype or spot location. This variation most likely reflects slight differences in the age of petals. For instance, the most consistent pattern among the downstream genes is that their levels tend to be lower in central spotted petals than in either basal-spotted or double spotted petals. This is likely due to the age of the floral buds. Central spots appear earlier in development than basal spots, which means that central spotted flowers were harvested at a slightly earlier stage of development than the other genotypes (Fig. S3). This is significant because it suggests that, at this early stage of development, downstream genes are not fully and consistently activated.
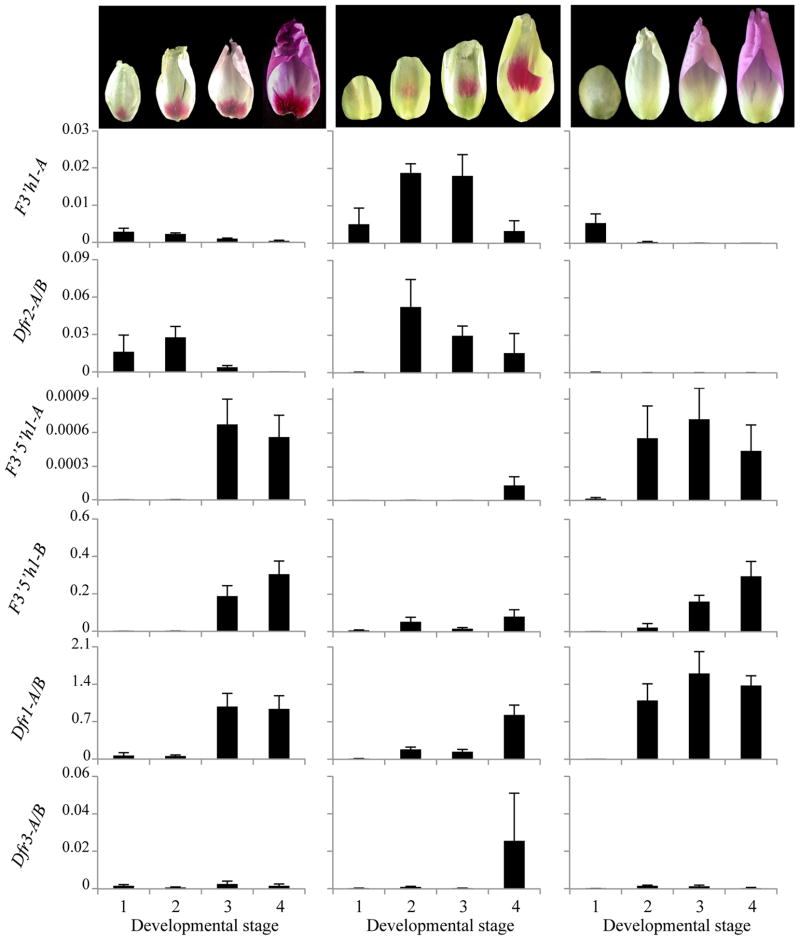
Fig. 4.
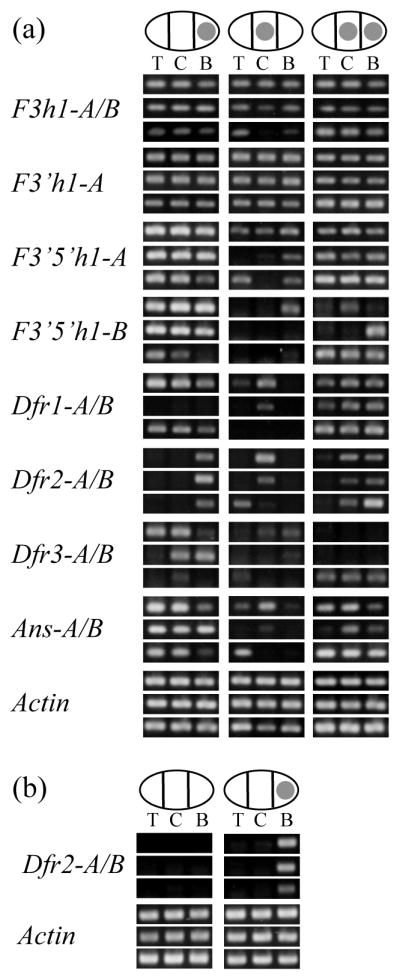
Spatial expression of anthocyanin biosynthetic genes in C. gracilis petals. (a) Expression patterns for three individual plants for each of the P-locus phenotypes. The same nine individuals were used for all genes, and they are placed in the same position for each panel. Ovals represent petals, and filled circles represent spot locations. T, C, and B refer to top, central, and basal sections of the petals. F3h, flavanone 3 hydroxylase; F3′h, flavanoid 3′ hydroxylase; F3′5′h, flavonoid 3′-5′ hydroxylase; Dfr, dihydroflavonol-4-reductase; Ans, anthocyanidin synthase; (b) Expression patterns for Dfr2 in three spotted and three unspotted plants from a F2 population segregating for I.
