Abstract
Alcohol use and hepatitis C virus (HCV) infection synergize to cause liver damage and microRNA-122 (miR-122) appears to play a key role in this process. Argonaute 2 (Ago2), a key component of the RNA-induced silencing complex, has been shown to be important in modulating miR-122 function during HCV infection. However, GW182, a critical component of processing bodies (GW-bodies) that is recruited by Ago2 to target mRNA has not been assessed in HCV infection. To characterize the role of GW182 in the pathogenesis of HCV infection, we determined its transcription and protein expression in an HCV J6/JFH1 culture system. Here we show that transcript and protein levels of GW182 as well as HCV RNA and protein expression increased with alcohol exposure. Specific silencing of mRNA expression by short interference (si) RNA against GW182 significantly decreased HCV RNA and protein expression. Over-expression of GW182 significantly increased HCV RNA and protein expression in HCV J6/JFH1 infected Huh-7.5 cells. Furthermore, GW182 co-localized and co-immunoprecipitated with HSP90 which increased on alcohol exposure with and without HCV infection and enhanced HCV gene expression. The use of an HSP90 inhibitor or knockdown of HSP90 decreased GW182 and miR-122 expression and significantly reduced HCV replication. Overall, our results suggest that GW182 protein that is linked to miR-122 biogenesis and HSP90, which has been shown to stabilize the RNA-induced silencing complex, are novel host proteins that regulate HCV infection during alcohol abuse.
Keywords: MicroRNA, GW-body, J6/JFH1 virus, alcohol, antiviral target
INTRODUCTION
Hepatitis C virus (HCV) is estimated to infect at least 2–3 % of the world’s population. Despite clinical observation and medical advice of the devastating consequence of alcohol use during HCV infection, some patients consume alcohol with the increased risk of rapid disease progression to liver cirrhosis, fibrosis and even hepatocellular carcinoma (1–6). The molecular mechanisms of how alcohol exacerbates HCV infection and worsens HCV disease outcome remain to be elucidated.
HCV, a positive-sense RNA virus of the Flaviviridae family can hijack host cofactors to facilitate its replication. Of those, microRNA-122 (miR-122), a miRNA representing 70% of all miRNAs in hepatocytes (7, 8), was recently identified to play a critical role in the HCV life cycle (9–11) and has been a promising target for antiviral drug development (12). Several groups including ours have demonstrated that ethanol can modulate microRNAs expression in the liver (13–16). Traditionally miRNAs function by binding to the 3′UTR of target genes suppressing gene transcription and translation. However, miR-122 binds to the 5′ UTR of the viral genome promoting HCV replication (9, 17, 18). It is unknown if miR-122 regulation of HCV RNA translation or RNA accumulation requires direct association with protein complexes similar to the miRNA-induced silencing complex (miRISC), or if the activity of miR-122 involves HCV RNA translocation to mRNA-processing bodies (P-bodies) (19). Recently, GW-body components and associated proteins such as DDX3 (16), PATL1 (20), DDX6 (20, 21), and Ago2 (22) a key component of the RNA-induced silencing complex have been shown as essential host cofactors for HCV replication. However, GW182, a critical component of GW bodies (GWBs) (23) distinct from P-bodies (24) and having binding pockets for Ago2 (25), has not been assessed in HCV infection.
In this study, we tested the hypothesis that ethanol facilitates HCV replication through modulation of GW182 expression. We found that ethanol increased expression of GW182 and HSP90 and that GW182 colocalized with HSP90 and promoted HCV gene expression. Specific silencing of mRNA expression by short interference (si) RNA against GW182 and HSP90 decreased miR-122, HCV RNA and protein expression. Our data suggest a role for HSP90 and GW182 which are linked to miR-122 biogenesis as novel important factors in the pathomechanism of alcohol-induced augmentation of HCV replication.
MATERIALS AND METHODS
Cells and virus
Huh-7.5 cells highly permissive for HCV infection and Huh-7.5 cells harboring Con1 (genotype 1b) full-length replicon were cultured as previously described (26). An infectious clone of HCV J6/JFH1, generated by plasmid pFL-J6/JFH1, was transfected into Huh-7.5 cells and cultured as previously described (27). Huh-7.5 cells and Con1/FL replicon cells were a gift of Dr. Charles Rice (Rockefeller University, New York, NY). Plasmid pFL-J6/JFH1 was a gift of Dr. Charles Rice and Dr. Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan).
Alcohol Treatment and HSP90 Inhibition
For ethanol exposure, cells were placed in culture chambers (C. B. S. Scientific Co., San Diego, CA) to maintain a stable alcohol concentration, as described previously (28). To inhibit HSP90 activity, J6/JFH1-infected Huh7.5 cells were treated with 17-DMAG HCl (Alvespimycin) (Selleckchem Cat. # S1142).
Transfection
Lipofectamine™ RNAiMAX (Invitrogen, cat. #13778-075) and FugeneHD (Roche, cat. #04709705001) was used for transfection of siRNA or over-expression plasmid according to the manufacturer’s specification. The siRNA (Santa Cruz Biotechnology Inc., Santa Cruz, CA) used in this study were as follows: Control siRNA (FITC Conjugate)-A sc-36869; Control siRNA-A sc-37007; Control shRNA Plasmid-A sc-108060; GW182 siRNA (h) sc-45516; HSP 90α/β siRNA (h) sc-35608. GW182 (pFRT/TO/FLAG/HA-DEST TNRC6A Gene Bank ID NM_014494) plasmid was purchased from Addgene (Addgene plasmid 19883).
Quantification of miRNA Expression
After specific treatment as indicated MicroRNAs were extracted with miRNeasy kit (Qiagen Sciences, Valencia, CA) according to the manufacturer’s specification. miR-122 expression was determined using the Taqman® microRNA assay (Applied Biosystems, Carlsbad, CA). To normalize the expression levels of miR-122, RNU6B was used as an endogenous control.
Virus Quantification in culture supernatant for MOI determination
Virus supernatants containing the HCV virus was quantified and supernatants used such that infections were done using an MOI of 1 or 0.5 as indicated. Briefly RNA was extracted from 200μL of virus supernatant using the RNeasy kit (Qiagen) according to the manufacturer’s protocol. Viral RNA was then eluted in 50μL of RNase free water. 10μL of viral RNA was then reverse transcribed to cDNA using the Promega Reverse Transcription System (Cat # A3500) in a 20μL final reaction volume. 5μL of viral DNA was then used for real time PCR along with 5uL of plasmid standard (pFL-J6/JFH1 plasmid) to contain 10; 100; 1000, 10,000; 100,000; 1000,000; 10,000,000 copies per 5μL. This standard allowed for the quantification of the amount of viruses in our supernatant.
