Abstract
c-Jun NH2-terminal kinases (JNKs) and phosphatidylinositol 3-kinase (PI3-K) play critical roles in chronic diseases such as cancer, type II diabetes, and obesity. We describe here the binding of quercetagetin (3,3′,4′,5,6,7-hydroxyflavone), related flavonoids, and SP600125 to JNK1 and PI3-K by ATP-competitive and IMAP-based FP assays and measure the effect of quercetagetin on JNK1 and PI3-K activities. Quercetagetin attenuated the phosphorylation of c-Jun and AKT, suppressed AP-1 and NF-κB promoter activities and also reduced cell transformation. It attenuated tumor incidence and reduced tumor volumes in a two-stage skin carcinogenesis mouse model.
Our crystallographic structure determination data show that quercetagetin binds to the ATP-binding site of JNK1. Notably, the interaction between Lys55, Asp169, and Glu73 of JNK1 and the catechol moiety of quercetagetin reorients the N-terminal lobe of JNK1, thereby improving compatibility of the ligand with its binding site. The results of a theoretical docking study suggest a binding mode of PI3-Kwith the hydroxyl groups of the catechol moiety forming hydrogen bonds with the side chains of Asp964 and Asp841 in the p110γ catalytic subunit. These interactions could contribute to the high inhibitory activity of quercetagetin against PI3-K. Our study suggests the potential use of quercetagetin in the prevention or therapy of cancer and other chronic diseases.
Introduction
The c-Jun NH2-terminal kinases (JNKs) are a group of serine/threonine protein kinases that are members of the mitogen-activated protein kinase (MAPK) family, which also includes the extracellular signal regulated kinases (ERKs) and p38 kinases. JNK1 and JNK2 have a broad tissue distribution, whereas JNK3 appears primarily to be localized to neuronal tissues and cardiac myocytes 1. JNKs are potently activated by various inflammatory signals and stressors, and expression of JNK proteins is frequently altered in human tumors and cancer cells 2. Although some debate exists regarding the roles of JNKs in cancer, they are up-regulated in several types of cancer, such as liver and prostate cancers. JNKs are best known for their role in the activation of the c-Jun/activator protein-1 (AP-1) transcription-factor complex. AP-1 activation is required for neoplastic transformation 6 and for skin tumor formation in mice 7. Tumor formation is inhibited in c-Jun-knockout mice 8. A recent study suggested that the interaction of the tumor suppressor p16INK4a with JNK1 can occur at the same site where c-Jun binds, and it interferes with the phosphorylation and activation of c-Jun in response to UV exposure 9. Additionally, JNKs are crucial mediators of obesity and insulin resistance and potential targets in type II diabetes 10. Therefore, inhibition of JNKs might provide clinical benefits in chronic disease.
The phosphatidylinositol 3-kinase (PI3-K)/AKT signaling pathway has been identified as a key player in human cancer, including skin cancer 11, and is considered an attractive target for cancer prevention or treatment. This pathway can also regulate JNKs. Sawyers and colleagues recently showed, using an elegant screening technique, that JNK pathway activation is a major consequence of PTEN loss, suggesting that PI3-K promotes cancer progression by inducing the parallel activation of AKT and JNKs 12. PTEN deficiency sensitizes cells to JNKs inhibition. Moreover, negative feedback regulation of PI3-K was impaired in PTEN-null cells. Thus, dual JNKs and PI3-K inhibition might be a novel and effective therapeutic approach in patients, preventing feedback and cross-talk.
Flavonoids have been known for some time for their general chemopreventive effects in human health, which might be explained partially by the identification of the molecular targets and their mechanism of action. An earlier, small-scale study examined the effects of 24 flavonoids on AP-1 transactivation and c-Jun phosphorylation in cell-based systems 15. To identify a novel natural inhibitor of JNK1, we examined the activity of four representative flavonoids (quercetagetin, quercetin, myricetin, and kaempferol) using an in vitro kinase screening system. Only quercetagetin strongly suppressed JNK1 activity. Here, we report the crystal structure of JNK1 bound to quercetagetin and the effects of quercetagetin in in vitro and in vivo models. The results of a docking study suggest that PI3-K is also a molecular target of quercetagetin.
Results
Crystal structure of the ternary JNK1–pepJIP1–quercetagetin complex
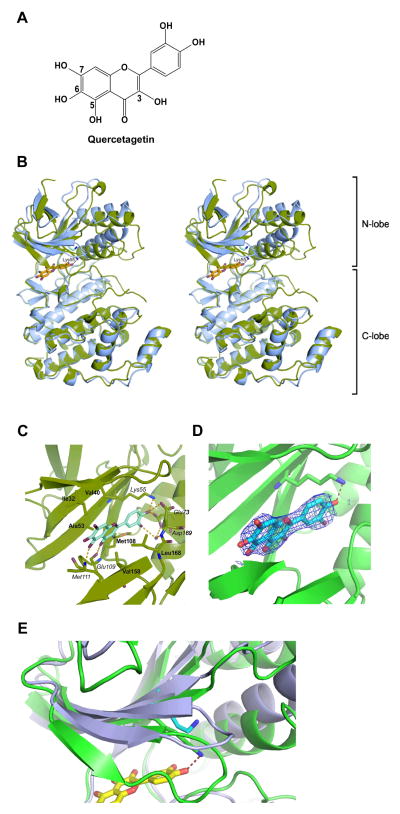
To investigate the molecular basis of the inhibition of JNK1 by quercetagetin (Figure 1A), we determined the crystal structure of the JNK1–pepJIP1–quercetagetin ternary complex, where pepJIP1 is a docking site peptide fragment of the scaffold protein JIP1. JNK1 consists of N- and C-terminal lobes linked through a loop, the ‘hinge region.’ Interestingly, the N-terminal lobe of JNK1 underwent substantial structural changes in our structure when compared with the apo form 16. The whole N-terminal lobe region is rotated toward the C-terminal lobe, causing shifts of approximately 2.5 Å in peripheral segments (Figure 1B). Similar rearrangement was observed in the structure of the JNK1-α1 isoform in complex with a biaryl tetrazol inhibitor (A-82118; PDB-code 3O2M 17), which does not utilize the ATP-binding site. Quercetagetin is located in the ATP-binding site and forms hydrogen bonds with the protein (Figure 1C, D). The side chains of Lys55, Asp169, and Glu73 form a network of hydrogen bonds with the 4-hydroxy group of the 3,4-dihydroxyphenyl part (the catechol moiety) of the ligand, while the benzopyran portion forms hydrogen bonds with the protein main chain of Glu109 and Met111 (in the hinge loop). The ligand forms additional hydrophobic interactions with both non-polar faces of the binding cleft (Ile32, Val40, Ala53, Met108, Val158, and Leu168 from the N- and C-terminal lobes). A water molecule in the hydrogen-bonding network connects the 3,4-dihydroxyphenyl part of quercetagetin with the Asp169 main-chain amide nitrogen and the Glu73 side-chain carboxy group. The movement of the N-terminal lobe narrows the binding site, which allows the Lys55-NH2 group to form a hydrogen bond with quercetagetin and moves Val158 and Leu168 approximately 2 Å closer to the ligand. Additionally, the Lys30–Ala42 region of the N-terminal lobe folds over and caps the binding site. The glycine-rich loop Gly33–Gly38 is substantially shifted in this region, with the Gly35 Cα atom moving 6 Å toward the C-terminal lobe from its position in the apo structure (Figure 1E). The adaptability of this region is a feature of many kinase structures 18. All changes in the N-terminal lobe of JNK1 seem to improve the compatibility of the ligand with the binding site and must be taken into account when designing JNK1-specific inhibitors based on the apo structure.