The two homeologs of one gene, Dfr2, had a pattern that was strikingly distinct from all the other genes surveyed, a pattern that is correlated with the presence of the spot (Fig. 4a). Dfr2 appeared to be expressed at high levels in sections of the petal that contained a spot, but not in unspotted sectors. In central-spotted individuals Dfr2 was expressed at high levels only in the center of the petal, but was expressed only at the base of basal-spotted petals. Similarly, double-spotted individuals showed high expression of Dfr2 in both central and basal sections of the petal. The low levels of expression that were sometimes seen in sections adjacent to the spot can be attributed to imprecise sectioning of the buds, since they are only a few millimeters in length when dissected. The sole exception to this pattern is that one central spotted individual, showed a brighter band for Dfr2 in the top as opposed to the central section of the petal (Fig. 4a). This could be partly due to initial RNA quality, as the top section for that petal yielded a brighter band than the central sections for the other genes surveyed, including Actin. The spot-specific pattern of expression of Dfr2 in petals (and lack in unspotted petals) was confirmed in nine other C. gracilis plants grown from seed derived from wild populations (3 unspotted and 2 central spotted C. g. sonomensis, 3 basal spotted C. g. albicaulis, and 1 basal spotted C. g. tracyi, Fig. S6). Therefore we conclude that Dfr2 (and only this gene) shows a spot-specific expression pattern.
The role of Dfr2 in spot formation was further evaluated by examining the expression of this gene in individuals segregating for the I locus. In basal-spotted plants (IPIP or IPIA), Dfr2 is expressed at high levels in the basal section whereas unspotted siblings (IAIA ) show very low or no Dfr2 expression (Fig. 4b). This corroborates the pattern seen in F2s from the P locus cross, suggesting that Dfr2 is necessary for spot production. It also indicates that suppression of the basal spot by the I locus is genetically upstream of Dfr2.
Dfr2 deviates from other downstream genes in showing expression early in petal development
The variable expression of genes encoding downstream enzymes suggested the possibility that temporal regulation plays a role in petal spot development. This seems plausible since petal spots, especially central spots, are visible several days before the appearance of background color (Fig. S3), implying that genes required for spot development might be expressed earlier than genes that are only required for the production of background color. To investigate this possibility, we used qPCR to obtain an expression time-course for Dfr and for the functional copies of F3′h1, F3′5′h1, which are situated at the branch-point between cyanidin/peonidin and malvidin biosynthesis in developing buds. We assayed all three copies of Dfr and both homeologs for all genes except the B copy of F3′h1, which did not appear to be expressed in petals.
As shown in Fig. 5, two genes, F3′h1-A and Dfr2, are expressed early in development, having high expression levels at the first time points (as the spot appears) and declining expression in more mature buds (which are developing background color). Although F3′h1-A shows a similar pattern in all three phenotypes, Dfr2 expression is reduced or absent in flowers that lack spots, consistent with the results obtained from non-quantitative PCR (Fig. 4). In contrast to F3′h1-A and Dfr2, F3′5′h1 and Dfr1 were expressed later in petal development, having low expression levels in early time points, but higher expression later in development.
Fig. 5.
Quantitative analysis of gene expression of anthocyanin biosynthetic genes through C. gracilis petal development. Photos above the graphs show a representative petal for each of the four developmental stages defined for the purpose of this experiment. From L to R: basal, central, and unspotted. All buds are shown in the upright position (apex at the top, base at the bottom). From left to right, basal spotted plants: spot first appearing, spot well defined, background beginning to show, mature petal. Central spotted plants: no color, spot first appearing, spot well formed, background color beginning to appear. Unspotted plants: young colorless, older colorless, background first appearing, mature petal. Y-axes show expression of genes normalized to Actin. F3′h, flavanoid 3′ hydroxylase; F3′5′h, flavonoid 3′-5′ hydroxylase; Dfr, dihydroflavonol-4-reductase. Columns represent averages across three replicates (except last time point for central spotted, n=2), error bars indicate standard error.
Finally, one gene Dfr3 shows no clear pattern, being expressed at very low levels in all tissues except for a peak in late, central-spotted flowers. The latter could be an experimental artifact (note error bars). The success in amplifying Dfr3 in RT-PCR experiments confirms that this gene is expressed, but the level of expression appears to be substantially lower than the other Dfr gene copies, indicating that this gene may lack a function in pigment production in the petal.