Real Time Reverse Transcription Polymerase Chain Reaction
Real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR) was performed with the CFX96 Real-Time System (Bio-Rad Laboratories, Inc, Hercules, CA) and SYBR Green PCR Master Mix (Eurogentec, Fremont, CA) using 18S for normalization of the relative gene expression. Data were analyzed using the comparative ΔΔCt method. Primers for detection of HCV RNA were described previously(29). Specific primers used included the following:
| DDX3X sense | gtggaacaaacactcgctt, |
| DDX3X antisense | acctttagtagcttctcggtt; |
| DDX6 sense | caggaacatcgaaatcgtg, |
| DDX6 antisense | tccaatacgatggagatagg; |
| EIF2C2 sense | cggacaatcagacctcaacca, |
| EIF2C2 antisense | cccagtcacgtctgtcatctc; |
| HSP90 sense | acaaggatctgcagccatt, |
| HSP90 antisense | gtcaagctttcataccggatt; |
| PATL1 sense | tcctgctccctatggtgagag, |
| PATL1 antisense | catggcagcaagtggactacc; |
| GW182 sense | ctgaacctccctcacggaa; |
| GW182 antisense | ggctttgtgcaaagaaacgac. |
Western Blotting and Immunoprecipitation
Anti-NS5A (9E10, kindly provided by Dr. Charles Rice); anti-NS3 (ViroStat, Portland, Me) or anti-CORE (ViroStat, Portland, Me); anti-HSP90 (Cell signaling cat. #4874); GW182 antibody (Aviva Systems Biology cat. # ARP40956_P050); anti-HA tag antibody (Abcam cat. #ab18181); anti–β-actin (Abcam, Cambridge, MA) were used as primary antibody, followed by horseradish peroxidase–labeled secondary antibody (Santa Cruz Biotechnology Inc.). For immunoprecipitation after specific treatment as indicated, cells where washed twice with ice-cold phosphate buffer saline (Gibco cat. #14190) lysed with immunoprecipitation (IP) lysis buffer (Thermo Scientific cat. #87788) supplemented with protease inhibitor cocktail (Roche Cat. #11836153001). 2μg of each specific immunoprecipitation antibody was then added to each specific sample and a control sample immunoprescipitated with 2μg of IgG control antibody from Santa Cruz (Mouse IgG cat. # SC2025 or Rabbit IgG cat. # 2027) to match the animal species in which the antibody of interest was generated from. After immunoprecipitation samples were subjected to western blot analysis with specific antibodies of interest as indicated.
Confocal Microscopy/Immunofluorescence microcopy
Intracellular staining was performed as previously described(29). Antibodies used in this study were listed as follows:
GW182 (2D6) (Mouse, monoclonal, SANTA CRUZ, sc-56313);
GW182 (Rabbit, polyclonal, Sigma-Aldrich, G5922);
HCV CORE (Mouse, monoclonal, Thermo Fisher Scientific, MA1-7368);
HCV NS3 (Mouse, monoclonal, ViroStat,1847);
HCV NS3 (Mouse monoclonal, Abcam Cat. #ab13830);
HCV NS5A (9E10, kindly provided by Dr. Charles Rice);
HSP 90α/β (H-114) (Rabbit, polyclonal, SANTA CRUZ, sc-7947);
HSP 90 (Rabbit polyclonal, Cell Signaling cat. #4874)
Confocal imaging was performed on a Leica TCS SP2 confocal microscope (Leica Microsystems, Wetzlar, Germany), and Leica confocal software (LCS) was used for the acquisition of images. Immunofluorescence images were acquired using an Olympus BX51 fluorescence microscope and the Nixon NIS-Element BR 3.10 software for image analysis. GW182-bodies sizes and numbers were quantified using the NIH ImageJ 1.46 software for 35 cells in each experimental condition selected randomly from different fields and slides from four independent repeat experiments.
Cytotoxicity Analysis of chemicals and siRNA used
Cytotoxicity of DMAG, HSP90 siRNA and GW182 siRNA was performed using the LDH-Cytotoxicity Assay Kit (Abcam, cat #65393) according to the manufacturer’s specification.
Statistical Analysis
Data presented as mean ± standard error of the mean (SEM) and statistically analyzed by the two tailed Student’s t test; and P values less than 0.05 were considered statistically significant. Analysis of variance (ANOVA) was used when comparing variation with more than two experimental means.
RESULTS
GW182, a GW-body component, is essential for HCV replication
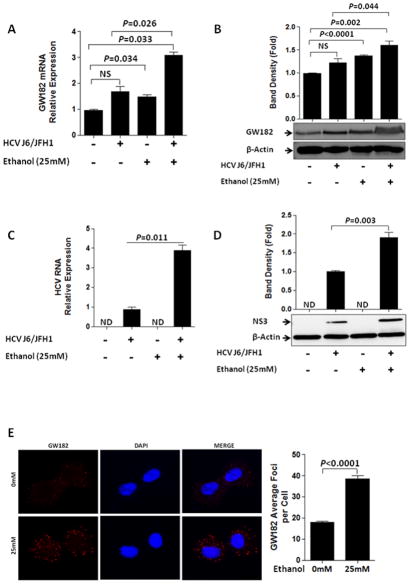
GW-body-associated proteins have recently been shown as essential host factors for HCV replication (16, 20–22). This prompted us to surmise that ethanol might modulate HCV replication by affecting the expression of GW-body proteins. We found that among the GW-body components, GW182 mRNA and protein was selectively up-regulated by ethanol exposure (Fig. 1A&B) compared to no changes in the expression of EIF2C2 (Ago2), DDX3X, DDX6 and PALT1 mRNA (Supplementary Fig. 1A, B, C, D&E). Ethanol treatment upregulated HCV RNA levels as well as HCV protein expression in genotype 2 J6/JFH 1 virus infection (Fig. 1C&D). Fluorescence microscopy analysis revealed that endogenous GW182 was localized to intensely stained, punctuate peri-nuclear cytoplasmic structures consistent with GW-bodies and within 24 hours of ethanol exposure the number of GW-bodies (GW182 antibody as marker) was significantly increased compared to untreated cells (Fig. 1E and supplementary Fig. 2A–D).
