Abstract
Calmodulin (CaM) is an important regulator of Kv7.x (KCNQx) voltage-gated potassium channels. Channels from this family produce neuronal M-currents and cardiac and auditory IKS currents, and harbor mutations that cause arrhythmias, epilepsy, and deafness. Despite extensive functional characterization, biochemical and structural details of the interaction between CaM and the channel have remained elusive. Here, we show that both apo-CaM and Ca2+/CaM bind to the C-terminal tail of the neuronal channel Kv7.4 (KCNQ4), which is involved both hearing and mechanosensation. Interactions between apo-CaM and the Kv7.4 tail involve two C-terminal tail segments, known as the A and B segments, whereas the interaction between Ca2+/CaM and the Kv7.4 C-terminal tail requires only the B segment. Biochemical studies show that the calcium dependence of the CaM:B segment interaction is conserved in all Kv7 subtypes. X-ray crystallographic determination of the structure of the Ca2+/CaM:Kv7.4 B segment complex shows that Ca2+/CaM wraps around the Kv7.4 B segment, which forms an α-helix, in an antiparallel orientation that embodies a variation of the classic 1-14 Ca2+/CaM interaction motif. Taken together with the context of prior studies, our data suggest a model for modulation of neuronal Kv7 channels involving a calcium-dependent conformational switch from an apo-CaM form that bridges the A and B segments to a Ca2+/CaM form bound to the B-helix. The structure presented here also provides a context for a number of disease causing mutations and for further dissection of the mechanisms by which CaM controls Kv7 function.
Introduction
Pore-forming subunits of the Kv7.x (KCNQx) potassium channel family (Kv7.1-Kv7.5) (KCNQ1-KCNQ5)1; 2; 3 form the classically studied M-current in neurons (Kv7.2-7.5)4; 5; 6 and IKS current (Kv7.1) in heart, vestibular, and auditory cells2; 7; 8; 9. Kv7 channels open at subthreshold membrane potentials, do not inactivate, and therefore, provide a strong brake on membrane excitation4; 10. Consequently, a variety of signaling pathways tune electrical excitability by regulating Kv7 function3; 4; 6. Commensurate with their pivotal role in controlling excitation, Kv7 channels are targets for the development of modulators to treat diseases involving neuronal hyperexcitability such as epilepsy and neuropathic pain11; 12; 13; 14. Further, mutations in Kv7 channels have been linked to various human diseases, including cardiac arrhythmias, deafness, and epilepsy2; 14; 15.
Kv7 channels have the canonical six-transmembrane architecture found throughout the voltage-gated ion channel family16 (Figure 1A). The Kv7 cytoplasmic C-terminal tail (~300-400 residues) comprises approximately half of the total residues of the pore-forming subunit and is composed of four conserved elements known as segments A-D (Figure 1B, Supplementary Figure S1)17; 18. This region forms an important, multifunctional entity that is central to channel assembly, gating, regulation, and the formation of complexes with regulatory factors17 including calmodulin (CaM)19; 20; 21; 22; 23. Although Kv7 function has been characterized extensively4; 6; 10; 14, structural knowledge for this channel class is limited to a portion of the cytoplasmic C-terminal tail, called the D-helix or A-domain tail 18; 24; 25, that is important for subunit specific assembly. Strikingly, over 60% of the known Kv7 disease mutations occur in the cytoplasmic C-terminal tail where their direct functional consequences are not obvious18; 26; 27; 28. Thus, defining the structure that underlies this hub of Kv7 regulation remains a key objective for understanding both the basic mechanisms by which Kv7 channels are regulated and assembled and for understanding how disease mutations cause channel dysfunction.
Figure 1. Kv7 C-terminal tail and interactions with calmodulin.
A, Kv7 cartoon diagram Two of the four transmembrane subunits are shown. Transmembrane segments are labeled S1-S6. The four C-terminal tail segments, A-D, are indicated in dark green, red, orange, and purple, respectively. The D segment/A domain Tail is shown in cartoon form using the Kv7.4 D segment/A domain Tail structure24. Dashed lines indicate non-conserved C-terminal end. B, Sequence comparisons of Kv7 tail A, B, C, and D segments. Colors are as in panel ‘A’. Secondary structure of the B segment is indicated. Blue and green circles indicate Ca2+/N-lobe and Ca2+/C-lobe anchors, respectively. Open black circles indicate the 1-10-14 motif. Filled grey and black circles under the B segments indicate the two suggested 1-5-10 motifs from23 C, Superdex 200 chromatography of the CaM:Q4AD complexes in the presence (top) and absence (bottom) of calcium. Inset shows peak fraction components. Cartoon represents the state investigated complex D, Exemplar equilibrium sedimentation data from 238 nm for the 20 μM of the apo-CaM:Q4AD complex at 7000 rpm. Inset shows residuals from a single species fit. Blue, green, yellow, and red curves show the expected distributions for 4:4, 3:4, 2:4, and 1:4 CaM:Q4AD complexes, respectively. Cartoon represents the state investigated complex. CaM binding orientations in all cartoons are for illustrative purposes only.
One of the key Kv7 modulation pathways involves interaction between the calcium sensor CaM19; 29; 30 and the Kv7 C-terminal cytoplasmic domain29. CaM affects the function of all Kv7 subtypes19; 20; 21; 29; 31; 32 by binding to distinct regions within the cytoplasmic C-terminal tail19; 22; 23. Two of these, the A and B segments, have sequence features that have been proposed as CaM binding motifs, an IQ motif in the A segment and two adjacent 1-5-10 motifs in the B segment23. Studies of the calcium dependency of the CaM interaction with Kv7.118; 20; 21 and Kv7.2, 7.3, and 7.433 C-terminal tails indicate that both forms of CaM, apo-CaM and Ca2+/CaM, bind to the channel. However, the details of the precise interactions and calcium dependencies remain controversial17; 22; 34 and exactly how CaM binds to the channel and how CaM-mediated calcium signals affect channel activity remains unclear.
Here, in an effort to unravel the structural details of the CaM-Kv7 association, we examined the interactions of CaM with the Kv7.4 C-terminal domain. Kv7.4 is a neuronal channel that has been shown to be important for mechanosensation in cochlear outer hair cells35; 36 and touch-sensitive dorsal root ganglia neurons37 and carries mutations causing autosomal dominant hearing loss known as nonsyndromic sensorineural deafness type 2 (DNFA2)36. Systematic delineation of the Ca2+/CaM interaction site shows that that the B segment alone constitutes a minimal binding module for Ca2+/CaM. Determination of the crystal structure of the Ca2+/CaM:Kv7.4 B helix complex at 2.60 Å resolution revealed that Ca2+/CaM binds the B helix in an antiparallel orientation that uses a novel variation of the 1-14 Ca2+/CaM binding motif. This structure together with our biochemical studies suggests that there is a conformational switch between the apo-CaM and Ca2+/CaM states that involves a change in binding mode from a crossbridged form using the A and B segments to an antiparallel binding interaction focused on the B-helix.
Results
CaM binds to the Kv7.4 C-terminal tail independently of calcium
To investigate the interaction of CaM with the Kv7.4 C-terminal tail, we co-expressed CaM and a construct bearing the Kv7.4 cytoplasmic C-terminus elements A-D (residues 319-645) fused C-terminal to a His6-Maltose binding protein-TEV protease tag, HMT, (termed “HMT-Q4AD”) in Escherichia coli and purified the complex under two conditions: high calcium (1 mM CaCl2) and low calcium (1 mM ethylene glycol tetraacetic acid, EGTA). Consistent with previous results for the purified Kv7.1 C-terminal tail18 and with pulldown experiments reported for various glutathione S-transferase (GST) fusions of Kv7.2, Kv7.3, and Kv7.4 C-terminal tails33, we found by size exclusion chromatography (SEC) measurements that the purified CaM:Q4AD complexes remained associated independent of the presence of calcium (Figure 1C).
In the SEC experiments, the purified complexes elute much earlier than what would be predicted for a globular protein of the same size (apparent molecular weight ~380 kDa vs. the expected molecular weight for a 4:4 complex of 211 kDa). Considering that the apparent molecular weight from SEC relies on the hydrodynamic radius and is thus, directly affected by the shape of the protein in question38, we set out to assess the oligomerization state of the complex more accurately by equilibrium sedimentation experiments, which provide shape-independent mass information39; 40. Under low calcium conditions (1 mM EDTA), we observed a mass distribution that could be fit by a single species model and that revealed an apparent molecular weight of 208 ± 17 kDa (Figure 1D), a value having excellent agreement with the expected molecular weight of a 4:4 CaM:Q4AD complex (221 kDa). This behavior was maintained over a range of starting concentrations (3-40 μM). The 4:4 nature of this complex and the similarity of the SEC behavior between the 1 mM EDTA and 1 mM Ca2+ and conditions suggest that both the apo and calcium bound forms of the CaM:Q4AD complex have a 4:4 stoichiometry.