Co-segregation analysis suggests that the P and I loci correspond to trans-regulators of Dfr2
Qualitative and quantitative gene expression data indicate that cyanidin/peonidin-based petal spots in C. gracilis correlate with the early and localized expression of Dfr2, coupled with the early expression of F3′h1-A. Under the assumption that localized, early expression of Dfr2 is the proximate cause of petal spot development, then the P and I loci must act by modulating the expression of Dfr2, and likely correspond to upstream regulators of Dfr2. However, it is possible that either P or I could correspond to Dfr2 itself if it had undergone a cis-regulatory change that resulted in altered expression.
To verify whether Dfr2 alleles co-segregate with spot location and/or presence, we genotyped several F2 individuals from both the P and I crosses, for both homeologs of Dfr2 (Table 3). The genotypes at neither of the Dfr2 homeologs co-segregated perfectly with spot location in the P cross, suggesting that the P locus does not encode Dfr2, but rather is a trans-regulator of Dfr2. However, the p-value for co-segregation between Dfr2-B and P approached significance (P = 0.0843), suggesting that perhaps there is some genetic linkage between P and Dfr2-B. Larger numbers of offspring would be necessary to confirm this hypothesis. Similarly, neither of the Dfr2 homeologs co-segregated with the absence/presence of basal spots in F2s from the I locus cross, suggesting that I is also a trans regulator of Dfr2.
Table 3.
Analysis of co-segregation of alleles at the DFR2-A and DFR2-B loci with phenotypes obtained from the I and P crosses
| Homeolog | Genotype | I Cross | P Cross | |||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Phenotype
|
P | Phenotype
|
P | |||||
| S | U | B | D | C | ||||
| Dfr2-A | A1A1 | 11 | 2 | 0.591 | 3 | 5 | 1 | 0.9314 |
| A1A2 | 19 | 5 | 5 | 8 | 5 | |||
| A2A2 | 12 | 1 | 2 | 2 | 1 | |||
| Dfr2-B | B1B1 | 10 | 4 | 0.2365 | 3 | 1 | 0 | 0.0843 |
| B1B2 | 18 | 3 | 0 | 6 | 2 | |||
| B2B2 | 14 | 1 | 6 | 8 | 5 | |||
Different alleles of Dfr2-A/B were identified using SNPs and indels in introns 2 or 3.
Abbreviations: S = spotted; U = unspotted; B = basal-spotted; D = double-spotted; C = central spotted. P are from Fisher’s exact test. Dfr, dihydroflavonol-4-reductase.
Discussion
A model for the development of pigmentation patterns in C. gracilis petals
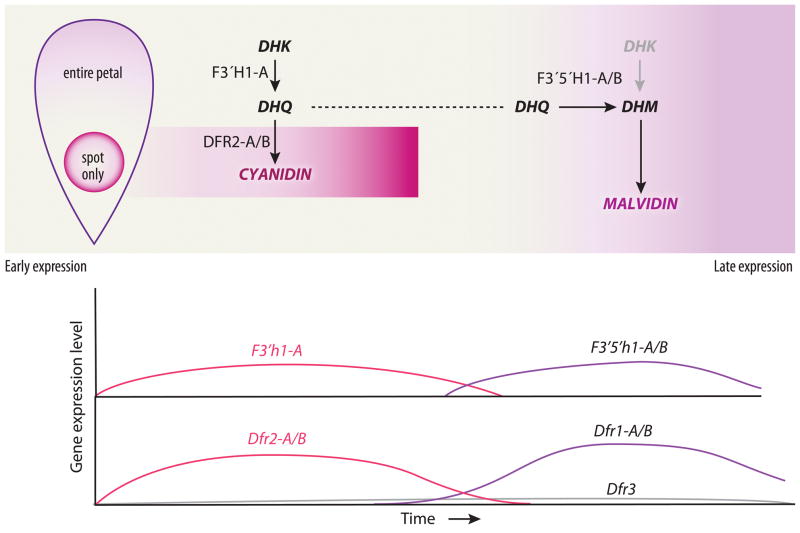
Based on the results presented here we propose a model for petal pigment development in C. gracilis that can explain the production of cyanidin/peonidin-based anthocyanins in spots and malvidin-based anthocyanins in the background (Fig. 6). This model invokes transcriptional regulation of anthocyanin biosynthetic genes as a function of time and space. Although post-translational regulation may occur, it is not necessary to explain the observed data. For this model, we assume that Dfr3 is expressed at such low levels that it plays no significant role in petal pigmentation. Likewise we will ignore the putatively non-functional copies of F3′h and F3′5′h.