Figure 1. Acute ethanol treatment modulated the expression of GW182.
(A–E) The expression of GW182 mRNA, GW182 protein, HCV RNA, HCV NS3 and GW-bodies was assessed as indicated. (A) GW182 mRNA (B) GW182 protein (C) HCV RNA (D) HCV NS3 protein were evaluated 24hr after ethanol exposure in J6/JFH1-infected and non-infected Huh-7.5 cells. (E) Huh-7.5 cells were treated with ethanol (0mM or 25mM). The expression of GW-bodies with GW182 as marker was examined by fluorescence microscopy after 24hr ethanol exposure. Fixed cells were stained with a rabbit polyclonal antibody to GW182 (red). GW182 foci levels were determined for at least 35 cells chosen randomly from different fields using the Olympus BX51 fluorescence microscope for imaging and the Nixon Nikon NIS-Element BR 3.10 software for acquisition and the NIH ImageJ 1.46 software quantification. Results presented are representative of 3–4 independent repeat experiments and data expressed as SEM, p< 0.05 were considered statistically significant (by two-tailed Student’s t test. Multiplicity of infection (MOI) of 1 was used for all infections.
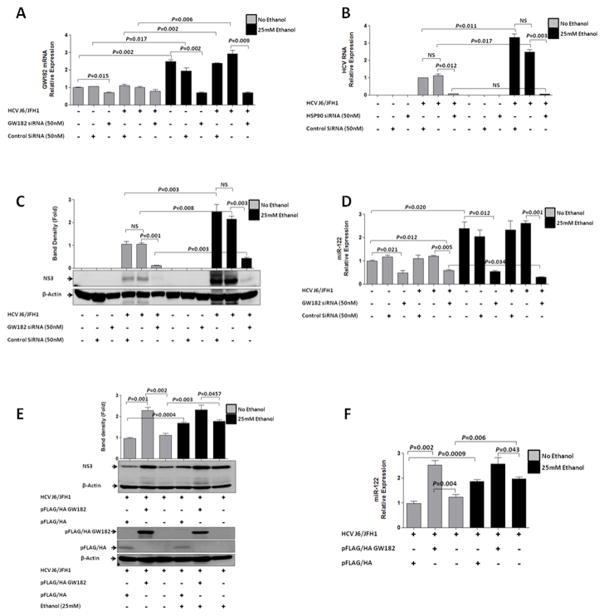
To determine the effect of GW182 on HCV RNA accumulation, we used FITC-conjugated control or GW182 siRNAs to ensure transfection efficiency (data not shown) and knock-down was confirmed by decreased GW182 mRNA(Fig. 2A). Compared to the siRNA control, GW182-specific siRNA-transfected J6/JFH1-infected Huh-7.5 cells showed significantly reduced expression of HCV RNA (Fig. 2B) and protein (Fig. 2C). Next we sought to evaluate the mechanisms by which GW182 could facilitate HCV replication and asked whether GW182 influenced miR-122 abundance. We found a significant reduction in miR-122 levels after transfection of J6/JFH1-infected Huh-7.5 cells with the GW182-specific siRNA compared to controls (Fig. 2D). These results indicated that the ethanol triggered induction of miR-122 might be mediated, at least partially, by GW182. We did not observe any changes in miR-370 which can regulate miR-122 expression with alcohol exposure in Huh-7.5 cell with and without HCV J6/JFH1 infection (Supplementary Fig. 1F).
Figure 2. GW182 is essential for HCV replication.
J6/JFH1-infected or uninfected Huh-7.5 cells with and without ethanol exposure were transfected with GW182 siRNA (50nM) or control siRNA (50nM). 48hr after transfection, (A) GW182 mRNA (RT-qPCR), (B) intracellular HCV RNA (RT-qPCR), (C) HCV NS3 protein (western blot) expression and (D) the miR-122 relative expression was quantified. (E) Over-expression of GW182 confirmed with an anti-HA antibody in J6/JFH1-infected cells and NS3HCV protein expression compared to controls with and without alcohol exposure. (F) miR-122 expression determined with and without alcohol exposure following GW182 over expression.
Results presented are representative of 3–4 independent repeat experiments and data expressed as SEM, p< 0.05 were considered statistically significant (by two-tailed Student’s t test). Multiplicity of infection (MOI) of 1 was used for all infections.
In contrast, over-expression of GW182 prior to HCV infection significantly increased HCV protein expression compared to the empty vector control in Huh-7.5 cells (Fig. 2E) with and without alcohol exposure. GW182 over-expression was also associated with an increase in miR-122 transcript levels with no significant difference observed in the presence or absence of alcohol (Fig. 2F). We also found upregulation of HCV RNA and not HCV proteins after 24hours acute ethanol treatment in Con1/FL replicon cells (data not shown) and acute alcohol treatment did not increase GW182 protein expression in Con/FL replicon cells (Supplementary Fig. 2E) though HCV RNA increased significantly (Supplemental Fig 2F). The observation that ethanol exposure had no effect on GW182 expression in replicon cells reflect differences among the two cell lines which may deserve further investigation.