Figure 1.
Crystal structure of the ternary JNK1-pepJIP1-quercetagetin complex. A, chemical structure of quercetagetin. B, structures of apo-JNK1 alone (blue) and in complex with quercetagetin (green), presented as stereo ribbon plots. In the complex, the N-terminal lobe is shifted substantially toward the C-terminal lobe and Lys55 assumes a different conformation to form a hydrogen bond with the ligand. The glycine-rich loop (Gly33-Gly38) relocates to cover the binding site. C, molecular interactions between quercetagetin and JNK1. Residues involved in hydrophobic interactions are highlighted in bold, and those involved in hydrogen bonding in italics. All hydrogen bonds are marked as dashed yellow lines. D, 2Fo-Fc omit-map of quercetagetin electron density contoured at 1σ. The map was created without quercetagetin in the starting model. The density data allowed unambiguous building of the ligand molecule. The hydrogen bond formed with Lys55 is shown. E, enlarged view of the Lys30-Ala42 region of the N-terminal lobe. Apo-JNK1 is shown in blue, and the complex with quercetagetin in green. The region loses its beta-strand configuration upon binding, and folds over and caps the binding site. The glycine-rich loop Gly33-Gly38 is substantially shifted in this region, with the Gly35 moving by 6 Å toward the C-terminal lobe from its position in the apo-structure.
Determination of IC50 values for JNK1 with the inhibitors quercetagetin and SP600125 using the IMAP system
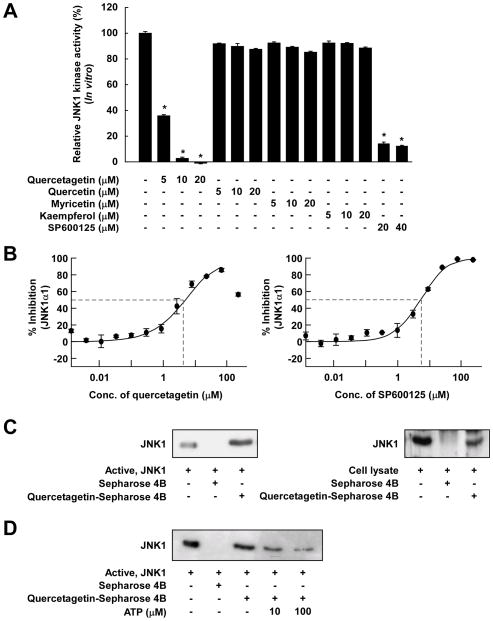
Next, we examined the effect of quercetagetin and other flavonoids on JNK1 activity. In vitro JNK1 kinase assays revealed that quercetagetin inhibited JNK1 activity more potently than did SP600125 (a pharmacological JNK1 inhibitor). Quercetin, myricetin, and kaempferol had no effect on JNK1 activity in vitro (Figure 2A). We next calculated IC50 values for quercetagetin and SP600125 using the IMAP assay system 19. The IC50 values for quercetagetin and SP600125 were 4.6 μM and 5.2 μM, respectively (Figure 2B). These values are in accord with the in vitro JNK1 kinase assay results.
Figure 2.
Effects of quercetagetin on JNK1 activity. A, comparison of the effects of various flavonoids and SP600125 on JNK1 activity. JNK1 kinase assays were performed as described in Materials and Methods. Kinase activity is presented as percent inhibition relative to the corresponding untreated control. Average 32P count was determined from the results of three independent experiments. Data are presented as means ± S.D. Asterisks (*) indicate significant differences in kinase activity between active JNK1 and individual compounds and active JNK1 alone (kinase assays; P < 0.05). B, inhibition of JNK1 by quercetagetin and SP600125 and comparison of the IC50 values in an IMAP-based FP assay. An IMAP assay was performed as described in Materials and Methods. Each experiment was performed in triplicate. The data point for quercetagetin titration above 100 μM is affected by limited solubility and not included in the calculation and curve fitting. C, JNK1-quercetagetin binding in vitro and ex vivo was confirmed by Western blotting using an antibody against JNK1. Lane 1 (input control), JNK1 protein standard or lysate; lane 2 (control), JNK1 or lysate pulled down using Sepharose 4B beads; lane 3, JNK1 or lysate pulled down using quercetagetin-Sepharose 4B affinity beads. D, quercetagetin competes with ATP for binding to JNK1. Lane 1, input control; lane 2, negative control, indicating that JNK1 cannot bind to Sepharose 4B; lane 3, positive control, indicating that JNK1 can bind to quercetagetin-Sepharose 4B. Each experiment was performed three times.