Fig. 6.
Model of pigment pattern development in C. gracilis flowers. Early pigment production in spots is explained by F3′h expression throughout the petal and Dfr2 expression in spots, which leads to dihydroquercetin (DHQ) production throughout the petal and cyanidin/peonidin production in spots only. Later in development expression of F3′5′h and Dfr1 converts DHQ (and any residual dihydrokaempferol (DHK)) in the petal background to dihydromyricetin (DHM) and then malvidin. F3′h, flavanoid 3′ hydroxylase; F3′5′h, flavonoid 3′-5′ hydroxylase; Dfr, dihydroflavonol-4-reductase.
According to the model (Fig. 6), most of the core enzyme-coding genes of the anthocyanin pathway (Chs, Chi, F3h, F3′h, Ans, and Uf3gt) are activated early in development and expressed throughout petal tissue. In addition, Dfr2 is activated early but is expressed only in the area destined to become a pigmented spot, implying that it is under different regulatory control. By contrast, neither F3′5′h1 nor Dfr1 are expressed at this time. Because Dfr is required for pigment production, the spatial restriction of Dfr2 explains why pigment is produced only as a spot. Further, because F3′5′h1 is not activated early, only anthocyanins derived from dihydroquercetin (DHQ) (i.e., cyanidin and peonidin) are produced in the spot domain.
At a later stage of development, the core enzymes are still active. This may represent continuous expression of these genes, or a second wave of expression. At this point, F3′h1-A and Dfr2 expression has ceased. Activation of both F3′5′h1 and Dfr1 throughout the developing petal means that the complete set of enzymes is present, causing the background pigment to be deposited. The expression of F3′5′h1 causes this pigment to be derived via DHM rather than DHK or DHQ, explaining the production of malvidin in the background. Thus, the difference in pigment type between background and spot, as well as the spatial restriction of the spot, is explained by the different temporal expression of F3′h1-A/Dfr2 versus F3′5′h1/Dfr1.
Regulation of enzyme-coding genes
In most species that have been examined, the core enzyme genes of the anthocyanin pathway (those depicted in Fig. 2) are coordinately regulated by common sets of transcription factors (Cone et al., 1986; Paz-Ares et al., 1986, 1987; Ludwig et al., 1989; Goodrich et al., 1992; de Vetten et al., 1997; Elomaa et al., 1998; Quattrocchio et al., 1998, 1999; Walker, 1999; Borevitz et al., 2000; Spelt et al., 2000, 2002; Carey et al., 2004; Schwinn et al., 2006; Gonzalez et al., 2008; Lin-Wang et al., 2010; Niu et al., 2010; Albert et al., 2011). In some taxa, such as Ipomoea and maize, all core genes are expressed at similar times in development and are activated by a single set of transcription factors (Cone et al., 1986; Paz-Ares et al., 1987; Ludwig et al., 1989; Morita et al., 2006). In other species, such as Petunia and Antirrhinum, genes are activated in two blocks, with those coding for upstream enzymes activated first, and those coding for downstream enzymes activated later, perhaps by a different set of transcription factors (Martin et al., 1991; Goodrich et al., 1992; Jackson et al., 1992; Quattrocchio et al., 1998; Spelt et al., 2000; Schwinn et al., 2006).
The pattern of regulation exhibited by C. gracilis differs from these canonical patterns. Although we have not demonstrated it directly, the production of anthocyanins in the petal implies that upstream and downstream genes, represented by F3h and Ans, are expressed from the time anthocyanins are first produced in spots to the time they are last produced in the remainder of the petal—although the existence of sub-blocks within these genes cannot be ruled out. However, Dfr, F3′h, and F3′5′h exhibit more restricted temporal distributions, with Dfr2 and F3′h1-A being expressed only in the early stages of development and Dfr1 and F3′5′h1 being expressed only later in development. This pattern suggests that factors regulating these genes differ from those regulating the remaining core enzymes, and that the factors regulating Dfr2 and F3′h1-A differ from those regulating Dfr1 and F3′5′h1. Moreover, two patterns indicate that Dfr2 is regulated differently from all other enzyme coding genes. First, only Dfr2 exhibits a spatially restricted expression domain, confined to the regions that develop spots. Second, variation at the P and I loci affect only expression of Dfr2. Because neither of these loci cosegregates with Dfr2, they likely correspond to transcription factors or cofactors that are unique to this gene. These patterns imply that the production of spots has required a substantial reworking of the anthocyanin regulatory network. We next discuss the evolutionary implications of this reworking.