Identification of Ca2+-CaM and apo-CaM binding sites on the Kv7 C-terminal tail
Previous studies indicated that the Kv7 C-terminal tail A and B segments are candidate CaM binding elements6; 18; 20; 21; 22; 23; 33, however, opposing observations about the specific role of these domains in the interaction have prevented the proposal of a consensus model. In order to address the contribution of each of the segments to CaM binding within the Kv7.4 (KCNQ4) C-terminal tail, we co-expressed CaM with Kv7.4 (KCNQ4) constructs bearing helices B-D (residues 522-645, denoted as ‘Q4BD’) or C-D (residues 546-645, denoted ‘Q4CD’) to compare their CaM binding behaviors with the Q4AD construct. Pulldown experiments of the HMT-Q4BD fusion co-expressed with CaM revealed an obvious difference from the Q4AD construct. Although capture of HMT-Q4BD in the presence of 1 mM CaCl2 brought down CaM (Figure 2A), similar experiments in the presence of 1 mM EDTA failed to pull down CaM even though sufficient amounts had been co-expressed with the HMT-Q4BD fusion (Figure 2B). Experiments with the HMT-Q4CD construct, which lacks both potential CaM binding elements, showed an absence of interaction with CaM in either the presence or absence of calcium (Figure 2C). These data indicate that the B segment is indispensable for the interaction and agree with similar results obtained for a previously reported GST-fusion construct of a similar portion of Kv7.223. Thus, together with the experiments on the Q4AD construct, our data support the notion that the B segment has a prominent role in Ca2+/CaM binding (Figure 2A and 2C), whereas the A element appears essential for apo-CaM binding (Figures 1C, D, and 2B) and are in agreement with prior studies of these elements19; 23.
Figure 2. Kv7.4 B-D interactions with calmodulin.
A, SDS-PAGE analysis of a pulldown of Ca2+/CaM with Kv7.4 B-D (HMT-Q4BD). B, SDS-PAGE analysis of pulldown of apo-CaM and Kv7.4 B-D (HMT-Q4BD). C, SDS-PAGE analysis of a pulldown of Ca2+/CaM and apo-CaM with Kv7.4 C-D (HMT-Q4CD). D, Superdex200 size exclusion chromatography showing the behavior of the CaM:Q4BD complex in the presence of 1 mM CaCl2 (blue) and 1 mM EDTA (black). Inset shows peak fractions. E, Exemplar equilibrium sedimentation data for the Ca2+/CaM:Q4BD complex. Inset shows residuals. Blue, green, orange, and red curves show expected mass distributions for a 4:4, 3:4, 2:4, and 1:4 Ca2+/CaM:Q4BD complex. F, Exemplar SDS-PAGE analysis of pulldowns showing the calcium dependent interaction of CaM with each of the Kv7 isoforms: Kv7.1 (Q1BD), Kv7.2 (Q2BD), Kv7.3 (Q3BD), and Kv7.5 (Q5BD). Cartoons indicate the species investigated in each panel.
We were able to purify the Ca2+/CaM:Q4BD complex and analyze its properties by SEC. These experiments showed that in the presence of calcium, the two components associated in an assembly that was a least as large as a 4:4 complex (Figure 2D). In contrast, addition of EDTA to the purified Ca2+/CaM:Q4BD complex just prior to SEC resulted in a single peak that was much smaller than the complex and that contained only CaM (Figure 2D). Under these conditions, the Q4BD portion of the sample precipitated once CaM was disengaged, explaining the absence of the Q4BD component. This behavior is decidedly different from the Q4AD construct in which CaM remained associated in both Ca2+/CaM and apo-CaM states (Figure 1C) and is consistent with the inability of the HMT-Q4BD fusion to pull down apo-CaM (Figure 2B). Assessment of the oligomerization state of the Ca2+/CaM:Q4BD complex by equilibrium sedimentation experiments using a range of starting concentrations (10-40 μM) revealed a molecular weight of 124 ± 4 kDa indicating a 4:4 Ca2+/CaM:Q4BD stoichiometry (expected 125 kDa) (Figure 2E). This stoichiometry matches that of the Ca2+/CaM:Q4AD complex and confirms that helix A is dispensable for tetramerization of the C-terminal tail. Pulldown experiments using similar BD constructs from the other Kv7 isoforms demonstrates that all are capable of interacting with Ca2+/CaM (Figure 2F) and indicates that the Ca2+/CaM:Kv7 B segment interaction is common, is calcium dependent in all Kv7 subtypes, and shared by the entire Kv7 family. Size exclusion chromatography of these constructs shows that similar to Q4BD, both Ca2+/CaM:Q2BD and Ca2+/CaM:Q5BD complexes produce clean, well-behaved, tetrameric material (Supplementary Figure S2).
Kv7 segment B is necessary and sufficient for Ca2+-CaM binding
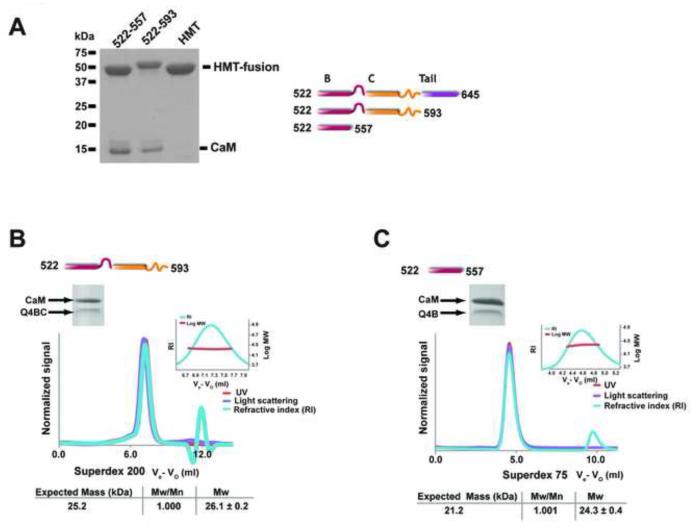
To define the Ca2+/CaM binding site better, we examined C-terminal deletions of the C and D regions for their ability to bind Ca2+/CaM. Truncation of either the D segment/A domain Tail (Q4BC, 522-593) or both the C segment and D segment/A domain Tail (Q4B, 522-557) failed to disrupt Ca2+/CaM binding (Figure 3A). Further investigation by size exclusion chromatography coupled with multiangle light scattering (SEC-MALS)41; 42 showed that at 35 μM the Ca2+/CaM:Q4BC (residues 522-593) complex forms a monodisperse complex having a 1:1 stoichiometry (Figure 3B). This result differs from a previous report that indicated that the Kv7.1 C helix alone is dimeric at concentrations below 100 μM18. This discrepancy may be due to the fact that the Kv7.1 C segment is different in sequence from that of Kv7.4 (Figure 1B) or that the presence of Ca2+/CaM perturbs the ability of the C segment to self-associate. SEC-MALS investigation of the Ca2+/CaM:Q4B (residues 522-557) complex found a monodisperse entity having a 1:1 Ca2+/CaM:Q4B stoichiometry (Figure 3C). The absence of higher order complexes provides further evidence that the C and D segments are necessary for oligomerization18; 24; 43; 44; 45 and shows that Ca2+/CaM binding is independent of the oligomeric state of the channel C-terminal tail. The data further demonstrate that the B segment is both necessary and sufficient for Ca2+/CaM binding.
Figure 3. Characterization of Ca2+/CaM:Kv7.4 B-D interactions.
A, SDS-PAGE analysis of a pulldown of Ca2+/CaM with indicated Kv7.4 BD truncations. Cartoons indicate the domain structure of the various constructs. B and C, Exemplar SEC-MALS characterization of B, Ca2+/CaM:Kv7.4 B-C (Q4BC) complex loaded at a concentration of 35 μM on a Superdex 200 column run in a buffer of 200 mM KCl, 1 mM CaCl2, 10 mM HEPES, pH 7.4. C, Ca2+/CaM:Kv7.4 B (Q4B) complex loaded at a concentration of 80 μM on a Superdex 75 column run in a buffer of 200 mM KCl, 1 mM CaCl2, 10 mM HEPES, pH 7.4. Insets show molecular weight distribution across the main peak and peak fraction SDS-PAGE analysis. MALS derived parameters and expected molecular masses for a 1:1 Ca2+/CaM:Kv7.4 complex are shown.
Structure of the Ca2+-CaM/Kv7.4 (KCNQ4) B helix complex
We were able to grow crystals from hanging drop setups containing the Ca2+/CaM:Q4BC (Kv7.4 residues 522-593) complex. The best of these was a hexagonal crystal form that diffracted X-rays to 2.60 Å resolution (Table 1). Structure determination by molecular replacement using the N-lobe polypeptide from the Ca2+/CaM:smooth muscle myosin light chain kinase (sMLCK) peptide complex46 showed that the asymmetric unit contained only a single Ca2+/CaM:peptide complex comprising one Ca2+/CaM bound to an α-helix formed by Kv7.4 B segment residues Asp524-Phe549 (Figure 4A and Supplementary Figure S3). Subsequent mass spectroscopy analysis of the crystallization mixture revealed that the Q4BC construct had been truncated at residue 551 during the process of crystallization leading to the hexagonal crystal. This deletion essentially removed the entire C segment (cf. Methods) and explains its absence from the structure.
Table 1.