Quercetagetin competes with ATP for binding to JNK1
Pull-down assays showed that active JNK1 binds to quercetagetin-conjugated Sepharose 4B beads (Figure 2C, left panel, lane 3) but not to unconjugated Sepharose 4B beads (Figure 2C, left panel, lane 2). The input lane (Figure 2C, left panel, lane 1), to which 50 ng of active JNK1 was loaded (as a marker), verified that the detected band represented JNK1. Cell-based pull-down assays further revealed that quercetagetin strongly binds to UVB-induced JNK1 in JB6 P+ cells (Figure 2C, right panel). Furthermore, the ability of quercetagetin to bind to JNK1 varied according to ATP levels(Figure 2D), suggesting that, as expected from its location in the ATP-binding site in the crystal structure, quercetagetin competes with ATP.
Evaluation of the binding affinity of quercetagetin for JNK1 by docking analysis
The results of the docking procedure (Supplemental Table 1) are represented as Glide docking scores. The Glide docking score is a semi-quantitative measure of binding energy. The lower the value, the higher the binding affinity. The goal of the docking procedure is to explain the preference of JNK1 for quercetagetin over the other flavonoids and SP600125. The results indicate that quercetagetin had the lowest docking score, and thus the highest binding affinity, under both sets of docking conditions (Supplemental Table 1). This can be explained by consideration of the entropic terms in the docking score. Although the interaction energy for the binding of SP600125 to JNK1 was lower than that for the tested flavonoids, SP600125 has a lower loss of entropy upon binding. The Glide scoring function applied higher entropic penalties to the flavonoids. Consequently, their predicted binding affinities were reduced. Considering this penalization, quercetagetin was predicted to have the highest binding affinity, followed by SP600125, in agreement with the results of the kinase assays. But it is clear that the differences in the flavonoid series are small, perhaps not surprising in view of their related chemistry. Detailed ranking on the basis of these calculations must obviously be viewed with caution.
Quercetagetin inhibits PI3-K activity through direct ATP-competitive binding
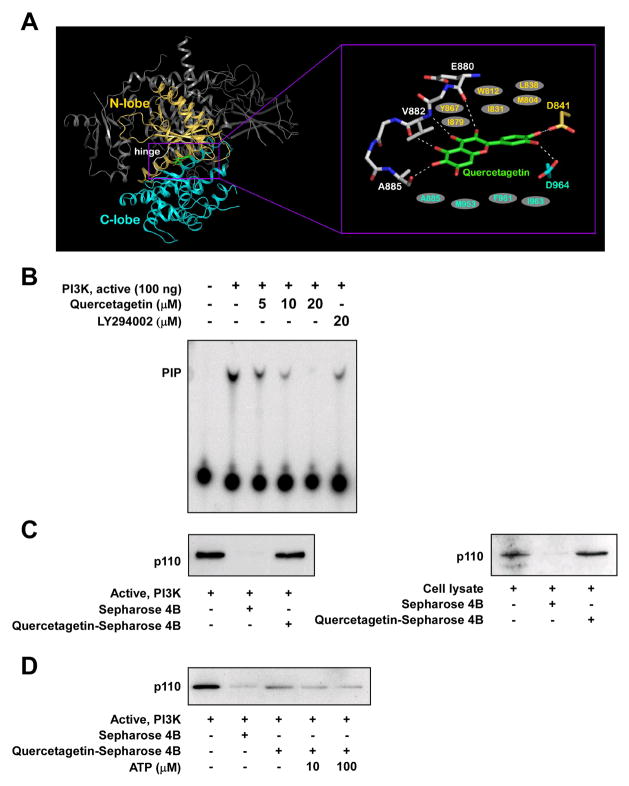
A previous study suggested that quercetin binds to PI3-K 22. Because the chemical structure of quercetin is very similar to that of quercetagetin, we tested the interaction of quercetagetin with PI3-K in a modeling study using the crystal structure of PI3-K in complex with quercetin 22 and found that quercetagetin docked well to the ATP-binding site of PI3-K (Figure 3A). The hydroxyl groups at the 3, 5, and 6 positions and the carbonyl group at the 4 position can form hydrogen bonds with the backbone atoms of the hinge region (amino acids 880–885). The hydroxyl groups at the 3′ and 4′ positions can form hydrogen bonds with the side chains of Asp964 and Asp841, respectively. Additionally, quercetagetin would be sandwiched by the side chains of the hydrophobic residues in the ATP-binding site, including Met804, Trp812, Ile831, Leu838, Tyr867, and Ile879 from the N-terminal lobe and Ala885, Met953, Phe961, and Ile963 from the C-terminal lobe. To confirm that the interaction between PI3-K and quercetagetin leads to the suppression of PI3-K activity, we tested PI3-K activity in vitro. At a concentration of 20 μM, quercetagetin almost completely blocked PI3-K activity, the inhibitory effect being greater than that observed with the specific PI3-K inhibitor LY294002 (Figure 3B). We performed a binding assay and verified that quercetagetin directly interacts with PI3-K and competes with ATP (Figure 3C and D). These findings suggest that PI3-K, like JNK1, is an important target of quercetagetin.
Figure 3.
Effects of quercetagetin on PI3-K activity. A, hypothetical model of PI3-K in complex with quercetagetin. The N-terminal lobe, C-terminal lobe, and hinge region of the catalytic domain are shown in yellow, cyan, and white, respectively. Quercetagetin (atomic color) binds to the ATP-binding site in the catalytic domain of PI3-K. In the close-up view, the hydrogen bonds are depicted as dashed lines, and the residues in gray ellipses are hydrophobic residues interacting with quercetagetin. B, quercetagetin inhibits PI3-K activity in vitro. The resulting 32P-labeled phosphatidylinositol-3-phosphate was measured as described in Materials and Methods. C, quercetagetin specifically binds to the p110 subunit of PI3-K in vitro and ex vivo, as confirmed by Western blotting with an antibody directed against the p110 subunit. Lane 1, PI3-K protein standard or whole-cell lysate (input control); lane 2, PI3-K or lysate precipitated with Sepharose 4B beads (control); lane 3, PI3-K or whole-cell lysate pulled down using quercetagetin–Sepharose 4B affinity beads. D, quercetagetin binds to PI3-K in an ATP-competitive manner. Lane 1, input control; lane 2, negative control showing lack of binding of PI3-K to Sepharose 4B beads; lane 3, binding of PI3-K to quercetagetin–Sepharose 4B (positive control); lanes 4 and 5, increasing concentrations of ATP alter the binding of quercetagetin to PI3-K. Each experiment was performed three times.