HSP90 interacts with GW182 and HCV viral proteins
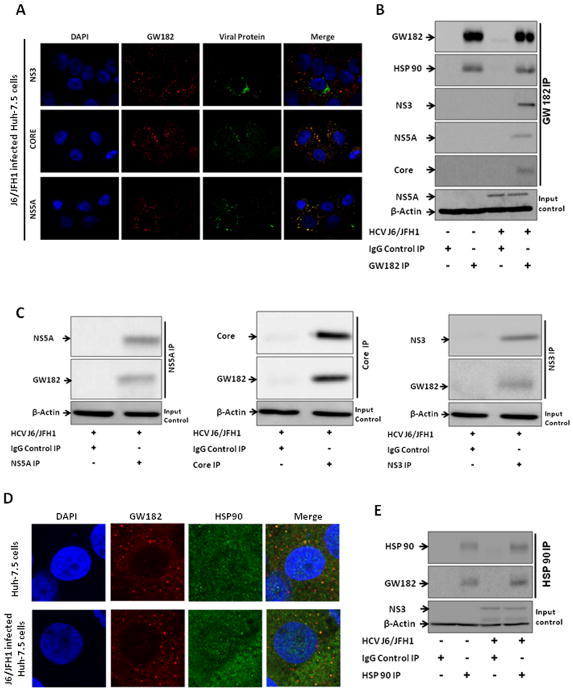
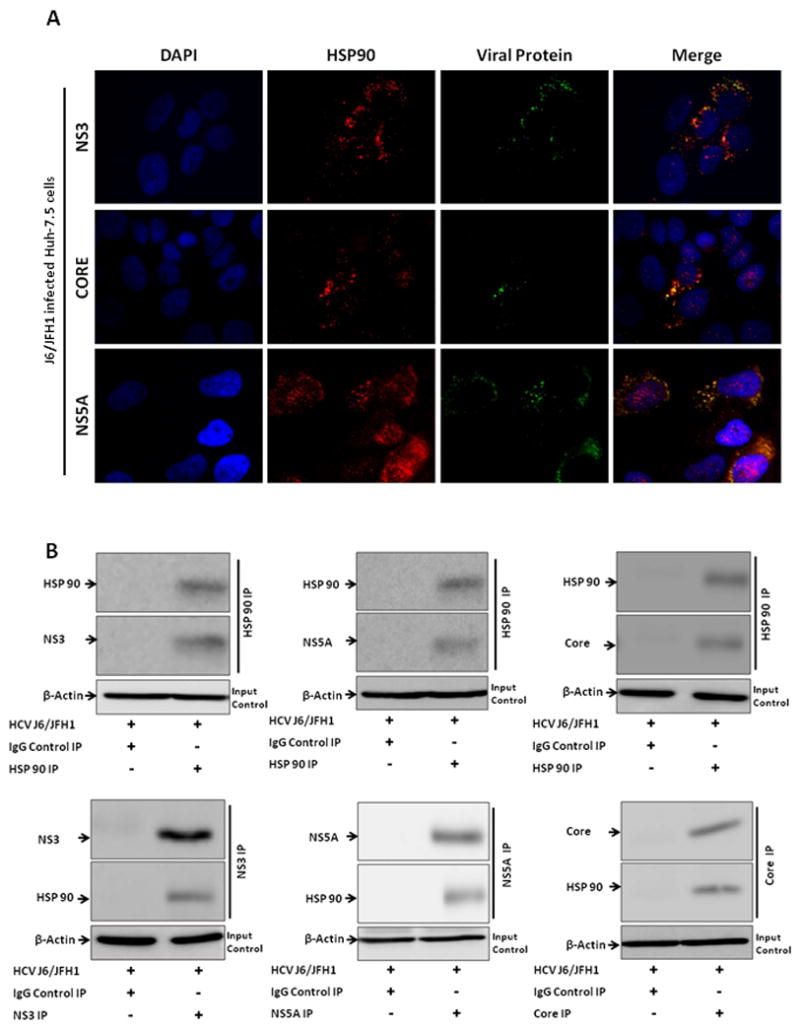
Previous studies have shown that heat shock protein 90 was important in mediating HCV replication through recruiting FKBP8 and NS5A (30), NS3 (31) and hB-ind1 (32) and that HSP90 inhibition decreases GW182 expression (33). Given that GW182 was increased by alcohol exposure, we evaluated the intracellular localization and interaction of endogenous GW182 with HCV and HSP90 proteins. We found differential extents of co-localization of GW182 with the viral NS3, Core and NS5A proteins in J6/JFH1-infected Huh-7.5 cells ranging between 40–80% using fluorescence microscopy (Fig. 3A and supplementary Fig. 3). These interactions were confirmed in co-immunoprecipitation experiments (Fig. 3B&C). HSP90 is one of the most conserved heat shock proteins (HSPs) that can stabilize Argonaute proteins associated with P-bodies as well as stress granules (SGs) in human Hela cells (33–35). Thus, we hypothesized that HSP90 might interact with GW182 in hepatoma cells. Indeed, we found that GW182 and HSP90 co-localized and co-immunoprecipitated in J6/JFH1-infected and uninfected Huh-7.5 cells (Fig. 3D&E).
Figure 3. GW182 co-localized and co-immunoprecipitated with viral proteins and HSP 90.
(A) The co-localization of GW182 (stained with rabbit polyclonal antibody to GW182 (Red) and HCV protein (Green) were evaluated by fluorescence microscopy in J6/JFH1-infected Huh-7.5 cells. (B & C) Co-immunoprecipitation of GW182 with HSP 90 and viral proteins was evaluated in J6/JFH1-infected Huh 7.5 cells. (D) The co-localization of GW182 (stained with a mouse monoclonal antibody to GW182 (Red) and HSP 90 protein (Green) were evaluated by confocal microscopy in J6/JFH1-infected and uninfected Huh-7.5 cells. (E) Co-immunoprecipitation of HSP 90 with GW182 was evaluated in J6/JFH1-infected and uninfected Huh 7.5 cells. Results presented are representative of 3–4 independent repeat experiments with at least 10 fields sequential analyzed for each microscopy slide to minimize spectral bleed-through artifacts. Multiplicity of infection (MOI) of 1 was used for IP infections and MOI of 0.5 for microscopy slides.