X-ray Data collection and Refinement Statistics
| Ca2+/CaM:Kv7.4 B-helix complex | |
|---|---|
| Data collection | |
| Wavelength (Å) | 1.006 |
| Resolution (Å) | 50-2.60 (2.74-2.60) |
| Space group | P6522 |
| Cell dimensions | |
| a, b, c (Å) | 104.01, 104.01, 113.72 |
| α, β, γ (°) | 90, 90, 120 |
| Rsym(%) | 10.3 (96.4) |
| Rpim(%) | 2.6 (28.3) |
| I/σI | 19.1 (2.8) |
| Completeness (%) | 100 (100) |
| Redundancy | 16.7(12.7) |
| Refinement | |
| Number of reflections (work/test) | 11699/562 |
| Rwork/R free | 24.0/26.5 |
| Number of atoms | |
| Protein | 1340 |
| Water | 18 |
| Average B-factor (Å2) | 27.2 |
| R.M.S. deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.778 |
| Ramachandran Plot | |
| Favored region (%) | 93.2 |
| Allowed region (%) | 6.8 |
Values in parentheses are those for the highest resolution shell
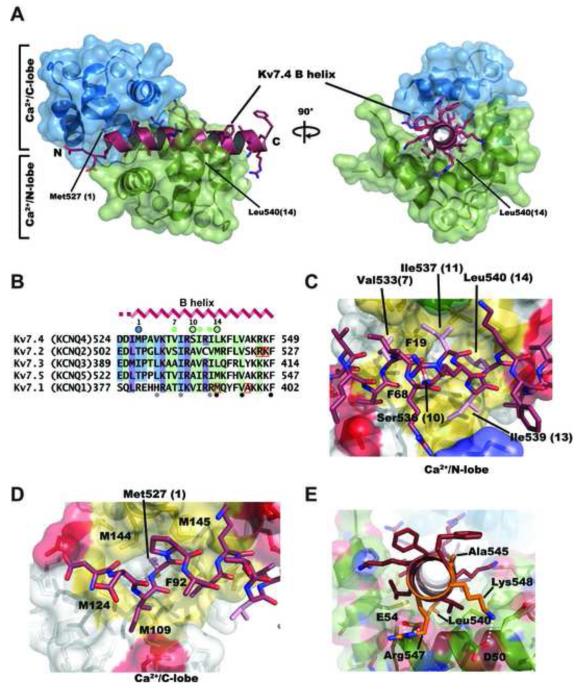
Figure 4. Ca2+/CaM:Kv7.4 B-helix complex structure.
A, Ca2+/CaM:Kv7.4 B-helix complex. Ca2+/CaM N-lobe and C-lobe are shown in green and blue, respectively. Kv7.4 B-helix is shown in firebrick with sidechains shown as sticks. N and C-terminal ends of the B-helix are labeled. Axial view is from the B-helix C-terminal end. Positions of the 1-14 anchors are indicated. B, Sequence comparison of Kv7 B-peptide segments. Residues shaded in blue, green, and purple indicate Kv7.4 positions contacted by Ca2+/C-lobe, Ca2+/N-lobe, and both lobes, respectively. Colors on other Kv7 sequences indicate conserved contact residues. B-helix secondary structure is indicated. Blue and green circles indicate Ca2+/N-lobe and Ca2+/C-lobe anchors, respectively. Open black circles indicate the 1-10-14 motif. Filled grey and black circles under the alignment indicate the two suggested B segment 1-5-10 motifs from23. Orange squares indicate positions of disease mutants in Kv7.1 and Kv7.2 B segments. C, and D, Details of C, Ca2+/C-lobe:Kv7.4 B-helix and D, Ca2+/N-lobe:Kv7.4 B-helix interactions. Ca2+/C-lobe and Ca2+/N-lobe are shown as semi-transparent surfaces and sticks. Hydrophobic, basic, acidic, and polar sidechains from CaM are colored yellow, blue, red, and green, respectively. Kv7.4 B helix is shown as sticks and colored firebrick with nitrogen and oxygen atoms are colored blue and red, respectively. Kv7.4 anchor sidechains are pink. In both panels, key residues of Ca2+/CaM are labeled by single letter code. Kv7.4 B-helix residues are labeled in three letter code followed by the position number within the 1-14 motif, indicated in parenthesis. E, Locations of Kv7.1 and Kv7.2 disease mutants, colored orange and indicated by three letter code, mapped on the Ca2+/CaM:Kv7.4 B helix structure. Ca2+/N-lobe is colored green with nitrogen and oxygen atoms colored blue and red, respectively. CaM residues are indicated by single letter code. White dashed lines indicate the electrostatic interactions for Arg547-Glu54 (2.1 and 2.7 Å), and Lys548-Asp50 (4.2Å).
The structure revealed that Ca2+/CaM wraps around the Kv7.4 B helix in an anti-parallel orientation in which the Ca2+/C-lobe and Ca2+/N-lobe bind to the N-terminal and C-terminal portions of the target helix, respectively. This interaction buries a substantial amount of surface area, 2831 Å2, of which 1570 Å2 is hydrophobic. Kv7.4 B helix residues Met527 and Leu540 form the most distant Ca2+/CaM anchors and comprise a 1-14 motif 47; 48. Hence, we indicate each of the residues in Helix B by both the residue number and the indicator number 1 through 14 with respect to this motif (Figure 4B). This binding motif was first described in Ca2+/CaM complexes with the M13 peptide from skeletal muscle myosin light chain kinase (sMLCK)46; 49 and is regarded as a motif that is specific to Ca2+/CaM binding rather than apo-CaM binding47. Prior studies had suggested that the B segment had two possible 1-5-10 Ca2+/CaM binding motifs (Figures 1B and 4B)23. Although some of the residues in these putative 1-5-10 motifs interact with Ca2+/CaM, neither of these motifs forms the heart of the Ca2+/CaM:Kv7.4 B helix interaction. Further, the conserved aromatic residues at the (16) and (23) positions (Kv7.4 Phe542 and Phe549), including the key 10 position of the second putative 1-5-10 motif (Figure 4B), do not interact with Ca2+/CaM.
Of the two Ca2+-CaM lobes, Ca2+/N-lobe makes more extensive interactions with the B-helix (1764 Å2 versus 1068 Å2 buried surface, for Ca2+/N-lobe and Ca2+/C-lobe, respectively). The C-terminal anchor of the 1-14 motif, Leu540 (14), resides in a pocket formed by Ca2+/N-lobe residues Phe19, Leu32, Met51, Phe68, and Met71 (Figure 4C). Ca2+/N-lobe also has extensive contacts with the B-helix residues Val533 (7), Ser536 (10), Ile537 (11), Ile539 (13), and Leu540 (14) (Supplementary Figure S4). Of these, Val533 (7), Ser536 (10), and Leu540 (14) are the most buried. Ca2+/C-lobe interactions are centered on Kv7.4 Met527 (1), which is buried in a methionine-rich pocket formed from CaM residues Phe92, Leu105, Met109, Met124, Met144, and Met145 (Figure 4D and Supplementary Figure S4). Additionally, a number of B-helix residues form salt bridges and hydrogen bond interactions with Ca2+/N-lobe (Lys 531, Arg547, and Lys548), Ca2+/C-lobe (Arg538), or both (Arg535).
In good agreement with the commonality of the Ca2+/CaM:B segment interaction (Figure 2), the size and hydrophobicity of the majority of key Ca2+/N-lobe contact residues (positions 7, 10, 11, and 14) are well conserved among the Kv7 family (Figure 4B). One Ca2+/N-lobe contacts, position 13 (Kv7.4 Ile539), is an arginine in Kv7.1, but as this residue is not completely buried, it seems possible that the both the hydrophobic and charged natures of the arginine could be accommodated by the binding mode seen for Kv7.4 (Figure 4D). Besides the 1-14 anchors, the other highly buried position is Ser536 (10). The γ-hydroxyl of this residue is buried in a hydrophobic environment (Supplementary Figure S5) in which the nearest polar contact is to the Val533 backbone carbonyl (2.7 Å). Notably, this position is conserved as a small residue among the Kv7s (Figure 4B). From this perspective, the Kv7 B-helix Ca2+/CaM binding motif could be considered a 1-10-14 variation on the 1-14 motif.
The observed interaction network is in line with reported effects of aspartate mutants in the Kv7.2 B segment23 in which a Ser→Asp change at position (7) (Kv7.4 V533) interfered with Ca2+/CaM binding23; 34, whereas an Ala→Asp change at position (19) (Kv7.4 Ala545), which makes no contacts, showed no effect. In contrast to the high conservation of Ca2+/N-lobe anchor sites, Ca2+/C-lobe contact sites (Figure 4B) are more varied. In particular, the identity of the (1) position is not well conserved, and is a positively charged residue in Kv7.1. Other Ca2+/C-lobe contact positions Asp524, Pro528 (2) and Val529 (4), are well conserved among the neuronal channels, Kv7.2-Kv7.5, but all have changes in charge in Kv7.1. It is possible that these differences underlie the apparent difference in the CaM response of Kv7.1 versus the other Kv7s19; 20; 21.
Location of B-helix disease mutants and regulatory sites
Kv7 channels bear a large number of human mutations that cause cardiovascular and hearing disorders and epilepsies15; 26; 50. The conservation of the key anchoring residues in the B-helix (Figure 4B) allows the identification of the positions of a number of known disease mutants from other Kv7 channels within the context of the Ca2+/CaM:Kv7.4 B helix complex structure. Of the four reported B helix disease mutants, three occur at positions that contact the Ca2+/N-lobe. The Kv7.1 mutant M520R51 corresponds to anchor position (14) in the 1-14 motif, Leu540 in Kv7.4 (Figure 4B, D). In accordance with the buried nature of this position, the M520R change has been reported to disrupt both channel function and CaM binding51. Two Kv7.2 mutations, R533Q26; 52 and K562N53 correspond to Kv7.4 positions Arg547 and Lys548 (Figure 4B) that make electrostatic interactions with Ca2+/N-lobe residues Glu54 and Asp50, respectively (Figure 4E) and would disrupt these electrostatic contacts. The fourth reported mutant, Kv7.1 A525T54 corresponds to Kv7.4 Ala545, a residue having no contacts with Ca2+/CaM (Supplementary Figure S4). Because the B segment is involved in both apo-CaM and Ca2+/CaM binding, it is not possible from the structure alone to know whether the disease mutations act by affecting apo-CaM binding, Ca2+/CaM binding, or both. Nevertheless, the structure provides an important context for further studies addressing this question.