Quercetagetin suppresses UVB-induced AP-1 and NF-κB transactivation and RasG12V- and H-ras-induced cell transformation in JB6 P+ cells by targeting JNK1 and PI3-K
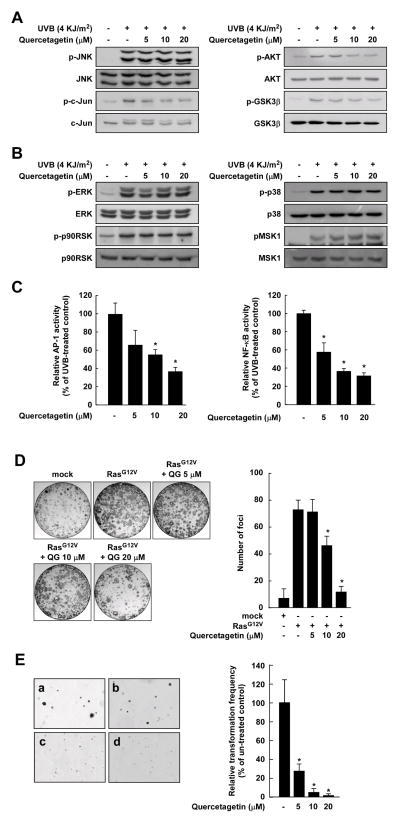
Consistent with the JNK1 and PI3-K kinase assay data, quercetagetin strongly suppressed UVB-induced phosphorylation of c-Jun, AKT, and GSK3β, but not JNKs, ERKs, p90RSK, p38, or MSK1 (Figure 4A, B). Quercetagetin inhibited UVB-induced transactivation of AP-1 and NF-κB in a dose-dependent manner (Figure 4C). Previous studies showed that H-Ras acts as a potent activator of the JNKs and PI3-K signaling pathways that lead to neoplastic transformation. To examine the effects of quercetagetin on cell transformation, we introduced Ras (RasG12V) expression vectors into NIH3T3 cells and conducted a focus-forming assay. As expected, quercetagetin inhibited H-Ras-induced cell transformation and RasG12V-induced focus formation in a dose-dependent manner (Figure 4D, E). At a concentration of 5 μM, quercetagetin inhibited H-Ras-induced neoplastic cell transformation by 73%. These findings suggest that quercetagetin suppresses cell transformation by targeting JNK1 and PI3-K.
Figure 4.
Effects of quercetagetin on UVB-induced JNKs and PI3-K signaling. A and B, quercetagetin inhibits UVB-induced phosphorylation of c-Jun, AKT, and GSK3β, but not JNKs, ERKs, p90RSK, p38, or MSK1. The data are representative of three independent experiments that gave similar results. C, quercetagetin inhibits UVB-induced AP-1 and NF-κB transactivation. Luciferase activity was assayed and AP-1 and NF-κB activities calculated relative to the values for control cells (without UVB). Data are presented as means ± S.D. of AP-1 and NF-κB luciferase activities obtained from three independent experiments. D, quercetagetin inhibits RasG12V-induced focus formation. The data are representative of three independent experiments that gave similar results. The graph shows the average number of foci. E, effects of quercetagetin on H-Ras-induced cell transformation in untreated control cells (a), and cells treated with quercetagetin at a concentration of 5 μM (b), 10 μM (c), or 20 μM (d). Data are presented as means ± S.D. of three independent experiments. The asterisk (*) indicates a significant difference (p < 0.05) between groups untreated or treated with quercetagetin. For D and E, cell colonies were counted under a microscope with the aid of Image-Pro Plus software (v. 4).
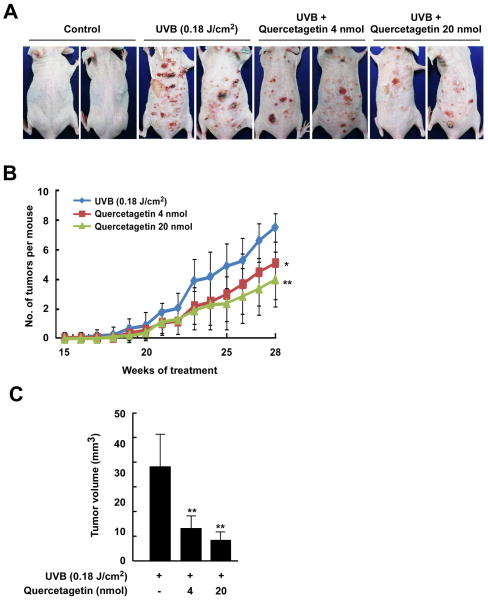
Quercetagetin inhibits UVB-induced skin tumorigenesis in an SKH-1 hairless mouse model
To investigate the pharmaceutical effect of quercetagetin, we used a two-stage mouse skin tumorigenesis model. We found that quercetagetin significantly inhibited UVB-induced skin cancer development (Figure 5A). Topical application of 4 or 20 nmol of quercetagetin to mouse skin reduced tumor incidence by 32.0% and 46.7%, respectively (p < 0.001 vs the UVB irradiation group, n = 10; Figure 5B). The volumes of the skin tumors that developed in UVB-exposed mice were significantly reduced by quercetagetin treatment (Figure 5C). Overall, these results indicate that quercetagetin might serve as an effective chemopreventive agent against UVB-mediated skin cancer.
Figure 5.
Effect of quercetagetin on UVB-induced skin carcinogenesis and COX-2 expression in SKH-1 hairless mice. A, external appearance of UVB-induced tumors. B, quercetagetin strongly reduces the incidence of UVB-induced tumors in SKH-1 hairless mice. A tumor was defined as an outgrowth of > 1 mm in diameter that persisted for 2 weeks or longer. Tumor incidence and multiplicity were recorded each week until the end of the experiment. C, quercetagetin strongly reduces UVB-induced tumor volume in mice. At the end of the study, the dimensions of each tumor in each mouse were recorded. Tumor volume was calculated using the hemiellipsoid model formula: Tumor volume = 1/2 (4π/3) (l/2) (w/2) h, where l is the length, w is the width, and h is the height. The data were analyzed using SAS software (SAS Institute, Cary, NC).