Possible substrate specificity in different copies of DFR
DFR2 and DFR1 appear to only encounter one substrate, DHQ or DHM, respectively, and, while the model we propose for formation of cyanidin/peonidin-based spots does not require evolution of substrate specificity among the different copies of DFR, it is possible that it has occurred. In C. gracilis, the two different copies differ in numerous sites, including in the active site, where they differ by nine amino acid substitutions. One such substitution, at residue 133 (Fig. S7), has been implicated in substrate specificity in other species (Shimada et al., 2005; DesMarais and Rausher, 2008; Johnson et al., 2001). Possible evolution of substrate specificity could be explored by biochemical characterization of these enzymes combined with more fine-scale temporal expression studies.
Evolutionary implications
Although the progenitors of C. gracilis have petal spots, as do other species in the section Rhodanthos, our analysis of spot formation in C. gracilis sheds light on aspects of how petal spots originally evolved in this group. Two major types of regulatory change appear to have been required for the evolution of spots. The first was a spatial restriction of the expression domain of Dfr2. It is reasonable to assume that pigment deposition throughout the petal was the ancestral state because species that are situated in the phylogeny as sister to the rest of the genus, namely C. breweri, C. concinna, and C. pulchella, are all pink and unspotted. Although it is unclear when the duplication of Dfr occurred, it is likely that initially both paralogs exhibited the same broad spatial expression pattern. Restriction of Dfr2 expression to the areas of spot formation subsequently occurred. Because Dfr2 seems no longer to be activated by the anthocyanin transcription factors that activate the other core anthocyanin enzymes, we suspect its cis-regulatory region evolved to respond to a new set of activators that are expressed at the time and position at which spot formation initiates. The gene products of the P and I loci are candidates for these new activators, since they influence expression of Dfr2 but not expression of the other core enzyme-coding genes.
The second major type of regulatory change necessary for the evolution of cyanidin-based spots separated the timing of expression of Dfr2 and F3′h on the one hand from Dfr1 and F3′5′h on the other. By itself, divergence in spatial expression domains of the two Dfr paralogs might produce a spot of increased pigment intensity, which could explain the existence of Clarkia species that produce spots but only malvidin in their flowers (Soltis, 1986), but cannot account for cyanidin/peonidin spots on a malvidin background, as seen in C. gracilis. Our analyses suggest that temporal separation of the expression of F3′h and F3′5′h is largely responsible for the production of cyanidin/peonidin-based anthocyanin in spots.
In most plant species that have been examined, both F3′h and F3′5′h are active simultaneously and result in the production of malvidin/delphinidin derivatives (Gerats et al., 1982; Tornielli et al. 2009, Hopkins & Rausher 2011). We therefore presume this was the pattern in ancestors of C. gracilis that lacked spots (only malvidin is produced in the unspotted flowers of C. breweri and C. concinna, Soltis, 1986). Cyanidin, peonidin and their derivatives can only be produced if F3′5′h activity is greatly reduced or eliminated. C. gracilis has apparently achieved this by delaying the expression of F3′5′h so that, at the time of spot development, only F3′h is expressed. By contrast, at the time of background pigment development, only F3′5′h is expressed, allowing the accumulation of malvidin derivatives. Finally, restriction of the expression of Dfr1 to the late stage of development was required to prevent cyanidin- and peonidin-based anthocyanins from being synthesized in the petal background.
Our analysis of the genetic control of spot pattern formation has revealed that the gene Dfr2 acts as a switch for spot production. Activating this gene completes the anthocyanin pathway and allows pigment deposition in spots. The evolution of this switch mechanism appears to have been facilitated by duplication of the ancestral Dfr gene, which allowed one paralog to evolve to serve as a spot regulator. Based on the broad occurrence of petal spots in Clarkia, we infer that the Dfr gene duplication occurred early in the radiation of the genus. Subsequent evolutionary remodeling of the regulatory network for both copies of Dfr, as well as for F3′h and F3′5′h, resulted in the novel ability to finely regulate the spatial pattern of contrasting colors in the petals of many Clarkia species. Further work in this group, particularly temporal and spatial expression assays of anthocyanin pathway genes in petals from species throughout the genus will help to illuminate the order of events that led to the evolution of petal spot.