In addition to the effects that B-helix mutations have on function, a pair of studies by Shapiro and co-workers has shown that N-ethylmaleimide (NEM) treatment, a channel opener that acts by covalent modification of a cysteine residue that is part of the B segment, blunts the effects of Ca2+/CaM suppression of Kv7.2, Kv7.4, and Kv7.5 currents29; 55. This effect is competitive with CaM action, a result that has suggested overlapping sites of action29. Although the cysteine responsible for this NEM effect was not present in the crystallized construct, it is just five residues away from the N-terminal end of the B-helix (Supplementary Figure S1). Examination of the structure suggests that given the covalent constraints of the polypeptide backbone, this cysteine should be near enough to contact Ca2+/C-lobe and thus, account for the interfering effects of NEM on Kv7 function, CaM binding, and antagonistic relationship between NEM and CaM29. Importantly, both Kv7.1 and Kv7.3, which are NEM insensitive56 lack a cysteine at the equivalent position29 (Supplementary Figure S1).
Comparison of the Ca2+/CaM:KV7.4 B-helix structure with canonical Ca2+/CaM:1-14 motif structures
The original analysis of the B segment sequence predicted two 1-5-10 motifs (Figure 4B)23, neither of which are used in Ca2+/CaM binding, but did not predict the 1-14 motif observed in the Ca2+/CaM:KV7.4 B-helix structure (Figure 4A-D). Hence, we were interested to compare the Ca2+/CaM:KV7.4 B-helix complex to other Ca2+/CaM:1-14 motif complexes to examine points of commonality and differences. Structure comparison with the Ca2+/CaM:smooth muscle myosin light chain kinase (sMLCK) peptide structure46 that originally defined the 1-14 motif, showed that faithful to the 1-14 motif, the anchor position fourteen residue spacing, Trp800 (1) and Leu813 (14) for sMLCK and Met527 (1) and Leu540 (14) for the KV7.4 B-helix, places the KV7.4 B-helix anchor positions on opposite faces of the α-helix where they can independently engage the two Ca2+/CaM lobes (Figure 5A). Notably, comparison of the structures reveals that the Kv7.4 B-helix is longer than the sMLCK helix by ~three turns and makes more Ca2+/N-lobe contacts. Although the overall RMSDCα for both complexes is 2.59 Å2 indicating some global differences that are due in part to the longer target peptide in the Kv7.4 structure, the conformations of the individual Ca2+/CaM lobes are very similar and have Cα RMSDs of 0.91 Å2 and 0.57 Å2 for Ca2+/C-lobe and Ca2+/N-lobe, respectively.
Figure 5. Comparison of the Ca2+/CaM:Kv7.4 B-helix complex with other 1-14 Ca2+/CaM complexes.
A, Superposition of Ca2+/CaM:Kv7.4 B-helix complex with the M13 peptide of smooth muscle myosin light chain kinase (cyan), sMLCK (1CDL)46. sMLCK complex is shown in light blue and cyan. Calcium ions are indicated as white (Kv7.4 complex) and grey (sMLCK) spheres. Residues from sMLCK are labeled in italics. Residues from the Kv7.4 complex are labeled in bold. B-E, Comparisons of Ca2+/CaM:Kv7.4 B-helix complex with other 1-14 Ca2+/CaM complexes. In each case, the (1) position or (14) position residue is shown in sticks and labeled. Residues in italics and surfaces come from the sMLCK, eNOS, DAPK structures. Kv7.4 residues are labeled in bold. Ca2+/CaM residues are indicated by single letter code. In all panels the Ca2+/CaM from the Kv7.4 complex is shown in marine and the Met527 sidechain is shown in maroon. Nitrogen, oxygen, and sulfur atoms are colored blue, red, and yellow, respectively. B, View of the Ca2+/C-lobe interactions surrounding the (1) position of the 1-14 motif for Ca2+/CaM:sMLCK complex. Ca2+/CaM:sMLCK is light blue. sMLCK Trp800 is cyan. sMLCK Ca2+/CaM C-lobe RMSDs are 0.568 and 1.375 for Cα and all atoms, respectively. C, Comparison of the (14) position binding pocket for Ca2+/CaM:sMLCK and Ca2+/CaM:Kv7.4 B-helix. Colors are as in ‘B’. sMLCK Leu813 is cyan. Ca2+/CaM N-lobe RMSDs are 0.914 and 1.636 for Cα and all atoms, respectively. D, Comparison of the (1) position binding pocket for the Ca2+/CaM:eNOS complex (1N1W)57 and Ca2+/CaM:Kv7.4 B-helix. Ca2+/CaM:eNOS complex is shown in orange. eNOS Phe465 is olive. Ca2+/CaM C-lobe RMSDs are 1.085 and 1.520 for Cα and all atoms, respectively. E, Comparison of the position 1 binding pocket for the Ca2+/CaM:DAPK (1YR5) complex and Ca2+/CaM:Kv7.4 B-helix58. Ca2+/CaM:DAPK is shown in white. DAPK Trp305 is tan. Ca2+/CaM C-lobe RMSDs are 0.903 and 1.648 for Cα and all atoms, respectively. F, Sequence comparison of target peptides from the Kv7.4, sMLCK, eNOS, and DAPK structures. Blue and green circles indicate Ca2+/N-lobe and Ca2+/C-lobe anchors. Open black circles indicate the 1-10-14 motif.
Examination of the local environments of the (1) and (14) anchor positions shows that these residues bind pockets formed from the same Ca2+/CaM lobe residues in both the Kv7.4 and sMLCK structures (Figure 5 B, C). The (1) position, Kv7.4 Met527 and sMLCK Trp 800, binds to a pocket formed by Ca2+/C-lobe residues Phe92, Met109, Met124, Met144, and Met145 (Figure 5B). Notably, there is a rather large difference in the way the (1) position residues occupy the Ca2+/C-lobe pocket. Kv7.4 Met527 is much closer to the wall of the pocket made by CaM residue Phe92, whereas sMLCK Trp800 is more central to the pocket. In concert with the changes seen at the Kv7.4 (1) position anchor, CaM residue Met144 adopts a sidechain rotamer that reduces the pocket size relative to the sMLCK complex. This conformational difference is consistent with the change from a large aromatic anchor, Trp800, to a smaller and less constrained hydrophobic residue, Met527 (Figure 5B). In contrast, the Kv7.4 and sMLCK (14) position anchors of, residues Leu813 and Leu540, respectively, occupy a pocket formed by Ca2+/N-lobe residues Phe68, Met71, Leu32 and Phe19 that has similar features in both structures (Figure 5C).
To see whether the differences in the 1-14 interaction were particular to the comparison with the classic sMLCK interaction or more specifically related to the Kv7.4 complex, we conducted a survey to identify all current known 1-14 complexes. This search identified ten Ca2+/CaM:1-14 complexes (Supplementary Table 1). The difference in the placement of the (1) position residue and the shape of the pocket around Ca2+CaM Met144 remain vis-à-vis the Kv7.4 B-helix interactions with respect to all complexes from this set. These differences are exemplified by the Ca2+/CaM:endothelial nitrogen oxide synthase peptide, eNOS, complex (PDB:1NIW)57 (Figure 5D) and an Ca2+/CaM complex with an autoregulatory domain peptide from the death associated protein kinase DAPK (PDB 1YR5)58 (Figure 5E). The structure having a non-aromatic at position (1), Leu in (PDB:3HR4)59, has a Ca2+/C-lobe pocket similar to the other structures, but has a very different pitch of the target helix, which deviates from the other complexes by ~30° and results in a placement of the (1) position residue midway between the aromatic position from sMLCK Trp800 and the position occupied by Kv7.4 Met 527. These observations provide additional support for the idea that the distinctive structural features we note in the Kv7.4 complex relative to other 1-14 complexes results from accommodation of a non-aromatic at the (1) position. The presence of the non-aromatic (1) position residue further stands out as aromatic residues occupy the Ca2+/C-lobe pocket in a large number of Ca2+/CaM complexes60. In light of this analysis, and consideration that the canonical pattern of residues at the 1 and 14 positions is defined by the presence of one of five hydrophobic residues (Phe/Ile/Leu/Val/Trp) at the 1 and 14 positions47, it is clear how the 1-14 pattern could be missed. Notably, none of the Kv7s have a canonical residue at the (1) position (Figure 4B). Undoubtedly, the well-known adaptability of CaM stands behind this observation and underscores the importance of experimental determination of Ca2+/CaM:peptide structures.
Discussion
The ability to open at membrane potentials that are below those required to initiate action potentials together with a lack of inactivation sets Kv7 channels in a key position to control electrical excitability in both the heart and nervous system4; 10. Hence, they are subject to a wide range of regulatory pathways3; 4; 6, have been identified as molecules in which misfunction underlies a variety of disorders in the cardiovascular, hearing, vestibular, and nervous systems2; 14; 15, and are attractive targets for a range of hyperexcitability disorders11; 12; 13; 14. Calmodulin (CaM) is a key regulator and affects the function of all Kv7 subtypes through interactions made in both its apo- and calcium-bound forms with the C-terminal tail of the channel19; 20; 21; 29; 31; 32. Although, the importance of CaM for Kv7 modulation has been long appreciated, the details of the interactions of CaM with the channel have remained unclear. Our data provide new biochemical and structural information that directly bear on the question of how CaM interacts with the channel in both apo- and calcium-bound forms.