Ethanol-induced HSP90 expression has a mechanistic role in regulation of HCV replication through miR-122
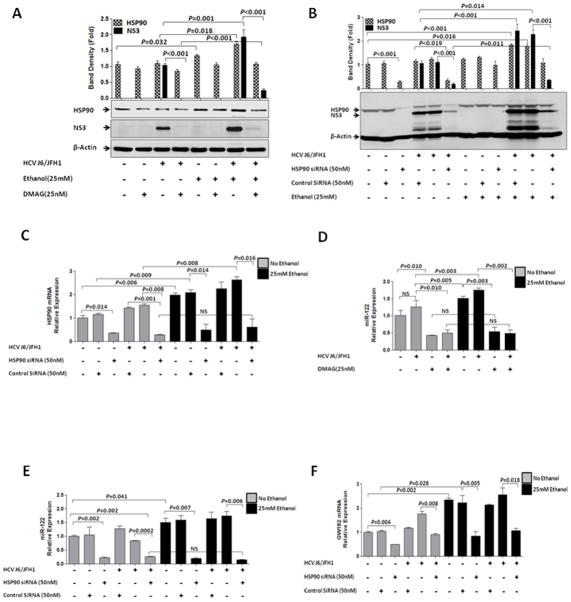
It has recently been shown that HSP90 could directly interact with HCV NS3 and NS5A to exert its role in HCV replication (36, 37). Also, inhibition of HSP90 by use of 17-DMAG has been shown to inhibit HCV replication by disrupting HSP90 stabilization of Argonaute complexes and P-body components (33). Given that HSP90 interacts with and can stabilize RISC complexes, we resorted to confirm if HSP90 can indeed interact with HCV viral proteins. We found that HSP90 and HCV proteins co-localized in 50–80% of HCV J6/JFH1 infected cells (Fig. 4A and supplementary Fig. 4). These interactions were confirmed by co-immunoprecipitation in J6/JFH1-infected Huh-7.5 cells (Fig. 4B) and Con1/FL replicon cells (Data not shown). Thus, we evaluated whether ethanol could modulate HCV replication via affecting HSP90 expression and if so, can HSP90 also modulate miR-122 expression that has been shown to regulate HCV infection. Interestingly, we found that ethanol synergized with HCV to significantly increase protein levels of HSP90 (Fig. 5A). Inhibition of HSP90 with 17-DMAG (Fig. 5A) or an HSP90-specific siRNA (Fig. 5B) reduced HCV protein (Fig. 5A&B) and RNA (Supplementary Fig. 5A) levels in J6/JFH1-infected Huh-7.5 cells as well as in Con1/FL replicon cells (Data not shown). The efficiency of HSP90 knockdown was confirmed in alcohol-naïve and alcohol-treated Huh 7.5 or J6/JFH1-Huh 7.5 cells at protein (Fig. 5B) and RNA (Fig. 5C) levels. DMAG treatment (Fig. 5D) or Knock-down of HSP90 (Fig. 5E) also significantly decreased miR-122 levels. HSP90 Knock-down was also associated with a decrease in GW182 RNA (Fig. 5F) and protein (Supplementary Fig. 5B) and this closely correlated with a significant reduction in intracellular HCV RNA (Supplementary Fig. 5A) and HCV NS3 protein (Fig. 5B). The concentrations of 17-DMAG, HSP90 siRNA and GW182 siRNA used showed no toxicity to cells (Supplementary Fig. 6A&B).
Figure 4. HSP90 co-localized and co-immunoprecipitated with HCV proteins.
(A) The co-localization of HSP90 (Red) and HCV proteins (Green) were evaluated by fluorescence microscopy and (B) co-immunoprecipitation in J6/JFH1-infected Huh-7.5 cells. (A) Representative slides with at least 10 fields sequential analyzed for each microscopy slide to minimize spectral bleed-through artifacts. Results presented are representative of 3 independent repeat experiments. Multiplicity of infection (MOI) of 1 was used for IP infections and MOI of 0.5 for microscopy slides.
Figure 5. HSP90 has a mechanistic role in regulation of HCV replication and miR-122.
(A) Huh-7.5 and J6/JFH1-infected Huh-7.5 cells were treated with ethanol, 17-DMAG or not for 24hr and HCV NS3 and HSP90 protein analyzed. (B) Huh-7.5 and J6/JFH1-infected Huh-7.5 cells with and without ethanol exposure were transfected with HSP90 (50nM), or control siRNA; HCV NS3 protein (B), HSP90 RNA (C), HSP90 protein (B), miR-122 (E) and GW182 RNA (F) was evaluated after 24hrs. (D) Huh-7.5 and J6/JFH1-infected Huh-7.5 cells were treated with ethanol, 17-DMAG or not for 24hr and miR-122 expression determined. (C, D, E&F) Data expressed as SEM, p< 0.05 were considered statistically significant (by two-tailed Student’s t test).
Acute ethanol treatment increases HCV replication via miR-122 induction
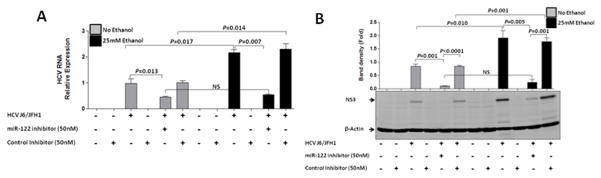
Using Huh-7.5 cells and the HCV J6/JFH system we found that acute ethanol (25 mM) treatment resulted in a significant increase in HCV RNA (Fig. 1C) and HCV NS3 protein expression (Fig. 1D) compared to ethanol naïve matching controls. The ethanol concentration used did not induce cytotoxicity as assessed by light microcopy cell morphology and LDH-Cytotoxicity assay (data not shown). MiR-122, a highly abundant microRNA in hepatocytes has been shown to modulate HCV replication(9) and we recently found that microRNA expression can be regulated by alcohol in Kupffer cells and in the liver tissue in vivo(13). Based on our earlier observation that ethanol treatment significantly upregulated miR-122 levels Huh-7.5 cells with and without HCV J6/JFH1 infection (Fig. 2D), we hypothesized that ethanol affects miR-122 expression and thereby regulates HCV replication. The functional role of the ethanol-induced miR-122 increase in HCV replication was evaluated by using an anti-miR-122 inhibitor. Our results show that the anti-miR-122 inhibitor, and not the anti-miR-122 negative control, attenuated HCV replication in ethanol-naïve cells and prevented the ethanol-induced increase in HCV RNA (Fig 6A) and HCV NS3 protein levels (Fig. 6B). These observations suggested that alcohol-induced miR-122 induction has a mechanistic role in regulating HCV replication.
Figure 6. Acute ethanol treatment induces miR-122 and activates hepatitis C virus replication.
(A&B) J6/JFH-infected and uninfected Huh-7.5 cells were transfected with a miR-122 inhibitor (50nM) or negative control (50nM) prior to ethanol treatment (0mM or 25mM). Intracellular HCV RNA (A) and NS3 protein (B) was quantified (RT-qPCR) 24hr after ethanol exposure. Results presented are representative of 3 independent repeat experiments and data expressed as SEM, p< 0.05 were considered statistically significant (by two-tailed Student’s t test). Multiplicity of infection (MOI) of 1 was used for these infections.
DISCUSSION
In this study, we report a novel mechanism in which ethanol regulates GW-body proteins and enhances HCV replication in human hepatoma cells involving GW182 and HSP90. We demonstrate here that alcohol increases HSP90, GW182 and miR-122 that are host factors in the regulation of HCV infection. Together with previous findings that GW182 is an important component in the RISC complex binding to miRNA/siRNA(38, 39) and HSP90 stabilizing RISC complexes(40), our observations support a model in which alcohol-induced increase of miR-122, GW182 and HSP90 abundance enhances HCV replication through increased HCV UTR binding sustained by increased miR-122-GW182 complex formations stabilized by HSP90. These observations provide new concepts that underlie host and HCV interactions and the mechanisms for alcohol-induced regulation of HCV replication.