Supplementary Material
Acknowledgments
We would like to thank the late Leslie Gottlieb for numerous helpful discussions and for providing us with his original seed materials. We also thank Brian Barringer and Norman Weeden for seeds, Ken Sytsma for DNA samples, and Vince Eckhart for primer sequences. Todd Barkman is thanked for helpful discussions. Kandis Elliot and Sara Friedrich are thanked for assistance with images. This work was supported by a NIH-Genetics Training Grant, a Sci-Med GRS Fellowship, a National Academy of Science Ford Foundation Dissertation Fellowship, and a NSF DBI-1103693 award to TRM and a Hamel Family Faculty Fellowship to DAB.
Footnotes
Additional supporting information may be found in the online version of this article.
Fig. S1 Phenotypes and sections of petals used for HPLC analyses.
Fig. S2 Phenotypes, age, and sections of petals used for qualitative RT-PCR analyses.
Fig. S3 Stages of C. gracilis petal development.
Fig. S4 Genotypes at the P and I loci and expected phenotypes.
Fig. S5 Phylogenetic trees of anthocyanin pathway genes in section Rhodanthos.
Fig. S6 Dfr2-A/B expression and Actin control in C. gracilis plants derived from seeds collected in natural populations.
Fig. S7 Alignment of the six different Dfr sequences found in C. gracilis.
Table S1 Voucher informatio
Table S2 Primers used in qualitative and quantitative PCR assay
Table S3 Primers used and expected sizes for Dfr2 genotype assays for P cros
Table S4 Primers used and expected sizes for Dfr2 genotype assays for I cros
Notes S1 Text files containing alignments used for phylogenetic analyse
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
References
- Abdel-Hameed F, Snow R. The origin of the allotetraploid Clarkia gracilis. Evolution. 1972;26:74–83. doi: 10.1111/j.1558-5646.1972.tb00175.x. [DOI] [PubMed] [Google Scholar]
- Albert NW, Lewis DH, Zhang H, Schwinn KE, Jameson PE, Davies KM. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. The Plant Journal. 2011;65:771–784. doi: 10.1111/j.1365-313X.2010.04465.x. [DOI] [PubMed] [Google Scholar]
- Beldade P, Brakefield PM, Long AD. Contribution of Distal-less to quantitative variation in butterfly eyespots. Nature. 2002;415:315–318. doi: 10.1038/415315a. [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia YJ, Blount J, Dixon RA, Lamb C. Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell. 2000;12:2383–2393. doi: 10.1105/tpc.12.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey CC, Strahle JT, Selinger DA, Chandler VL. Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell. 2004;16:450–464. doi: 10.1105/tpc.018796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB, Gates J, Keys DN, Paddock SW, Panganiban GB, Selegue JE, Williams JA. Pattern formation and eyespot determination in butterfly wings. Science. 1994;265:109–114. doi: 10.1126/science.7912449. [DOI] [PubMed] [Google Scholar]
- Cone K, Burr F, Burr B. Molecular analysis of the maize anthocyanin regulatory locus C1. Proc Natl Acad Sci USA. 1986;83:9631–9635. doi: 10.1073/pnas.83.24.9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesMarais D, Rausher MD. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature. 2008;454:762–766. doi: 10.1038/nature07092. [DOI] [PubMed] [Google Scholar]
- Dorn P, Bloom W. Anthocyanin variation in an introgressive complex in Clarkia. Biochemical Systematics and Ecology. 1984;12:311–314. [Google Scholar]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- Eckhart M, Rushing S, Hart M. Frequency dependent pollinator foraging in polymorphic Clarkia xantiana ssp. xantiana populations: implications for flower colour evolution and pollinator interactions. Oikos. 2006;112:412–421. [Google Scholar]
- Elomaa P, Mehto M, Kotilainen M, Helariutta Y, Nevalainen L, Teeri T. A bHLH transcription factor mediates organ, region and flower type specific signals on dihydroflavonol-4-reductase (dfr) gene expression in the inflorescence of Gerbera hybrida (Asteraceae) The Plant Journal. 1998;16:93–99. doi: 10.1046/j.1365-313x.1998.00273.x. [DOI] [PubMed] [Google Scholar]
- Fischer T, Halbwirth H, Meisel B, Stich K. Molecular cloning, substrate specificity of the functionally expressed dihydroflavonol 4-reductases from Malus domestica and Pyrus communis cultivars and the consequences for flavonoid metabolism. Archives of Biochemistry and Biophysics. 2003;412:223–230. doi: 10.1016/s0003-9861(03)00013-4. [DOI] [PubMed] [Google Scholar]