In agreement with previous evidence22; 23, we found that apo-CaM binding to the Kv7.4 C-terminal tail strictly requires the presence of both the A and B segments (Figures 1C, 1D, and 2B). Importantly, we show directly by sedimentation equilibrium studies that the interaction between apo-CaM and the Kv7.4 construct spanning the A-D segments (Q4AD) has a stoichiometry of 4:4 (Figure 1D), a result that is consistent with prior sedimentation equilibrium18 and chemical crosslinking studies of a similar Kv7.1 C-terminal domain construct20. This situation is striking as more than one hundred amino acids separate the A and B segments. Such a large insertion precludes a contiguous binding site and as first noted by Villarroel and colleagues23 and suggests that apo-CaM may use a bi-dentate binding mode to interact with these non-contiguous sites. There are a number of precedents that indicate that CaM is able to make this sort of bridged interaction61; 62; 63, most notably, those from the interaction of CaM and the intracellular domains of small-conductance (SK) potassium channel intracellular domains62; 63. Although there are no structural examples of a complete bridged structure formed by apo-CaM, such an interaction would be very much in line with the ability of the independent apo-CaM lobes to engage targets61; 64; 65; 66. It should be noted that our data do not distinguish whether the apo-CaM interaction with the A and B segments is intrasubunit or intersubunit. Prior studies of the oligomerization behavior of an Kv7.1 construct spanning just the A-B segments indicated the presence of a 1:1 complex in both the presence and absence of calcium and suggest that the apo-CaM crossbridge is intrasubunit18. Nevertheless, in the context of the full length channel, both configurations are reasonable possibilities given the proximity of the intracellular domains, which are constrained by both the interactions of the pore forming regions in the membrane at one end and those from the D segment/A domain Tail18; 24 at the other. Thus, based on the available data, we favor the interpretation that the apo-CaM bridging occurs within a single subunit. Nevertheless, further tests in the context of the full-length channel will be required to address this issue definitively.
In contrast to the complex interactions made by apo-CaM, both our structural and biochemical data indicate that Ca2+/CaM requires only the B segment (Figures 3C and 4). This interaction involves a classic, contiguous Ca2+/CaM binding site formed by a single α-helix (Figure 4). Ca2+/CaM wraps around the B segment helix in an antiparallel orientation that uses a variation of the well-described 1-14 binding motif47; 48 in which the two most distant anchoring positions are Met527 and Leu540. This binding mode contrasts with the original suggestion that Ca2+/CaM might interact via 1-5-10 motifs found in the B segment23 (Figure 4B) and has a pattern of buried and exposed residues that matches the reported effects of aspartate mutants on CaM binding23.
Consistent with the observation that constructs bearing the B-helix from all Kv7 isoforms are able to bind Ca2+/CaM (Figure 2F), the main determinants of this interaction are conserved throughout the Kv7 family (Figure 4B). One feature that stands out from other 1-14 Ca2+/CaM target structures is that rather than an aromatic residue at the 1 position, the Kv7.4 B-helix has a methionine. This substitution of an aromatic for long hydrophobic sidechain is readily accommodated by a small rearrangement in the Ca2+/C-lobe binding pocket. Interestingly, all Kv7 isoforms have a non-aromatic residue at the (1) position of the 1-14 motif (Figure 4B).
Functional studies indicate that the neuronal channels Kv7.2, Kv7.4, and Kv7.5, share a common mechanism of calcium-induced current suppression that involves CaM19; 29. However, this property is not shared by all members of the family and most notably, the calcium sensitivity of Kv7.1 has been a point of controversy17. Some studies have reported CaM involvement in calcium-dependent activation of Kv7.1 channels20; 21; 67, whereas other studies have failed to find this response29. Because the modulation of Kv channels is complex3; 4; 6, it is possible that the reported differences are due to the effects of other CaM-dependent pathways, such as the one involved in S-nitrosylation signals32; 67. Regardless, it is clear that Kv7.1 behaves differently with respect to CaM than the neuronal Kv7s. It is notable that even though Ca2+/CaM binds C-terminal constructs bearing the B-helix regardless of the Kv7 isoform tested (Figure 2F), there is a substantial divergence in the residues that would form the Ca2+/C-lobe contacts in Kv7.1 (KCNQ1), including a number of charged residues. Such differences may underlie the functional differences between the CaM-dependent responses of Kv7.1 versus the neuronal Kv7s. Additionally, there are differences at the (1) position that could play a role in tuning the CaM responses of the neuronal Kv7s in a subunit-specific manner. Our new structural data should help to guide further investigation of this phenomenon.
Studies of the calcium dependency of CaM interaction with Kv7.2, 7.3, and 7.4 C-terminal tails indicate that both apo-CaM and Ca2+/CaM bind22; 23; 33. The presence of apo-CaM binding sites in the A22; 23 and B23 segments of Kv7.2, together with the observations that the A/B region binds apo-CaM better than the individual segments23 and that Ca2+/CaM favors binding to the B segment in Kv7.2-Kv7.519 match well with our observations of the interaction seen using purified components. A number of previous proposals have suggested a cross-bridged binding mode for apo-CaM22; 23. Consideration of our new data together with this prior work leads us to propose an extension of the CaM modulation hypothesis to include a conformational switch between the apo-CaM and Ca2+/CaM states that involves a change from a cross-bridged form in the apo-CaM state to a classic Ca2+/CaM-helix interaction in the calcium bound form as seen in our crystal structure of the Ca2+-CaM/Kv7.4 B-helix complex (Figure 6). Because one of the parts of the bi-dentate binding site, the A segment, is very near the likely end of the pore-lining transmembrane segment S6, it is reasonable to posit that such a conformational change would affect channel function. For example, the interaction of apo-CaM with the A segment may effect S6 in a way that favors channel opening and, in contrast, when CaM switches to Ca2+-CaM site on the B-helix, loss of the A segment interaction causes the current to be suppressed. This hypothesis would explain one mechanism for calcium-dependent suppression of neuronal Kv7s. As mentioned above, the response of Kv7.1 to calcium is complicated and unlike that of the neuronal Kv7s. Given that the Kv7.1 B-helix binds Ca2+/CaM, even though the Ca2+/C-lobe site is quite different, Kv7.1 may use some variant of this proposal.
Figure 6. Model for the calcium dependent interactions of neuronal Kv7 channels and CaM.
Cartoon model depicts the proposed calcium dependent switch from the cross-bridged apo-CaM binding state, left, to the Ca2+/CaM bound state. CaM lobes are indicated as green (C-lobe) and blue (N-lobe). Only two of the four subunits are shown. It should be noted that the exact binding mode for the apo-CaM state is not known. Sequences following the D helix/A-domain Tail are omitted from the cartoon for clarity.
The idea that there would be a structural transition between a bi-dentate apo-CaM clamp to a calcium bound form in which the Ca2+/CaM interactions are restricted to the B-helix fits well with the observed, antagonistic effects of NEM alkylation at a B-helix cysteine55 29; 55 and with the data showing the calcium-sensitivity of the N-lobe is crucial29. Such a model would also be consistent with the known importance of the C-terminal tail region that spans the A and B segments for modulation of Kv7 channels by phosphatidylinositol 4,5-bisphosphate (PIP)68 and S-nitrosylation32,2, and suggests that these various inputs can be coupled in a way that makes the A-B region a signal integration domain. In this regard, it is striking that the sequence that links the A and B segments varies among the Kv7 isoforms in comparison to the A, B, C, and D segments (Figure S1), and is the site of Kv7.4 splice variation that affects function69.
Overall, our data are in agreement with the existence of two CaM binding sites on Kv7 channels, one Ca2+-independent and one Ca2+-dependent that confers Ca2+ sensitivity to the channel17. The structure of the Ca2+/CaM:Kv7.4 B-helix complex shows that a number of Kv7 disease mutations fall at positions that make direct contacts with Ca2+/CaM. However, as the B segment is also involved in apo-CaM binding22; 23 and interactions with CaM may have important effects in channel folding and trafficking20; 21; 70; 71, such mutations may interfere with multiple layers of CaM control of the channel. The structure presented here provides a key context for further dissection of the complex effects of Kv7 disease mutants. Finally, the novel variation we observe in the classic 1-14 Ca2+/CaM binding mode underscores the exceptionally adaptive nature of Ca2+/CaM for target recognition.
Materials and Methods
Cloning, expression and purification
Fragments of human Kv7.1-Kv7.5 (KCNQ1-KCNQ5) C-terminal domain were subcloned into either pSV272 (N. Pokala and T.M. Handel, unpublished) or pET28-HMT vectors (Novagen) using NarI/HindIII or NdeI/HindIII sites. Both vectors contain the sequence for a hexahistidine tag followed by a maltose binding protein (MBP) and a tobacco etch virus (TEV) protease site before the gene of interest. The only difference between the two vectors is that the NarI site following the TEV cleavage site in pSV272 is replaced by an NdeI site in pET28-HMT. Full-length native calmodulin and mutants were cloned into pEGST without an affinity tag as previously described72.
Kv7 fragments co-expressed with CaM in Escherichia coli BL21(DE3)pLysS grown in 2YT media at 37°C for between 3-8 hours. Cells containing constructs containing B-D segments were lysed by sonication in buffer containing 100 mM Tris (pH 8.8), 200 mM KCl, 10% sucrose, 1mM MgCl2, 1mM PMSF, 20 μg ml-1 lysozyme, and 25 μg ml-1 DNaseI. 1mM CaCl2 or 1 mM ethylene glycol tetraacetic acid (EGTA) was also added before cell lysis depending on whether calcium-bound or calcium-free forms were the subject of purification. Lysed cells were centrifuged at 25,000 g for 30 minutes to remove insoluble material. For constructs spanning the A-D segments, the 10% sucrose in the lysis buffer was replaced by 10% glycerol and the cell lysis was done using an Emulsiflex C-5 homogenizer (Avestin).