First, we discovered that GW182, a GW-body marker, is up-regulated after alcohol exposure (25mM) in Huh-7.5 cells with and without HCV infection suggesting a possible role for GW182 as an important mediator of the biological effects of alcohol to increase HCV replication. We show for the first time that knockdown of GW182 by an RNAi approach reduces intracellular HCV RNA and protein levels even in the absence of alcohol exposure. GW182 (TNRC6A) is a 182-kDa protein characterized by multiple glycine (G) and tryptophan (W) motifs and an essential component of GW-bodies (41–43). It was controversial whether these structures are required for small RNA-mediated gene silencing or whether they simply form as a consequence of silencing (25, 44). Recent evidence suggests the latter scenario; it was observed that P-body formation was a consequence of RNA-mediated gene silencing indicating that GW-body components such as GW182 may increase the efficiency or kinetics of miRNA-mediated gene silencing despite their spatial concentration in discrete domains termed P-bodies(45).
Modulation of HCV replication by GW182 might involve multiple pathways. Our observations raise the possibility of a cross-regulation between GW182 and miR-122 expression because we found a significant reduction of miR-122 abundance after transfection of hepatoma cells with a GW182-specific siRNA similar to findings by Roberts et al (46). Previous reports indicated that some P-body components had no effect on microRNA expression in Hela cells (34); however, our results imply that GW182, a GW-body component can modulate miR-122 expression in human hepatoma cells. This speculation is also supported by the observation of reduced endogenous miR-122 levels following Ago1–4 RNAi administration (47). Another consideration is that modulation of HCV replication by GW182 may occur through the presence of small amounts of GW182 at the membrane-associated replication complex with NS3 leading to new HCV RNA synthesis. This possibility is supported by our observation of significant co-immunoprecipitation of GW182 with the viral NS3 proteins in J6/JFH1 infected Huh7.5 cells.
Based on the co-localization and co-immunoprecipitation of GW182 and HSP90 in naïve and J6/JFH1 infected Huh-7.5 and in Con1/FL replicon cells (data not shown), we identified that GW182 might be a new client protein of the chaperone, HSP90. We found that HSP90 activity was important for GW182 as well as HCV gene expression because the HSP90 knock-down or chemical inhibition significantly decreased both GW182 and HCV protein levels implying a mechanistic role for HSP90 in regulation of HCV replication. The results suggested that the alcohol-induced increase in expression of HSP90 might augment HCV replication by stabilizing miR-122 binding to the HCV genome and or enhancing GW182 gene expression. We speculate that HSP90 regulation of GW182 expression depends on the chaperone and/or the stabilizing effect of HSP90 and not by direct transcriptional regulation by HSP90 as the GW182 promoter has no consensus binding motif for heat shock elements (NCBI GW182 Reference Sequences: NM_018996.3 and NM_001142640.1). Additionally, inhibition of HSP90 activity, similar to GW182 inhibition, affected the abundance of miR-122 supporting the hypothesis that HSP90 could promote GW182 expression which can regulate miR-122 expression. These observations cannot rule out participation of other co-chaperones. Our results together with previous reports (37, 48) suggest that HSP90 has multiple roles in HCV replication including direct interaction with viral proteins, regulating GW182 gene expression, and the abundance of miR-122. We demonstrated that HSP90 works as a regulator of miR-122 abundance since inhibiting HSP90 activity with an inhibitor or HSP90-specific siRNAs, significantly reduced miR-122 expression. These data suggested that ethanol as a cellular stress inducer acts through the stress-responsive transcription factor, HSP90, to regulate miR-122 expression. Interestingly, we also found GW182, a GW-body component, to affect the expression of miR-122 and future studies may reveal roles of other GW-body components in miR-122 abundance or other miRNAs. However, we found that ethanol exposure had no effects on miR-370 (supplementary Fig. 1F) that also modulates miR-122 expression (49) indicating that miR-370 might not be involved in this process.
In summary, our data suggest that ethanol facilitates HCV replication involving GW182 and HSP90 modulation. In the context of alcohol abuse, we show for the first time that GW182 and HSP90 are host factors that spur disease progression in HCV infection. Our studies provide experimental evidence for the possible use of GW182 and HSP90 inhibitors as feasible targets to interfere with HCV replication and the undesirable effects of alcohol use in HCV infection.
Supplementary Material
(A–E) The relative mRNA expression (RT-qPCR) and or protein expression of (A) EIF2C2 (Ago2) mRNA, (B) EIF2C2 (Ago2) protein (C)DDX3X mRNA, (D) DDX6 mRNA and (E) PATL1 mRNA were evaluated 24hr after ethanol exposure or not in J6/JFH1-infected and uninfected Huh-7.5 cells. (F) miR-370 evaluated 24hr after ethanol exposure or not in J6/JFH1-infected and uninfected Huh-7.5 cells Results presented are representative of 3 independent repeat experiments and data expressed as SEM, p< 0.05 were considered statistically significant (by two-tailed Student’s t test). Multiplicity of infection (MOI) of 1 was used for all infections.
(A&B) GW182 expression intensity surface snapshots for Huh-7.5 cells and (C&D) Ethanol exposed Huh-7.5 cells for images in Fig. 1E.
(E) GW182 protein levels and (F) HCV RNA were determined in Con1/FL replicon cells 24hr after ethanol exposure
(A) The co-localization of GW182 (stained with rabbit polyclonal antibody to GW182 (Red) and HCV proteins (Green) were evaluated by fluorescence microscopy in J6/JFH1-infected Huh-7.5 cells.
The co-localization of HSP90 (Red) and HCV proteins (Green) were evaluated by fluorescence microscopy in J6/JFH1-infected Huh-7.5 cells.
(A) Huh-7.5 and J6/JFH1-infected Huh-7.5 cells were treated with HSP90 siRNA, or not for 24hr followed by ethanol exposure or not and HCV RNA analyzed. (B) Huh-7.5 and J6/JFH1-infected Huh-7.5 cells were treated with HSP90 siRNA, GW182 siRNA or not for 24hr followed by ethanol exposure for a further 24hrs or not and HCV NS3, HSP90 and GW182 proteins analyzed (C) Huh-7.5 and J6/JFH1-infected Huh-7.5 cells were treated with GW182 siRNA, or not for 24hr followed by ethanol exposure or not and HSP90 mRNA analyzed. Results presented are representative of 3 independent repeat experiments and data expressed as SEM, p< 0.05 were considered statistically significant (by two-tailed Student’s t test). Multiplicity of infection (MOI) of 1 was used for these infections.