Discussion
The development of clinically useful small-molecule kinase inhibitors has been a seminal event in the world of chronic disease. Although many natural compounds have been shown to regulate the activity of kinases in cell-based assays, fewer data exist to show that these molecules can directly bind to and inhibit specific target proteins in vitro. In the present study, we have shown, by X-ray crystallography, that JNK1 crystallizes as the apo form in the more open configuration16. We observed the same crystal form without bound ligand, even when ligand was present in the crystallization medium. The closed conformation, however, only crystallizes in the presence of ligand, which has a well defined electron density. Figure 1B is an overlay of the two experimental structures, the apo-form16 and the complex described and documents the pronounced relative movement of the N-and C-terminal domains upon ligand binding. Hinge bending in kinases is indeed well known and has been seen already in the first cAMP-dependant protein kinase structures18. The mechanism by which ligand binding induces hinge bending, however, is unclear. A distinction between the two limiting cases, induced fit or conformational selection, requires kinetic data for different ligand concentrations with a sufficient time resolution to identify the initial reaction step, which would be unimolecular for conformational selection and bimolecular for induced fit. We are not aware of any kinase system where these experiments have been performed.
Quercetagetin is a direct ligand for the ATP-binding pocket of JNK1 (Figure 1). Interestingly, the interaction between the Lys55–NH2 group and quercetagetin allows movement and changes in the N-terminal lobe of JNK1. This feature improves compatibility of the ligand with the binding site of JNK1 and is a unique binding mode. Quercetagetin reaches much deeper into the ATP-binding pocket with its catechol side group than the JNK1 inhibitor SP600125, making prominent contacts with Lys 55 and Glu73 through their hydroxyl groups, which may trigger the aforementioned N-terminal lobe movement. SP600152 binds to the P212121 crystal form (apo form) of JNK1 without causing substantial structural rearrangements 16. Quercetagetin binds to JNK1 similar to quercetin-PIM1 kinase (PDB ID: 2O3P 14). However the catechol moiety with its meta-hydroxyl pointing inside the binding cleft in our JNK1-quercetagetin complex is rotated by 180° to that observed by Holder et al. (PDB ID: 2O64 14) in the quercetagetin-PIM1 kinase complex. This difference might be caused by different environments of the catechol group. That quercetagetin can “adjust” to bind both PIM1 and JNK1 kinases by rotation of its only rotatable bond is interesting. As a note of caution, the electron density of the quercetagetin ligand is best interpreted as shown (meta-hydroxyl group ‘in’ (Figure 1D), but we cannot exclude partial occupation by the alternative conformer (meta-hydroxyl group ‘out’). The interaction mode in JNK1-quercetagetin is also similar to that shown in the structure of quercetin with phosphatidylinositol 3-kinase γ (PDB ID: 1E8W 22). Thus, the 4-hydroxy group of the catechol forms the majority of the hydrogen bonding, whereas in other structures, the hydrogen bonds are formed by the 3-hydroxy group. Another unique feature of our structure is the presence of a water molecule trapped below the ligand. Its presence suggests that increasing ligand selectivity and affinity might be realized by introducing a polar group to replace the water molecule. This was tested by changing the catechol moiety in silico to pyrogallol, whereby the additional hydroxyl group occupies approximately the space of the observed water molecule. The calculation of the glide score indicates a decrease (affinity improvement) of 0.7 (Glide score −13.2) compared to quercetagetin. A hydroxymethyl group would be even a closer mimic of the bound water and the corresponding quercetagetin derivative indeed has a glide score of −14.1, the highest in the series.
Quercetagetin inhibited JNK1 activity more strongly than did quercetin, myricetin, kaempferol, or SP600125. The IC50 value of quercetagetin, as determined by IMAP assay, was 4.6 μM in agreement with the in vitro JNK1 kinase assay. The binding of JNK1 and quercetagetin in an ATP-competitive manner was confirmed in a pull-down assay (Figure 2C, D). To further evaluate the binding affinity of quercetagetin, quercetin, myricetin, kaempferol, and SP600125, a series of docking simulations was created (Supplemental Table 1). The docking confirmed the unusually deep placement of the quercetagetin molecule in the pocket, which agreed with the observed structural and kinetic data. In the predicted binding conformation of quercetagetin, however, the catechol ring was flipped by 180° compared with the X-ray crystal structure. Obviously, the complicated nature of the H-bond pattern in this region, which is also influenced by the structural water molecule, is difficult to model. Nevertheless, quercetagetin is predicted to have the highest binding affinity for JNK1, followed by SP600126. Walker et al. presented X-ray crystallographic structures of the PI3-K γ-quercetin complex, which binds in the ATP-binding pocket 22. According to their structure, quercetin binds to PI3-K very differently from the JNK1 and PIM1 kinase complexes. Because the chemical structures of quercetagetin and quercetin have a high degree of similarity, we hypothesized that quercetagetin could also interact with PI3-K. The results of a docking study indicated that quercetagetin is an ATP-competitive inhibitor of PI3-K. Several hydrogen bonds and hydrophobic interactions are involved in the binding of quercetagetin. Notably, the hydroxyl groups at the 3′ and 4′ positions form hydrogen bonds with the side chains of Asp964 and Asp841 (Figure 3A). As shown experimentally (Figure 3B, C, D), quercetagetin strongly inhibits PI3-K activity through ATP-competitive binding, consistent with the above docking results.
In cell based-systems and an animal model, we examined the functional significance of the binding of quercetagetin to JNK1 and PI3-K. In these studies, quercetagetin inhibited UVB-induced phosphorylation of c-Jun and AKT in JB6 P+ cells, but had no effect on the phosphorylation of ERKs or p38. Inhibition of JNKs and PI3-K signaling led to the suppression of neoplastic transformation through inhibition of AP-1 and NF-κB (Figure 4). Quercetagetin delayed the development of tumors and reduced tumor volumes in an SKH-1 hairless mice model (Figure 5). These cancer chemopreventive effects of quercetagetin might be explained by its inhibitory effects on JNK1 and PI3-K activities. Overall, our crystallographic findings, the accompanying biochemical data, and the cell based and animal model data show that quercetagetin is a strong inhibitor of JNK1 and PI3-K and might have practical implications for the prevention or therapy of cancer and other chronic diseases.