For all complexes, the soluble fraction containing the Kv7 fragment and CaM was purified on a Poros20MC (Perseptive Biosystems) column equilibrated in buffer A (250 mM KCl, 1 mM CaCl2, 10 mM HEPES buffer, pH 7.4). When EGTA was used in the lysis buffer, a buffer of identical composition but lacking CaCl2 replaced buffer A. After loading the sample, the column was washed in two steps: first using 3 column volumes of buffer A, and then using 3 column volumes of buffer A plus 30 mM imidazole to remove weakly bound contaminants. Bound protein was then eluted using an imidazole gradient from 30 mM to 300 mM over 6 column volumes. After repeated spin concentration using a 50 kDa cutoff Amicon ultra-15 concentrator (Millipore) and dilution with buffer A to remove imidazole, the eluate from the Poros20MC column was loaded on to an amylose (New England Biolabs) column and washed with buffer A. The bound protein was then eluted in buffer A plus 10 mM maltose. The purified protein was digested with histidine tagged TEV protease73 in the amylose column elution buffer overnight at room temperature and was subsequently passed over a Poros20MC and amylose column run in Buffer A plus 1 mM CaCl2 to remove the protease and uncleaved material. Flowthough was collected and concentrated using Amicon ultra-15 concentrator with 10 kDa cut-off.
During this process the buffer was exchanged to a buffer of 10 mM KCl, 1mM CaCl2, 10mM Tris, pH8.8 through a cycle of dilution and concentration. The sample was the further purified using Resource Q or HiLoadQ (GE healthcare) anion exchange column and a gradient from 100 mM KCl to 350 mM KCl over 15 column volumes. This step is important for removal of residual His-MBP. Concentration of purified complex determined by measuring the absorption at 280 nm74.
Pulldown experiments
Various HMT tagged Kv7 constructs were co-expressed with CaM as described above. Cells were lysed by sonication and centrifuged at 25000 g for 30 minutes. 500 μl of supernant was incubated with the same amount of buffer A (250 mM KCl, 1 mM CaCl2 or EGTA, 10 mM HEPES, pH 7.4) and 500 μl of amylose resin and incubated for 30 minutes at 4°C. The unbound proteins were washed out using three washes with 5 ml buffer A. Bound proteins were eluted by buffer A plus 10 mM maltose and analyzed by SDS-PAGE.
Crystallization and data collection
The Ca2+/CaM:Kv7.4 B-C complex, containing Kv7.4 residues 522-593, yielded crystals suitable for structural studies grown using hanging drop vapor diffusion from a drop containing equal volumes (1μl each) of purified protein (~20 mg/ml) and a reservoir solution of 1.6M ammonium sulfate, 0.2 M sodium citrate (pH 6.0), 0.1 M sodium tartrate and 4% isopropanol. Rectangular rod shaped crystals appeared in 1-3 weeks at room temperature. Crystals having this morphology diffracted to ~2.8 Å but suffered from high mosaicity, anisotropy, and a large unit cell. One crystal having hexagonal plate morphology appeared in the same conditions subsequent to a round of crystal harvesting from the well in which it grew. This crystal diffracted sufficiently to be used for structure determination. Mass spec analysis revealed that the protein in this particular well had been cleaved C-terminal to residue 551, leaving only the B segment. In contrast, mass spectroscopy analysis of wells containing rod shaped crystals showed the presence of the complete Kv7.4 BC construct. Diffraction data were collected from frozen crystals at Beamline 12-2 (SSRL) data were indexed and processed in the space group P6522 using MOSFLIM and SCALA in the CCP4 package75.
Structure determination
The structure was determined by a molecular replacement search using the N-lobe without calcium ions from the sMLCK complex (PDB code: 1CDL) using PHASER76. Electron density indicating the Kv7.4 B-helix and Ca2+/C-lobe was readily visible in the initial maps. Structure was built by iterative cycles of manual building and refinement using REFMAC77 and PHENIX78. TLS refinement using three groups (Ca2+/N-lobe, Ca2+/C-lobe, and Kv7.4- helix) was added in the last few cycles. Residues 524-549 of Kv7.4 and all of CaM except for a few residues at the N-terminus and in the loop interdomain linker (residues 78-82) were resolved. Structural quality was evaluated using MolProbity79.
Size exclusion Chromatography and MALS-RI analysis
Purified Kv7.4 C-terminal domain:CaM complexes were run through either a Superdex200 10/300 GL or a Superdex75 HR column depending on the size of the complex. The columns were calibrated using at least four standard protein molecular mass markers (HMW and LMW calibration kits, GE Healthcare). The running buffer contained 250mM KCl, 1 mM CaCl2 or 1 mM ethylene diamine tetraacetic acid (EDTA), 10mM HEPES, pH 7.4. When EDTA was used, EDTA was added to the sample to a final concentration of 5 mM and the sample was incubated for 10 minutes before the run.
MALS-RI analysis41; 42 was performed using miniDAWN™ TREOS (LS) and Optilab® T-rEX™ (RI) (Wyatt Technology) for simultaneous detection of UV, refractive index, and multi-angle light scattering signals run in line with a Superdex200 10/300 GL or Superdex75 HR column in a buffer of 200 mM KCl, 1 mM CaCl2, 10mM HEPES, pH 7.4, calibrated using 0.2 mg BSA as an internal standard. ASTRA® 6 software package (Wyatt Technology) was used to compute protein molecular mass (Mw) and protein monodispersity (Mw/Mn) from the UV, refractive index, and multi-angle light scattering outputs.
Equilibrium Sedimentation
Equilibrium sedimentation experiment was carried out at 4° C using a Beckman Optima XL-A analytical ultracentrifuge (Beckman Coulter). For the CaM:Q4AD complex, the buffer contained 250mM KCl, 1mM EDTA, 10mM Tris, pH8.0. For the CaM:Q4BD complex, the buffer contained 1mM CaCl2 instead of EDTA. The purified protein was dialyzed overnight in the final buffer. The dialysis buffer was then used to dilute the protein to the intended concentration and as the reference cell sample. The data processing followed previously described procedures24. The molecular mass of the complex was obtained through a single exponential fit to the distribution of the complex over the radius.
Survey of Ca2+/CaM 1-14 structures
Structures of the Ca2+/CaM:Kv7.4 B helix (4GOW) and Ca2+/CaM:sMLCK (1CDL) 1-14 complexes were used as search queries using the DALI server80 to identify similar Ca2+/CaM complexes. Redundant structures were removed from the comparison and the remaining structures were aligned and manually examined to identify those using a 1-14 binding motif.
Supplementary Material
Ca2+/CaM is a key regulator of Kv7 (KCNQ) function
apo-CaM and Ca2+/CaM bind to the C-terminal tail of the neuronal channel Kv7.4 (KCNQ4)
Ca2+/CaM:B-helix interaction occurs in all Kv7 isoforms
Ca2+/CaM:Kv7.4 B-helix uses a novel variation of the 1-14 interaction
apo-CaM to Ca2+/CaM transition involves a switch from a crossbridged to a non-crossbridged state
Acknowledgements
This work was supported by grants to D.L.M. from NIH R01 DC007664 and the American Heart Association 0740019N and A.T from an EMBO long-term fellowship. We thank Rachel Gate and Susan Shim for excellent technical support, F. Findeisen for helpful comments on the manuscript, and members of the Minor lab for support throughout these studies. D.L.M. is an AHA Established Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
Coordinates and structure factors have been deposited in the Protein Data Bank with the following accession code: 4GOW.