Cellular toxicity of DMAG (A); HSP90 and GW182 siRNAs (B) were analyzed 24 hours after DMAG or siRNA treatment using the LDH-cytotoxicity assay by Abcam according to the manufacturers’ specifications. Results presented are representative of 3 independent repeat experiments.
Acknowledgments
Financial Support: This work was supported by grant R37AA014372 (to G.S.).
The authors are grateful to Drs Charles M. Rice and Takaji Wakita for kindly providing reagents and Dr. Tuschl T for the GW182 over-expression plasmid.
Abbreviations
- HCV
hepatitis C Virus
- NS3
HCV nonstructural protein 3
- NS5
HCV nonstructural protein 5
- MOI
multiplicity of infection
- miR-122
microRNA-122
- GW-body
GW182 mRNA-processing bodies
- HSP90
heat-shock protein 90
- RISC
RNA-induced silencing complex
Footnotes
Competing Interests: The authors declare that no competing interests exist.
References
- 1.Szabo G, Aloman C, Polyak SJ, Weinman SA, Wands J, Zakhari S. Hepatitis C infection and alcohol use: A dangerous mix for the liver and antiviral immunity. Alcohol Clin Exp Res. 2006;30:709–719. doi: 10.1111/j.1530-0277.2006.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Szabo G, Wands JR, Eken A, Osna NA, Weinman SA, Machida K, Joe Wang H. Alcohol and hepatitis C virus--interactions in immune dysfunctions and liver damage. Alcohol Clin Exp Res. 2010;34:1675–1686. doi: 10.1111/j.1530-0277.2010.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang T, Li Y, Lai JP, Douglas SD, Metzger DS, O’Brien CP, Ho WZ. Alcohol potentiates hepatitis C virus replicon expression. Hepatology. 2003;38:57–65. doi: 10.1053/jhep.2003.50295. [DOI] [PubMed] [Google Scholar]
- 4.McCartney EM, Semendric L, Helbig KJ, Hinze S, Jones B, Weinman SA, Beard MR. Alcohol metabolism increases the replication of hepatitis C virus and attenuates the antiviral action of interferon. J Infect Dis. 2008;198:1766–1775. doi: 10.1086/593216. [DOI] [PubMed] [Google Scholar]
- 5.Seronello S, Ito C, Wakita T, Choi J. Ethanol enhances hepatitis C virus replication through lipid metabolism and elevated NADH/NAD+ J Biol Chem. 2010;285:845–854. doi: 10.1074/jbc.M109.045740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romero-Gomez M, Grande L, Nogales MC, Fernandez M, Chavez M, Castro M. Intrahepatic hepatitis C virus replication is increased in patients with regular alcohol consumption. Dig Liver Dis. 2001;33:698–702. doi: 10.1016/s1590-8658(01)80048-7. [DOI] [PubMed] [Google Scholar]
- 7.Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA biology. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 8.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Current biology: CB. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 9.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 10.Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, et al. microRNA-122 stimulates translation of hepatitis C virus RNA. The EMBO journal. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:941–946. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bala S, Marcos M, Kodys K, Csak T, Catalano D, Mandrekar P, Szabo G. Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor {alpha} (TNF{alpha}) production via increased mRNA half-life in alcoholic liver disease. J Biol Chem. 2011;286:1436–1444. doi: 10.1074/jbc.M110.145870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, Szabo G. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res. 2009;33:1704–1710. doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miranda RC, Pietrzykowski AZ, Tang Y, Sathyan P, Mayfield D, Keshavarzian A, Sampson W, et al. MicroRNAs: master regulators of ethanol abuse and toxicity? Alcohol Clin Exp Res. 2010;34:575–587. doi: 10.1111/j.1530-0277.2009.01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ariumi Y, Kuroki M, Abe K, Dansako H, Ikeda M, Wakita T, Kato N. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J Virol. 2007;81:13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jangra RK, Yi M, Lemon SM. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. Journal of virology. 2010;84:6615–6625. doi: 10.1128/JVI.00417-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beckham CJ, Parker R. P bodies, stress granules, and viral life cycles. Cell Host Microbe. 2008;3:206–212. doi: 10.1016/j.chom.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scheller N, Mina LB, Galao RP, Chari A, Gimenez-Barcons M, Noueiry A, Fischer U, et al. Translation and replication of hepatitis C virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc Natl Acad Sci U S A. 2009;106:13517–13522. doi: 10.1073/pnas.0906413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jangra RK, Yi M, Lemon SM. DDX6 (Rck/p54) is required for efficient hepatitis C virus replication but not for internal ribosome entry site-directed translation. J Virol. 2010;84:6810–6824. doi: 10.1128/JVI.00397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilson JA, Zhang C, Huys A, Richardson CD. Human Ago2 is required for efficient microRNA 122 regulation of hepatitis C virus RNA accumulation and translation. J Virol. 2011;85:2342–2350. doi: 10.1128/JVI.02046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z, Jakymiw A, Wood MR, Eystathioy T, Rubin RL, Fritzler MJ, Chan EK. GW182 is critical for the stability of GW bodies expressed during the cell cycle and cell proliferation. Journal of cell science. 2004;117:5567–5578. doi: 10.1242/jcs.01477. [DOI] [PubMed] [Google Scholar]
- 24.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nature cell biology. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 25.Lian SL, Li S, Abadal GX, Pauley BA, Fritzler MJ, Chan EK. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA. 2009;15:804–813. doi: 10.1261/rna.1229409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 28.Mandrekar P, Bala S, Catalano D, Kodys K, Szabo G. The opposite effects of acute and chronic alcohol on lipopolysaccharide-induced inflammation are linked to IRAK-M in human monocytes. J Immunol. 2009;183:1320–1327. doi: 10.4049/jimmunol.0803206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou W, Aoki C, Yu L, Wen X, Xue Y, Gao B, Liu W, et al. A recombinant replication-competent hepatitis C virus expressing Azami-Green, a bright green-emitting fluorescent protein, suitable for visualization of infected cells. Biochem Biophys Res Commun. 2008;377:7–11. doi: 10.1016/j.bbrc.2008.08.145. [DOI] [PubMed] [Google Scholar]