Materials and Methods
Protein Expression and Purification
To express JNK1 for structural analysis, the C-terminal truncated form of human JNK1α1 (residues 1–364) was amplified by a standard PCR-based cloning strategy. The PCR product was inserted into the pET21b expression vector (Novagen) with a 6 His-tag at the C-terminus and this plasmid was transformed into BL21(DE3) Escherichia coli cells. The transformant cells were grown in LB medium at 37°C up to an A600 nm of 0.6. Protein expression was induced by adding 1 mM IPTG, and the cells were grown for 15 h. Cells were then harvested by centrifugation, resuspended in buffer A containing 50 mM HEPES (pH 7.2), 10% glycerol, 100 mM NaCl, 2 mM β-mercaptoethanol, and protease inhibitors (0.1 mM phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml pepstatin), and frozen quickly by immersion in liquid nitrogen. Then, JNK1 was purified. Briefly, cells were thawed, sonicated, and centrifuged. The supernatant fraction was passed through a 10 ml Ni-NTA Superflow Column. The column was washed with resuspension buffer A and washed again with buffer A plus 10 mM imidazole. The protein was then eluted with buffer A plus 250 mM imidazole. The eluted protein was dialyzed against buffer B (20 mM HEPES pH 7.0, 10% glycerol, 50 mM NaCl, 2 mM DTT) and applied to an SP-Sepharose cation-exchange column. The column was washed with seven column volumes of buffer B, and bound protein was then eluted with a ten-column-volume linear gradient of 50–400 mM NaCl. The eluted protein from the SP-Sepharose column was concentrated and passed over a gel filtration column (Superdex 200) pre-equilibrated with buffer C (25 mM HEPES (pH 7.0), 5% glycerol, 50 mM NaCl, 10 mM DTT). Peak fractions were concentrated to 10 mg/ml as measured by the Bradford method. Purity was judged to be > 98 % by Coomassie Blue-stained SDS-PAGE.
Crystallization and Data Collection
Before the crystallization trial, the purified protein was mixed with a peptide fragment of JIP1 (pepJIP1) with the sequence RPKRPTTLNLF at a molar ratio of 1:5 and incubated on ice for 3 h to allow complex formation. To obtain the JNK1–pepJIP1–quercetagetin ternary complex, the JNK1–pepJIP1 complex was mixed with a 10-fold excess of quercetagetin and concentrated to approximately 10 mg/ml. Crystallization was achieved at 4°C by vapor diffusion using the sitting drop method and a protein-to-well solution ratio of 1:1 with well solution containing 2.1 M (NH4)2SO4 and 0.1 M Bis-Tris (pH 5.5). Single crystals grew within 1 week to an average size of 0.3 × 0.1 × 0.1 mm. Crystals were transferred to cryoprotectant solution containing well solution plus 25% (v/v) ethylene glycol for a few seconds and then flash-frozen in liquid nitrogen. Datasets to 2.60 Å resolution were collected at 100 K on the PXII beamline at the SLS synchrotron (Paul Scherrer Institute, Switzerland) and processed using XDS and XSCALE software 23. Two different crystal forms were measured, one of which belonged to the space group P212121. The unit cell parameters were a = 61.24 Å, b = 80.31 Å, c = 83.36 Å. The second belonged to space group I422, and its cell parameters were a = b = 172.37, c = 86.04. Both crystal types contained one complex per asymmetric unit. Table I summarizes the statistics for data collection and refinement.
Table 1.
Data Collection and refinement statistics
| Data collection | ||
| Space group | I422 | |
| Cell constants (Å) | a=b= 172.37 c= 86.04 |
|
| Resolution range (Å) | 50 - 2.7 | |
| Wavelength (Å) | 1.0 | |
| Observed reflections | 138356 | |
| Unique reflections | 13950 | |
| Whole range | ||
| Completeness (%) | 77.0* | |
| Rmerge | 3.9 | |
| I/σ(I) | 26.33 | |
| Last shell | ||
| Resolution range (Å) | 2.7 – 2.8 | |
| Completeness (%) | 43.6 | |
| Rmerge | 15.5 | |
| I/σ(I) | 2.98 | |
|
| ||
| Refinement | ||
|
| ||
| No. of reflections | 13278 | |
| Resolution (Å) | 20 - 2.7 | |
| R-factor (%) | 22.6 | |
| Rfree (%) | 26.5 | |
| Average B (A2) | 63.6 | |
| R.m.s. bond length (Å) | 0.013 | |
| R.m.s. angles (°) | 1.458 | |
|
| ||
| Content of asymmetric unit | ||
|
| ||
| No. of protein-ligand complexes | 1 | |
| No. of protein residues/atoms | 362/2898 | |
| No. of solvent molecules | 27 | |
The data statistics are reported with signal to noise cutoff equal to 2. Using this cutoff, the data are 77% complete. Without cutoff, the data are 99% complete.
Structure Determination and Refinement
The structures of the JNK1–pepJIP1–quercetagetin ternary complex was solved with the molecular replacement program Phaser 24 using the N-and C-terminal domains of the structure of the binary complex JNK1–pepJIP1 separately 16, PDB accession code 1UKH). The crystals, which belonged to space group P212121, were isomorphous to the 1UKH unit cells and did not contain interpretable ligand density. The other space group, I422, contained an interpretable ligand density in the ATP-binding site. The model was subsequently improved by rigid body refinement of the individual domains and restrained refinement using Refmac software 25 and rebuilt using the Coot and X-fit programs 26. Water molecules were added by Arp/Warp 27. The refinement statistics are summarized in Table I.
Accession Numbers
The coordinates of the crystal structure of the ternary complex JNK1-pepJIP1- quercetagetin have been deposited in the Protein Data Bank under accession code 3V3V.