Author contributions
Q.X., A.C, A.T., and D.L.M. conceived the study. Q.X, A.C., and A.T. performed the experiments and analyzed data. D.L.M. analyzed data and provided guidance and support throughout. Q.X., A.C., A.T., and D.L.M. wrote the paper.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Gutman GA, Chandy KG, Adelman JP, Aiyar J, Bayliss DA, Clapham DE, Covarriubias M, Desir GV, Furuichi K, Ganetzky B, Garcia ML, Grissmer S, Jan LY, Karschin A, Kim D, Kuperschmidt S, Kurachi Y, Lazdunski M, Lesage F, Lester HA, McKinnon D, Nichols CG, O’Kelly I, Robbins J, Robertson GA, Rudy B, Sanguinetti M, Seino S, Stuehmer W, Tamkun MM, Vandenberg CA, Wei A, Wulff H, Wymore RS. International Union of Pharmacology. XLI. Compendium of voltage-gated ion channels: potassium channels. Pharmacol Rev. 2003;55:583–6. doi: 10.1124/pr.55.4.9. [DOI] [PubMed] [Google Scholar]
- 2.Jentsch TJ. Neuronal KCNQ potassium channels: Physiology and role in disease. Nature Reviews: Neuroscience. 2000;1:21–30. doi: 10.1038/35036198. [DOI] [PubMed] [Google Scholar]
- 3.Jespersen T, Grunnet M, Olesen SP. The KCNQ1 potassium channel: from gene to physiological function. Physiology (Bethesda) 2005;20:408–16. doi: 10.1152/physiol.00031.2005. [DOI] [PubMed] [Google Scholar]
- 4.Delmas P, Brown DA. Pathways modulating neural KCNQ/M (Kv7) potassium channels. Nat Rev Neurosci. 2005;6:850–62. doi: 10.1038/nrn1785. [DOI] [PubMed] [Google Scholar]
- 5.Wang H-S, Pan Z, Shi W, Brown BS, Wymore RS, Cohen IS, Dixon JE, McKinnon D. KCNQ2 and KCNQ3 potassium channel subunits: Molecular correlates of the M-Channel. Science. 1998;282:1890–1893. doi: 10.1126/science.282.5395.1890. [DOI] [PubMed] [Google Scholar]
- 6.Hernandez CC, Zaika O, Tolstykh GP, Shapiro MS. Regulation of neural KCNQ channels: signalling pathways, structural motifs and functional implications. J Physiol. 2008;586:1811–21. doi: 10.1113/jphysiol.2007.148304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barhanin J, Lesage F, Guillemare E, Fink M, Lazdunski M, Romey G. KVLQT1 and IsK (minK) proteins associate to form the IKs cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 8.Sanguinetti MC, Curran ME, Zou A, Shen J, Spector PS, Atkinson DL, Meating MT. Coassembly of KVLQT1 and minK (IsK) proteins to form cardiac IKs potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Curran ME, Splawski I, Burn TC, Millholland JM, VanRaay TJ, Shen J, Timothy KW, Vincent GM, de Jager T, Schwartz PJ, Toubin JA, Moss AJ, Atkinson DL, Landes GM, Connors TD, Keating MT. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 10.Brown DA, Passmore GM. Neural KCNQ (Kv7) channels. Br J Pharmacol. 2009;156:1185–95. doi: 10.1111/j.1476-5381.2009.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barrese V, Miceli F, Soldovieri MV, Ambrosino P, Iannotti FA, Cilio MR, Taglialatela M. Neuronal potassium channel openers in the management of epilepsy: role and potential of retigabine. Clin Pharmacol. 2010;2:225–36. doi: 10.2147/CPAA.S15369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fritch PC, McNaughton-Smith G, Amato GS, Burns JF, Eargle CW, Roeloffs R, Harrison W, Jones L, Wickenden AD. Novel KCNQ2/Q3 agonists as potential therapeutics for epilepsy and neuropathic pain. J Med Chem. 2010;53:887–96. doi: 10.1021/jm901497b. [DOI] [PubMed] [Google Scholar]
- 13.Wickenden AD, McNaughton-Smith G. Kv7 channels as targets for the treatment of pain. Curr Pharm Des. 2009;15:1773–98. doi: 10.2174/138161209788186326. [DOI] [PubMed] [Google Scholar]
- 14.Soldovieri MV, Miceli F, Taglialatela M. Driving with no brakes: molecular pathophysiology of Kv7 potassium channels. Physiology (Bethesda) 2011;26:365–76. doi: 10.1152/physiol.00009.2011. [DOI] [PubMed] [Google Scholar]
- 15.Maljevic S, Wuttke TV, Lerche H. Nervous system KV7 disorders: breakdown of a subthreshold brake. J Physiol. 2008;586:1791–801. doi: 10.1113/jphysiol.2008.150656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu FH, Yarov-Yarovoy V, Gutman GA, Catterall WA. Overview of molecular relationships in the voltage-gated ion channel superfamily. Pharmacol Rev. 2005;57:387–95. doi: 10.1124/pr.57.4.13. [DOI] [PubMed] [Google Scholar]
- 17.Haitin Y, Attali B. The C-terminus of Kv7 channels: a multifunctional module. J Physiol. 2008;586:1803–10. doi: 10.1113/jphysiol.2007.149187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiener R, Haitin Y, Shamgar L, Fernandez-Alonso MC, Martos A, Chomsky-Hecht O, Rivas G, Attali B, Hirsch JA. The KCNQ1 (Kv7.1) COOH terminus, a multitiered scaffold for subunit assembly and protein interaction. J Biol Chem. 2008;283:5815–30. doi: 10.1074/jbc.M707541200. [DOI] [PubMed] [Google Scholar]
- 19.Gamper N, Shapiro MS. Calmodulin mediates Ca2+-dependent modulation of M-type K+ channels. J Gen Physiol. 2003;122:17–31. doi: 10.1085/jgp.200208783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh S, Nunziato DA, Pitt GS. KCNQ1 assembly and function is blocked by long-QT syndrome mutations that disrupt interaction with calmodulin. Circ Res. 2006;98:1048–54. doi: 10.1161/01.RES.0000218863.44140.f2. [DOI] [PubMed] [Google Scholar]
- 21.Shamgar L, Ma L, Schmitt N, Haitin Y, Peretz A, Wiener R, Hirsch J, Pongs O, Attali B. Calmodulin is essential for cardiac IKS channel gating and assembly: impaired function in long-QT mutations. Circ Res. 2006;98:1055–63. doi: 10.1161/01.RES.0000218979.40770.69. [DOI] [PubMed] [Google Scholar]
- 22.Wen H, Levitan IB. Calmodulin is an auxiliary subunit of KCNQ2/3 potassium channels. J Neurosci. 2002;22:7991–8001. doi: 10.1523/JNEUROSCI.22-18-07991.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yus-Najera E, Santana-Castro I, Villarroel A. The Identification and Characterization of a Noncontinuous Calmodulin-binding Site in Noninactivating Voltage-dependent KCNQ Potassium Channels. J Biol Chem. 2002;277:28545–53. doi: 10.1074/jbc.M204130200. [DOI] [PubMed] [Google Scholar]
- 24.Howard RJ, Clark KA, Holton JM, Minor DL., Jr. Structural insight into KCNQ (Kv7) channel assembly and channelopathy. Neuron. 2007;53:663–75. doi: 10.1016/j.neuron.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Q, Minor DL., Jr. Crystal structure of a trimeric form of the K(V)7.1 (KCNQ1) A-domain tail coiled-coil reveals structural plasticity and context dependent changes in a putative coiled-coil trimerization motif. Protein Sci. 2009;18:2100–14. doi: 10.1002/pro.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soldovieri MV, Miceli F, Bellini G, Coppola G, Pascotto A, Taglialatela M. Correlating the clinical and genetic features of benign familial neonatal seizures (BFNS) with the functional consequences of underlying mutations. Channels (Austin) 2007;1:228–33. doi: 10.4161/chan.4823. [DOI] [PubMed] [Google Scholar]
- 27.Gardiner M. Molecular genetics of infantile nervous system channelopathies. Early Hum Dev. 2006;82:775–9. doi: 10.1016/j.earlhumdev.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 28.Singh NA, Westenskow P, Charlier C, Pappas C, Leslie J, Dillon J, Anderson VE, Sanguinetti MC, Leppert MF. KCNQ2 and KCNQ3 potassium channel genes in benign familial neonatal convulsions: expansion of the functional and mutation spectrum. Brain. 2003;126:2726–37. doi: 10.1093/brain/awg286. [DOI] [PubMed] [Google Scholar]
- 29.Gamper N, Li Y, Shapiro MS. Structural requirements for differential sensitivity of KCNQ K+ channels to modulation by Ca2+/calmodulin. Mol Biol Cell. 2005;16:3538–51. doi: 10.1091/mbc.E04-09-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaika O, Tolstykh GP, Jaffe DB, Shapiro MS. Inositol triphosphate-mediated Ca2+ signals direct purinergic P2Y receptor regulation of neuronal ion channels. J Neurosci. 2007;27:8914–26. doi: 10.1523/JNEUROSCI.1739-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roden DM. A new role for calmodulin in ion channel biology. Circ Res. 2006;98:979–81. doi: 10.1161/01.RES.0000221822.22971.8c. [DOI] [PubMed] [Google Scholar]
- 32.Asada K, Kurokawa J, Furukawa T. Redox- and calmodulin-dependent S- nitrosylation of the KCNQ1 channel. J Biol Chem. 2009;284:6014–20. doi: 10.1074/jbc.M807158200. [DOI] [PubMed] [Google Scholar]
- 33.Bal M, Zaika O, Martin P, Shapiro MS. Calmodulin binding to M-type K+ channels assayed by TIRF/FRET in living cells. J Physiol. 2008;586:2307–20. doi: 10.1113/jphysiol.2008.152777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomez-Posada JC, Aivar P, Alberdi A, Alaimo A, Etxeberria A, Fernandez-Orth J, Zamalloa T, Roura-Ferrer M, Villace P, Areso P, Casis O, Villarroel A. Kv7 channels can function without constitutive calmodulin tethering. PLoS ONE. 2011;6:e25508. doi: 10.1371/journal.pone.0025508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kharkovets T, Hardelin JP, Safieddine S, Schweizer M, El-Amraoui A, Petit C, Jentsch TJ. KCNQ4, a K+ channel mutated in a form of dominant deafness, is expressed in the inner ear and the central auditory pathway. Proc Natl Acad Sci U S A. 2000;97:4333–8. doi: 10.1073/pnas.97.8.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubisch C, Schroeder BC, Friedrich T, Lütjohan B, El-Amaraoui A, Marlin S, Petit C, Jentsch TJ. KCNQ4, a novel potassium channel expressed in sensory outer hair cells, is mutated in dominant deafness. Cell. 1999;96:437–446. doi: 10.1016/s0092-8674(00)80556-5. [DOI] [PubMed] [Google Scholar]