- Gerats AGM, de Vlaming P, Doodeman M, Al B, Schram AW. Genetic control of the conversion of dihydroflavonols into flavonols and anthocyanins in flowers of Petunia hybrida. Planta. 1982;155:364–368. doi: 10.1007/BF00429466. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Zhao M, Leavitt JM, Lloyd AM. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. The Plant Journal. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- Goodrich J, Carpenter R, Coen ES. A common gene regulates pigmentation pattern in diverse plant species. Cell. 1992;68:955–964. doi: 10.1016/0092-8674(92)90038-e. [DOI] [PubMed] [Google Scholar]
- Gottlieb L, Ford V. Genetic studies of the pattern of floral pigmentation in Clarkia gracilis. Heredity. 1988;60:237–246. [Google Scholar]
- Goulson D, Mcguire K, Munro EE, Adamson S, Colliar L, Park KJ, Tinsley MC, Gilburn AS. Functional significance of the dark central floret of Daucus carota (Apiaceae) L.; is it an insect mimic? Plant Species Biology. 2009;24:77–82. [Google Scholar]
- Harborne JB. Phytochemical methods: a guide to modern techniques of plant analysis. 2. London, UK: Chapman and Hall; 1984. [Google Scholar]
- Hopkins R, Rausher MD. Indentification of two genes causing reinforcement in the Texas wildflower Phlox drummondii. Nature. 2011;469:411–414. doi: 10.1038/nature09641. [DOI] [PubMed] [Google Scholar]
- Jackson D, Roberts K, Martin C. Temporal and spatial control of expression of anthocyanin biosynthetic genes in developing flowers of Antirrhinum majus. The Plant Journal. 1992;2:425–434. [Google Scholar]
- Johnson E, Ryu S, Yi H, Shin B, Cheong H, Choi G. Alteration of a single amino acid changes the substrate specificity of dihydroflavonol-4-reductase. The Plant Journal. 2001;25:325–333. doi: 10.1046/j.1365-313x.2001.00962.x. [DOI] [PubMed] [Google Scholar]
- Johnson S, Midgley J. Fly pollination of Gorteria diffusa (Asteraceae), and a possible mimetic function for dark spots on the capitulum. American Journal of Botany. 1997;84:429–436. [PubMed] [Google Scholar]
- Jones K. Pollinator behavior and postpollination reproductive success in alternative floral phenotypes of Clarkia gracilis (Onagraceae) International Journal of Plant Sciences. 1996;157:733–738. [Google Scholar]
- Keys DN, Lewis DL, Selegue JE, Pearson BJ, Goodrich LV, Johnson RL, Gates J, Scott MP, Carroll SB. Recruitment of a hedgehog regulatory circuit in butterfly eyespot evolution. Science. 1999;283:532–534. doi: 10.1126/science.283.5401.532. [DOI] [PubMed] [Google Scholar]
- Lin-Wang K, Bolitho K, Grafton K, Kortstee A, Karunairetnam S, McGhie TK, Espley RV, Hellens RP, Allan AC. An R2R3 MYB transcription factor associated with regulation of the anthocyanin biosynthetic pathway in Rosaceae. BMC plant biology. 2010;10:50. doi: 10.1186/1471-2229-10-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig SR, Habera LF, Dellaporta SL, Wessler SR. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the Myc-homology region. Proc Natl Acad Sci USA. 1989;86:7092–7096. doi: 10.1073/pnas.86.18.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. MacClade 4: analysis of phylogeny and character evolution. Sunderland, USA: Sinauer Associates; 2000. [DOI] [PubMed] [Google Scholar]
- Martin C, Prescott A, Mackay S, Bartlett J, Vrijlandt E. Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. The Plant Journal. 1991;1:37–49. doi: 10.1111/j.1365-313x.1991.00037.x. [DOI] [PubMed] [Google Scholar]
- Morita Y, Saitoh M, Hoshino A, Nitasaka E, Iida S. Isolation of cDNAs for R2R3-MYB, bHLH and WDR transcriptional regulators and identification of c and ca mutations conferring white flowers in the Japanese morning glory. Plant Cell Physiol. 2006;47:457–470. doi: 10.1093/pcp/pcj012. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. Pattern formation on Lepidopteran wings: determination of an eyespot. Developmental Biology. 1980;80:267–274. doi: 10.1016/0012-1606(80)90403-0. [DOI] [PubMed] [Google Scholar]
- Niu S-S, Xu C-J, Zhang W-S, Zhang B, Li X, Lin-Wang K, Ferguson IB, Allan AC, Chen K-S. Coordinated regulation of anthocyanin biosynthesis in Chinese bayberry (Myrica rubra) fruit by a R2R3 MYB transcription factor. Planta. 2010;231:887–899. doi: 10.1007/s00425-009-1095-z. [DOI] [PubMed] [Google Scholar]