IMAP-based FP assay
An IMAP-based FP (Immobilized Metal Ion Affinity-Based Fluorescence Polarization) assay was carried out in accordance with the instructions provided by Molecular Devices. The IMAP reaction was carried out with recombinant JNK1 in 384-well black plates containing serially diluted test compounds. The reaction contained 7.46 μM ATP, 100 nM JNK1, 400 nM FITC-labeled JNK1 substrate peptide (LVEPLTPSGEAPNQK-5FAM-COOH), 20 mM MOPS (pH 6.5), 1 mM DTT, 10 mM MgCl2, and 0.01% Brij35. It was incubated for 1 h at room temperature with the addition of IMAP Binding Buffer (a 1:1200 dilution of IMAP Progressive Binding Reagent in 65% IMAP Progressive Binding Buffer A/45% IMAP Progressive Binding Buffer B). Then the plate was read using a PHERAstar Plus microplate reader from BMG Labtech. The excitation and emission wavelengths were 485 nm with a bandwidth of 20 nm and 530 nm with a bandwidth of 25 nm, respectively.
Docking simulations
To further evaluate the binding affinity of quercetagetin in comparison with other flavonoids, a series of docking simulations were performed. A set of 5 inhibitors (the flavonoids quercetagetin, quercetin, myricetin, and kaempferol and the commercially available inhibitor SP600126) was docked in the ATP-binding sites of two JNK1 structures: 1) JNK1 in complex with quercetagetin (reported in this manuscript) and 2) the apo-protein structure (PDB ID 1UKH). A 25 Å simulation box was defined around the binding pocket. The geometric center of the docking box was chosen to coincide with the molecule’s center of mass. The docking procedure consisted of 3 stages: 1) inhibitor-receptor pose generation; 2) pose minimization; and 3) scoring of the final pose. After the first stage, 400 poses were selected for energy minimization (100 steps of Steepest Descendant). The XP-scoring function of Glide version 5.6 was used to evaluate the final pose. No constraints were imposed on the system.
Molecular modeling and docking
Insight II (Accelrys, Inc., San Diego, CA) was used for docking studies and structure analysis with the crystal coordinates of PI3-K in complex with quercetin (accession code 1E8W), available from the Protein Data Bank (http://www.rcsb.org/pdb/).Docking was subsequently performed using the XP-scoring function of Glide version 5.6 (Schrödinger, LLC, New York, NY, 2010). No constraints were imposed on the system. For each ligand, 400 poses were selected for energy minimization (100 steps Steepest Descent) and scoring.
In vitro JNK1 kinase assays
The in vitro kinase assay was conducted in accordance with the instructions provided by Upstate Biotechnology. Briefly, each reaction contained 20 μL of assay dilution buffer [20 mmol/L MOPS (pH 7.2), 25 mM β-glycerophosphate, 5 mM EGTA, 1 mM sodium orthovanadate (Na3VO4), 1 mM DTT] and a magnesium-ATP cocktail buffer. For JNK1, the activating transcription factor 2 substrate peptide was included at a concentration of 3 μM. Active JNK1 protein (20 ng) and 10 μL of diluted [γ-32P]ATP solution were incubated at 30°C for 10 min with the above assay buffer and substrate peptide, and then 15 μL aliquots were transferred onto p81 paper, and washed three times with 0.75% phosphoric acid (5 min per wash) and once with acetone (5 min). The incorporation of radioactivity was determined using a scintillation counter (LS6500, Beckman Coulter). Each experiment was performed in triplicate.
In vitro PI3-K kinase assay
Active PI3-K protein (100 ng) was incubated with quercetagetin for 10 min at 30°C. The mixture was then incubated with 20 μL of 0.5 mg/ml phosphatidylinositol (Avanti Polar Lipids, Alablaster, AC) for 5 min at room temperature and then incubated in reaction buffer [100 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (pH 7.6), 50 mM MgCl2, 250 μM ATP] containing 10 μCi of [γ-32P]ATP for an additional 10 min at 30°C. The reaction was stopped by adding 15 μL of 4 N HCl and 130 μL of chloroform:methanol (1:1). After vortexing, 30 μL of the lower chloroform phase were spotted onto a 1% potassium oxalate-coated silica gel plate, which had previously been activated through incubation for 1 h at 110°C. The resulting 32P-labeled phosphatidylinositol-3-phosphate was separated by thin-layer chromatography, and the radiolabeled spots were visualized by autoradiography.
Statistical analysis
As necessary, data are expressed as means ± S.D. or S.E., and significant differences were determined using one-way ANOVA. A probability value of p < 0.05 was used as the criterion for statistical significance. All analyses were performed using Statistical Analysis Software (SAS, Inc.).
Supplementary Material
Highlights.
Quercetagetin is a direct ligand for the ATP-binding pocket of JNK1.
Quercetagetin is also an ATP-competitive inhibitor of PI3-K.
Quercetagetin inhibits JNK1 and PI3K activities, and suppresses cell transformation.
Quercetagetin delayed the development of tumors in an SKH-1 hairless mice model.
We report the potential use of quercetagetin in the prevention or therapy of cancer.