- 37.Heidenreich M, Lechner SG, Vardanyan V, Wetzel C, Cremers CW, De Leenheer EM, Aranguez G, Moreno-Pelayo MA, Jentsch TJ, Lewin GR. KCNQ4 K(+) channels tune mechanoreceptors for normal touch sensation in mouse and man. Nat Neurosci. 2012;15:138–45. doi: 10.1038/nn.2985. [DOI] [PubMed] [Google Scholar]
- 38.Minor DL., Jr The neurobiologist’s guide to structural biology: a primer on why macromolecular structure matters and how to evaluate structural data. Neuron. 2007;54:511–33. doi: 10.1016/j.neuron.2007.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laue TM. Sedimentation equilibrium as a thermodynamic tool. Meth. Enzymol. 1995;259:427–453. doi: 10.1016/0076-6879(95)59055-2. [DOI] [PubMed] [Google Scholar]
- 40.Laue TM, Bhairavi BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry; Cambridge: 1992. [Google Scholar]
- 41.Folta-Stogniew E, Williams KR. Determination of molecular masses of proteins in solution: Implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J Biomol Tech. 1999;10:51–63. [PMC free article] [PubMed] [Google Scholar]
- 42.Folta-Stogniew E. Oligomeric states of proteins determined by size-exclusion chromatography coupled with light scattering, absorbance, and refractive index detectors. Methods Mol Biol. 2006;328:97–112. doi: 10.1385/1-59745-026-X:97. [DOI] [PubMed] [Google Scholar]
- 43.Maljevic S, Lerche C, Seebohm G, Alekov AK, Busch AE, Lerche H. C-terminal interaction of KCNQ2 and KCNQ3 K+ channels. J Physiol. 2003;548:353–60. doi: 10.1113/jphysiol.2003.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schwake M, Athanasiadu D, Beimgraben C, Blanz J, Beck C, Jentsch TJ, Saftig P, Friedrich T. Structural determinants of M-type KCNQ (Kv7) K+ channel assembly. J Neurosci. 2006;26:3757–66. doi: 10.1523/JNEUROSCI.5017-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwake M, Jentsch TJ, Friedrich T. A carboxy-terminal domain determines the subunit specificity of KCNQ K+ channel assembly. EMBO Rep. 2003;4:76–81. doi: 10.1038/sj.embor.embor715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meador WE, Means AR, Quiocho FA. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992;257:1251–5. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 47.Rhoads AR, Friedberg F. Sequence motifs for calmodulin recognition. Biochem. J. 1997;11:331–340. doi: 10.1096/fasebj.11.5.9141499. [DOI] [PubMed] [Google Scholar]
- 48.Hoeflich KP, Ikura M. Calmodulin in action: diversity in target recognition and activation mechanisms. Cell. 2002;108:739–42. doi: 10.1016/s0092-8674(02)00682-7. [DOI] [PubMed] [Google Scholar]
- 49.Ikura M, Barbato G, Klee CB, Bax A. Solution structure of calmodulin and its complex with a myosin light chain kinase fragment. Cell Calcium. 1992;13:391–400. doi: 10.1016/0143-4160(92)90052-t. [DOI] [PubMed] [Google Scholar]
- 50.Maljevic S, Wuttke TV, Seebohm G, Lerche H. KV7 channelopathies. Pflugers Arch. 2010;460:277–88. doi: 10.1007/s00424-010-0831-3. [DOI] [PubMed] [Google Scholar]
- 51.Schmitt N, Calloe K, Nielsen NH, Buschmann M, Speckmann EJ, Schulze-Bahr E, Schwarz M. The novel C-terminal KCNQ1 mutation M520R alters protein trafficking. Biochem Biophys Res Commun. 2007;358:304–10. doi: 10.1016/j.bbrc.2007.04.127. [DOI] [PubMed] [Google Scholar]
- 52.Moulard B, Picard F, le Hellard S, Agulhon C, Weiland S, Favre I, Bertrand S, Malafosse A, Bertrand D. Ion channel variation causes epilepsies. Brain Res Brain Res Rev. 2001;36:275–84. doi: 10.1016/s0165-0173(01)00104-7. [DOI] [PubMed] [Google Scholar]
- 53.Borgatti R, Zucca C, Cavallini A, Ferrario M, Panzeri C, Castaldo P, Soldovieri MV, Baschirotto C, Bresolin N, Dalla Bernardina B, Taglialatela M, Bassi MT. A novel mutation in KCNQ2 associated with BFNC, drug resistant epilepsy, and mental retardation. Neurology. 2004;63:57–65. doi: 10.1212/01.wnl.0000132979.08394.6d. [DOI] [PubMed] [Google Scholar]
- 54.Larsen LA, Fosdal I, Andersen PS, Kanters JK, Vuust J, Wettrell G, Christiansen M. Recessive Romano-Ward syndrome associated with compound heterozygosity for two mutations in the KVLQT1 gene. Eur J Hum Genet. 1999;7:724–8. doi: 10.1038/sj.ejhg.5200323. [DOI] [PubMed] [Google Scholar]
- 55.Li Y, Gamper N, Shapiro MS. Single-channel analysis of KCNQ K+ channels reveals the mechanism of augmentation by a cysteine-modifying reagent. J Neurosci. 2004;24:5079–90. doi: 10.1523/JNEUROSCI.0882-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roche JP, Westenbroek R, Sorom AJ, Hille B, Mackie K, Shapiro MS. Antibodies and a cysteine-modifying reagent show correspondence of M current in neurons to KCNQ2 and KCNQ3 K+ channels. Br J Pharmacol. 2002;137:1173–86. doi: 10.1038/sj.bjp.0704989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aoyagi M, Arvai AS, Tainer JA, Getzoff ED. Structural basis for endothelial nitric oxide synthase binding to calmodulin. Embo J. 2003;22:766–75. doi: 10.1093/emboj/cdg078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Diego I, Kuper J, Bakalova N, Kursula P, Wilmanns M. Molecular basis of the death-associated protein kinase-calcium/calmodulin regulator complex. Sci Signal. 2010;3:ra6. doi: 10.1126/scisignal.2000552. [DOI] [PubMed] [Google Scholar]
- 59.Xia C, Misra I, Iyanagi T, Kim JJ. Regulation of interdomain interactions by calmodulin in inducible nitric-oxide synthase. J Biol Chem. 2009;284:30708–17. doi: 10.1074/jbc.M109.031682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ataman ZA, Gakhar L, Sorensen BR, Hell JW, Shea MA. The NMDA receptor NR1 C1 region bound to calmodulin: structural insights into functional differences between homologous domains. Structure. 2007;15:1603–17. doi: 10.1016/j.str.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sarhan MF, Tung CC, Van Petegem F, Ahern CA. Crystallographic basis for calcium regulation of sodium channels. Proc Natl Acad Sci U S A. 2012;109:3558–63. doi: 10.1073/pnas.1114748109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang M, Abrams C, Wang L, Gizzi A, He L, Lin R, Chen Y, Loll PJ, Pascal JM, Zhang JF. Structural basis for calmodulin as a dynamic calcium sensor. Structure. 2012;20:911–23. doi: 10.1016/j.str.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schumacher MA, Rivard AF, Bächinger HP, Adelman JP. Structure of the gating domain of a Ca2+ activatived K+ channel complexed with Ca2+/calmodulin. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- 64.Feldkamp MD, Yu L, Shea MA. Structural and energetic determinants of apo calmodulin binding to the IQ motif of the Na(V)1.2 voltage-dependent sodium channel. Structure. 2011;19:733–47. doi: 10.1016/j.str.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chagot B, Chazin WJ. Solution NMR structure of Apo-calmodulin in complex with the IQ motif of human cardiac sodium channel NaV1.5. J Mol Biol. 2011;406:106–19. doi: 10.1016/j.jmb.2010.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schumacher MA, Crum M, Miller MC. Crystal structures of apocalmodulin and an apocalmodulin/SK potassium channel gating domain complex. Structure. 2004;12:849–60. doi: 10.1016/j.str.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 67.Bai CX, Namekata I, Kurokawa J, Tanaka H, Shigenobu K, Furukawa T. Role of nitric oxide in Ca2+ sensitivity of the slowly activating delayed rectifier K+ current in cardiac myocytes. Circ Res. 2005;96:64–72. doi: 10.1161/01.RES.0000151846.19788.E0. [DOI] [PubMed] [Google Scholar]
- 68.Hernandez CC, Zaika O, Shapiro MS. A carboxy-terminal inter-helix linker as the site of phosphatidylinositol 4,5-bisphosphate action on Kv7 (M-type) K+ channels. J Gen Physiol. 2008;132:361–81. doi: 10.1085/jgp.200810007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu T, Nie L, Zhang Y, Mo J, Feng W, Wei D, Petrov E, Calisto LE, Kachar B, Beisel KW, Vazquez AE, Yamoah EN. Roles of alternative splicing in the functional properties of inner ear-specific KCNQ4 channels. J Biol Chem. 2007;282:23899–909. doi: 10.1074/jbc.M702108200. [DOI] [PubMed] [Google Scholar]
- 70.Shahidullah M, Santarelli LC, Wen H, Levitan IB. Expression of a calmodulin-binding KCNQ2 potassium channel fragment modulates neuronal M-current and membrane excitability. Proc Natl Acad Sci U S A. 2005;102:16454–9. doi: 10.1073/pnas.0503966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Etxeberria A, Aivar P, Rodriguez-Alfaro JA, Alaimo A, Villace P, Gomez-Posada JC, Areso P, Villarroel A. Calmodulin regulates the trafficking of KCNQ2 potassium channels. FASEB J. 2008;22:1135–43. doi: 10.1096/fj.07-9712com. [DOI] [PubMed] [Google Scholar]
- 72.Van Petegem F, Chatelain FC, Minor DL., Jr Insights into voltage-gated calcium channel regulation from the structure of the CaV1.2 IQ domain-Ca2+/calmodulin complex. Nat Struct Mol Biol. 2005;12:1108–15. doi: 10.1038/nsmb1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 74.Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967;6:1948–54. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 75.Collaborative Computational Project, N. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 76.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallogr D Biol Crystallogr. 2004;60:432–8. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 77.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 78.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.