- 30.Okamoto T, Nishimura Y, Ichimura T, Suzuki K, Miyamura T, Suzuki T, Moriishi K, et al. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. The EMBO journal. 2006;25:5015–5025. doi: 10.1038/sj.emboj.7601367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ujino S, Yamaguchi S, Shimotohno K, Takaku H. Heat-shock protein 90 is essential for stabilization of the hepatitis C virus nonstructural protein NS3. The Journal of biological chemistry. 2009;284:6841–6846. doi: 10.1074/jbc.M806452200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taguwa S, Okamoto T, Abe T, Mori Y, Suzuki T, Moriishi K, Matsuura Y. Human butyrate-induced transcript 1 interacts with hepatitis C virus NS5A and regulates viral replication. Journal of virology. 2008;82:2631–2641. doi: 10.1128/JVI.02153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell. 2010;21:1462–1469. doi: 10.1091/mbc.E09-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pare JM, Tahbaz N, Lopez-Orozco J, LaPointe P, Lasko P, Hobman TC. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol Biol Cell. 2009;20:3273–3284. doi: 10.1091/mbc.E09-01-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki Y, Minami M, Suzuki M, Abe K, Zenno S, Tsujimoto M, Matsumoto K, et al. The Hsp90 inhibitor geldanamycin abrogates colocalization of eIF4E and eIF4E-transporter into stress granules and association of eIF4E with eIF4G. J Biol Chem. 2009;284:35597–35604. doi: 10.1074/jbc.M109.036285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okamoto T, Nishimura Y, Ichimura T, Suzuki K, Miyamura T, Suzuki T, Moriishi K, et al. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J. 2006;25:5015–5025. doi: 10.1038/sj.emboj.7601367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ujino S, Yamaguchi S, Shimotohno K, Takaku H. Heat-shock protein 90 is essential for stabilization of the hepatitis C virus nonstructural protein NS3. J Biol Chem. 2009;284:6841–6846. doi: 10.1074/jbc.M806452200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, et al. Disruption of GW bodies impairs mammalian RNA interference. Nature cell biology. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- 39.Eulalio A, Tritschler F, Izaurralde E. The GW182 protein family in animal cells: new insights into domains required for miRNA-mediated gene silencing. RNA. 2009;15:1433–1442. doi: 10.1261/rna.1703809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Molecular biology of the cell. 2010;21:1462–1469. doi: 10.1091/mbc.E09-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eystathioy T, Chan EK, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol Biol Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eystathioy T, Chan EK, Mahler M, Luft LM, Fritzler ML, Fritzler MJ. A panel of monoclonal antibodies to cytoplasmic GW bodies and the mRNA binding protein GW182. Hybrid Hybridomics. 2003;22:79–86. doi: 10.1089/153685903321947996. [DOI] [PubMed] [Google Scholar]
- 43.Tritschler F, Huntzinger E, Izaurralde E. Role of GW182 proteins and PABPC1 in the miRNA pathway: a sense of deja vu. Nat Rev Mol Cell Biol. 2010;11:379–384. doi: 10.1038/nrm2885. [DOI] [PubMed] [Google Scholar]
- 44.Pauley KM, Eystathioy T, Jakymiw A, Hamel JC, Fritzler MJ, Chan EK. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 2006;7:904–910. doi: 10.1038/sj.embor.7400783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Molecular and cellular biology. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic acids research. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts AP, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39:7716–7729. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Galardi S, Mercatelli N, Farace MG, Ciafre SA. NF-kB and c-Jun induce the expression of the oncogenic miR-221 and miR-222 in prostate carcinoma and glioblastoma cells. Nucleic Acids Res. 2011;39:3892–3902. doi: 10.1093/nar/gkr006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. Journal of lipid research. 2010;51:1513–1523. doi: 10.1194/jlr.M004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A–E) The relative mRNA expression (RT-qPCR) and or protein expression of (A) EIF2C2 (Ago2) mRNA, (B) EIF2C2 (Ago2) protein (C)DDX3X mRNA, (D) DDX6 mRNA and (E) PATL1 mRNA were evaluated 24hr after ethanol exposure or not in J6/JFH1-infected and uninfected Huh-7.5 cells. (F) miR-370 evaluated 24hr after ethanol exposure or not in J6/JFH1-infected and uninfected Huh-7.5 cells Results presented are representative of 3 independent repeat experiments and data expressed as SEM, p< 0.05 were considered statistically significant (by two-tailed Student’s t test). Multiplicity of infection (MOI) of 1 was used for all infections.
(A&B) GW182 expression intensity surface snapshots for Huh-7.5 cells and (C&D) Ethanol exposed Huh-7.5 cells for images in Fig. 1E.
(E) GW182 protein levels and (F) HCV RNA were determined in Con1/FL replicon cells 24hr after ethanol exposure
(A) The co-localization of GW182 (stained with rabbit polyclonal antibody to GW182 (Red) and HCV proteins (Green) were evaluated by fluorescence microscopy in J6/JFH1-infected Huh-7.5 cells.
The co-localization of HSP90 (Red) and HCV proteins (Green) were evaluated by fluorescence microscopy in J6/JFH1-infected Huh-7.5 cells.
(A) Huh-7.5 and J6/JFH1-infected Huh-7.5 cells were treated with HSP90 siRNA, or not for 24hr followed by ethanol exposure or not and HCV RNA analyzed. (B) Huh-7.5 and J6/JFH1-infected Huh-7.5 cells were treated with HSP90 siRNA, GW182 siRNA or not for 24hr followed by ethanol exposure for a further 24hrs or not and HCV NS3, HSP90 and GW182 proteins analyzed (C) Huh-7.5 and J6/JFH1-infected Huh-7.5 cells were treated with GW182 siRNA, or not for 24hr followed by ethanol exposure or not and HSP90 mRNA analyzed. Results presented are representative of 3 independent repeat experiments and data expressed as SEM, p< 0.05 were considered statistically significant (by two-tailed Student’s t test). Multiplicity of infection (MOI) of 1 was used for these infections.
Cellular toxicity of DMAG (A); HSP90 and GW182 siRNAs (B) were analyzed 24 hours after DMAG or siRNA treatment using the LDH-cytotoxicity assay by Abcam according to the manufacturers’ specifications. Results presented are representative of 3 independent repeat experiments.