- Parker HG, VonHoldt BM, Quignon P, Margulies EH, Shao S, Mosher DS, Spady TC, Elkahloun A, Cargill M, Jones PG, et al. An expressed Fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science. 2009;325:995–998. doi: 10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J, Wienand U, Peterson P, Saedler H. Molecular cloning of the c locus of Zea mays: a locus regulating the anthocyanin pathway. EMBO J. 1986;9:315–321. doi: 10.1002/j.1460-2075.1986.tb04291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987;6:3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky E, Raguso R, Lewinsohn E. Floral scent production in Clarkia (Onagraceae) (I. Localization and developmental modulation of monoterpene emission and linalool synthase activity) Plant Physiology. 1994;106:1533–1540. doi: 10.1104/pp.106.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Mol J, Koes R. Analysis of bHLH and MYB domain proteins: species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. The Plant journal. 1998;13:475–488. doi: 10.1046/j.1365-313x.1998.00046.x. [DOI] [PubMed] [Google Scholar]
- Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol J, Koes R. Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell. 1999;11:1433–1444. doi: 10.1105/tpc.11.8.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RD, Serfas MS. Butterfly wing pattern evolution is associated with changes in a Notch/Distal-less temporal pattern formation process. Current Biology. 2004;14:1159–1166. doi: 10.1016/j.cub.2004.06.046. [DOI] [PubMed] [Google Scholar]
- Robinson G, Robinson R. CLXXXII. A SURVEY OF ANTHOCYANINS. I. Biochem J. 1931;25:1687–1705. doi: 10.1042/bj0251687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito R, Fukuta N, Ohmiya A, Itoh Y, Ozeki Y, Kuchitsu K, Kakayama M. Regulation of anthocyanin biosynthesis involved in formation of marginal picotee petals in Petunia. Plant Science. 2006;170:828–834. [Google Scholar]
- Schwinn K, Venail J, Shang Y, Mackay S, Alm V. A small family of MYB-regulatory genes controls floral pigmentation intensity and patterning in the genus Antirrhinum. The Plant Cell. 2006;18:831–851. doi: 10.1105/tpc.105.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Venail J, Mackay S, Bailey PC, Schwinn KE, Jameson PE, Martin CR, Davies KM. The molecular basis for venation patterning of pigmentation and its effect on pollinator attraction in flowers of Antirrhinum. New Phytologist. 2011;189:602–615. doi: 10.1111/j.1469-8137.2010.03498.x. [DOI] [PubMed] [Google Scholar]
- Shimada N, Sasaki R, Sato S. A comprehensive analysis of six dihydroflavonol 4-reductases encoded by a gene cluster of the Lotus japonicus genome. Journal of Experimental Botany. 2005;56:2573–2585. doi: 10.1093/jxb/eri251. [DOI] [PubMed] [Google Scholar]
- Smith S, Rausher MD. Gene loss and parallel evolution contribute to species difference in flower color. Mol Biol Evol. 2011;28:2799–2810. doi: 10.1093/molbev/msr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis P. Anthocyanidin variation in Clarkia. Biochemical Systematics and Ecology. 1986;14:487–489. [Google Scholar]
- Spelt C, Quattrocchio F, Mol JNM, Koes R. Anthocyanin1 of petunia encodes a basic helix-loop-helix protein that directly activates transcription of structural anthocyanin genes. Plant Cell. 2000;12:1619–1631. doi: 10.1105/tpc.12.9.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelt C, Quattrocchio F, Mol J, Koes R. ANTHOCYANIN1 of petunia controls pigment synthesis, vacuolar pH, and seed coat development by genetically distinct mechanisms. Plant Cell. 2002;14:2121–2135. doi: 10.1105/tpc.003772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sunderland, USA: Sinauer Associates; 2002. [Google Scholar]
- Tornielli G, Koes R, Quattrocchio F. The genetics of flower color. In: Gerats T, Strommer J, editors. Petunia, Evolutionary, Developmental and Physiological Genetics. New York, USA: Springer; 2009. pp. 269–299. [Google Scholar]
- Van Kleunen M, Nänni I, Donaldson J, Manning J. The role of beetle marks and flower colour on visitation by monkey beetles (Hopliini) in the greater cape floral region, South Africa. Annals of Botany. 2007;100:1483–1489. doi: 10.1093/aob/mcm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten N, Quattrocchio F, Mol J, Koes R. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 1997;11:1422–1434. doi: 10.1101/gad.11.11.1422. [DOI] [PubMed] [Google Scholar]
- Walker AR. The TRANSPARENT TESTA GLABRA1 Locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 Repeat protein. Plant Cell. 1999;11:1337–1349. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319:1527–1530. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.