Acknowledgments
This work was supported by The Hormel Foundation and grants from the National Institutes of Health CA027502, CA120388, R37 CA081064 and ES016548, USA; by the World Class Institute Program (2009-002), World Class University Program (R31-2008-00-10056-0) and Leap Research Program (2010-0029233), the Ministry of Education, Science and Technology, and by the Next-Generation BioGreen 21 Program (Plant Molecular Breeding Center No. PJ008187-02-2011), Rural Development Administration, Republic of Korea; and by a Deutsche Krebshilfe Grant (108354) and the CONACYT-DAAD, Germany (M. A.). We thank Tonya M. Poorman at The Hormel Institute, University of Minnesota, USA for help submitting our manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Martin JH, Mohit AA, Miller CA. Developmental expression in the mouse nervous system of the p493F12 SAP kinase. Brain Res Mol Brain Res. 1996;35:47–57. doi: 10.1016/0169-328x(95)00181-q. [DOI] [PubMed] [Google Scholar]
- 2.Wagner EF, Nebreda AR. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer. 2009;9:537–49. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 3.Chang Q, Zhang Y, Beezhold KJ, Bhatia D, Zhao H, Chen J, Castranova V, Shi X, Chen F. Sustained JNK1 activation is associated with altered histone H3 methylations in human liver cancer. J Hepatol. 2009;50:323–33. doi: 10.1016/j.jhep.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ouyang X, Jessen WJ, Al-Ahmadie H, Serio AM, Lin Y, Shih WJ, Reuter VE, Scardino PT, Shen MM, Aronow BJ, Vickers AJ, Gerald WL, Abate-Shen C. Activator protein-1 transcription factors are associated with progression and recurrence of prostate cancer. Cancer Res. 2008;68:2132–44. doi: 10.1158/0008-5472.CAN-07-6055. [DOI] [PubMed] [Google Scholar]
- 5.Sakurai T, Maeda S, Chang L, Karin M. Loss of hepatic NF-kappa B activity enhances chemical hepatocarcinogenesis through sustained c-Jun N-terminal kinase 1 activation. Proc Natl Acad Sci U S A. 2006;103:10544–51. doi: 10.1073/pnas.0603499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong Z, Birrer MJ, Watts RG, Matrisian LM, Colburn NH. Blocking of tumor promoter-induced AP-1 activity inhibits induced transformation in JB6 mouse epidermal cells. Proc Natl Acad Sci U S A. 1994;91:609–13. doi: 10.1073/pnas.91.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young MR, Li JJ, Rincon M, Flavell RA, Sathyanarayana BK, Hunziker R, Colburn N. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc Natl Acad Sci U S A. 1999;96:9827–32. doi: 10.1073/pnas.96.17.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zenz R, Scheuch H, Martin P, Frank C, Eferl R, Kenner L, Sibilia M, Wagner EF. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell. 2003;4:879–89. doi: 10.1016/s1534-5807(03)00161-8. [DOI] [PubMed] [Google Scholar]
- 9.Choi BY, Choi HS, Ko K, Cho YY, Zhu F, Kang BS, Ermakova SP, Ma WY, Bode AM, Dong Z. The tumor suppressor p16(INK4a) prevents cell transformation through inhibition of c-Jun phosphorylation and AP-1 activity. Nat Struct Mol Biol. 2005;12:699–707. doi: 10.1038/nsmb960. [DOI] [PubMed] [Google Scholar]
- 10.Hirosumi J, Tuncman G, Chang L, Gorgun CZ, Uysal KT, Maeda K, Karin M, Hotamisligil GS. A central role for JNK in obesity and insulin resistance. Nature. 2002;420:333–6. doi: 10.1038/nature01137. [DOI] [PubMed] [Google Scholar]
- 11.Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–7. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 12.Vivanco I, Palaskas N, Tran C, Finn SP, Getz G, Kennedy NJ, Jiao J, Rose J, Xie W, Loda M, Golub T, Mellinghoff IK, Davis RJ, Wu H, Sawyers CL. Identification of the JNK signaling pathway as a functional target of the tumor suppressor PTEN. Cancer Cell. 2007;11:555–69. doi: 10.1016/j.ccr.2007.04.021. [DOI] [PubMed] [Google Scholar]
- 13.Edwards ML, Stemerick DM, Sunkara PS. Chalcones: a new class of antimitotic agents. J Med Chem. 1990;33:1948–54. doi: 10.1021/jm00169a021. [DOI] [PubMed] [Google Scholar]
- 14.Holder S, Zemskova M, Zhang C, Tabrizizad M, Bremer R, Neidigh JW, Lilly MB. Characterization of a potent and selective small-molecule inhibitor of the PIM1 kinase. Mol Cancer Ther. 2007;6:163–72. doi: 10.1158/1535-7163.MCT-06-0397. [DOI] [PubMed] [Google Scholar]
- 15.Ichimatsu D, Nomura M, Nakamura S, Moritani S, Yokogawa K, Kobayashi S, Nishioka T, Miyamoto K. Structure-activity relationship of flavonoids for inhibition of epidermal growth factor-induced transformation of JB6 Cl 41 cells. Mol Carcinog. 2007;46:436–45. doi: 10.1002/mc.20292. [DOI] [PubMed] [Google Scholar]
- 16.Heo YS, Kim SK, Seo CI, Kim YK, Sung BJ, Lee HS, Lee JI, Park SY, Kim JH, Hwang KY, Hyun YL, Jeon YH, Ro S, Cho JM, Lee TG, Yang CH. Structural basis for the selective inhibition of JNK1 by the scaffolding protein JIP1 and SP600125. EMBO J. 2004;23:2185–95. doi: 10.1038/sj.emboj.7600212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Comess KM, Sun C, Abad-Zapatero C, Goedken ER, Gum RJ, Borhani DW, Argiriadi M, Groebe DR, Jia Y, Clampit JE, Haasch DL, Smith HT, Wang S, Song D, Coen ML, Cloutier TE, Tang H, Cheng X, Quinn C, Liu B, Xin Z, Liu G, Fry EH, Stoll V, Ng TI, Banach D, Marcotte D, Burns DJ, Calderwood DJ, Hajduk PJ. Discovery and characterization of non-ATP site inhibitors of the mitogen activated protein (MAP) kinases. ACS Chem Biol. 6:234–44. doi: 10.1021/cb1002619. [DOI] [PubMed] [Google Scholar]
- 18.Bossemeyer D, Engh RA, Kinzel V, Ponstingl H, Huber R. Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5–24) EMBO J. 1993;12:849–59. doi: 10.1002/j.1460-2075.1993.tb05725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sportsman JR, Gaudet EA, Boge A. Immobilized metal ion affinity-based fluorescence polarization (IMAP): advances in kinase screening. Assay Drug Dev Technol. 2004;2:205–14. doi: 10.1089/154065804323056549. [DOI] [PubMed] [Google Scholar]
- 20.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–49. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 21.Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem. 2006;49:6177–96. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- 22.Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Mol Cell. 2000;6:909–19. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
- 23.Kabsch W. Automatic Processing of Rotation Diffraction Data from Crystals of Initially Unknown Symmetry and Cell Constants. Journal of Applied Crystallography. 1993;26:795–800. [Google Scholar]
- 24.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Lamzin VS, Wilson KS. Automated refinement of protein models. Acta Crystallogr D Biol Crystallogr. 1993;49:129–47. doi: 10.1107/S0907444992008886. [DOI] [PubMed] [Google Scholar]
- 26.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–65. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 27.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.