Abstract
During the past few years, crystallography of G protein-coupled receptors (GPCRs) has experienced exponential growth, resulting in structure determination of 14 distinct receptors, 7 of them in 2012 alone. Including closely related subtype homology models, this currently amounts to about 10% coverage of the GPCR superfamily. Agonist-bound active-state structures are now known for the adrenergic, rhodopsin, and adenosine receptor systems, including a structure of the receptor-G-protein complex for the β2-adrenergic receptor. Biochemical and biophysical techniques, such as nuclear magnetic resonance (NMR) and hydrogen/deuterium exchange-mass spectrometry (HDX-MS) are providing complementary insights on ligand-dependent dynamic equilibrium between different functional states. High resolution structures such as the 1.8 A adenosine A2a receptor are illistrating the receptor as allosteric machines, controlled not only by ligands, but by sodium, lipids, cholesterol, and water. This wealth of data is helping to redefine our knowledge of how GPCRs recognize such a diverse array of ligands and transmit signals 30 angstroms across the cell membrane, also shedding light on a structural basis of GPCR allosteric modulation and biased signaling.
Keywords: GPCR, recognition, diversity, activation, signaling
Introduction
G protein-coupled receptors (GPCRs) comprise the largest protein superfamily in mammalian genomes. They share a common seven-transmembrane topology and mediate cellular response to a variety of extracellular signals ranging from photons and small molecules to peptides and proteins (1). Diversity of the extracellular ligands is reflected in the structural diversity of more than 800 human GPCRs, which can be grouped in five major families and numerous subfamilies based on their amino acid sequence (2). Signal transduction by GPCRs is fundamental for most physiological processes, spanning from vision, smell and taste to neurological, cardiovascular, endocrine, and reproductive functions, thus, making the GPCR superfamily a major target for therapeutic intervention (3, 4). Current drug discovery efforts aim at both improving therapies for more than 50 established GPCR targets, and at expanding the list of targeted GPCRs (5, 6). In addition to regulating GPCR signal activation with agonists and inhibition with antagonists and inverse agonists, trends in modern pharmacology include discovery of allosteric and/or functionally selective modulators (7) that bias downstream signaling towards specific G-protein- or arrestin- activated pathways (8).
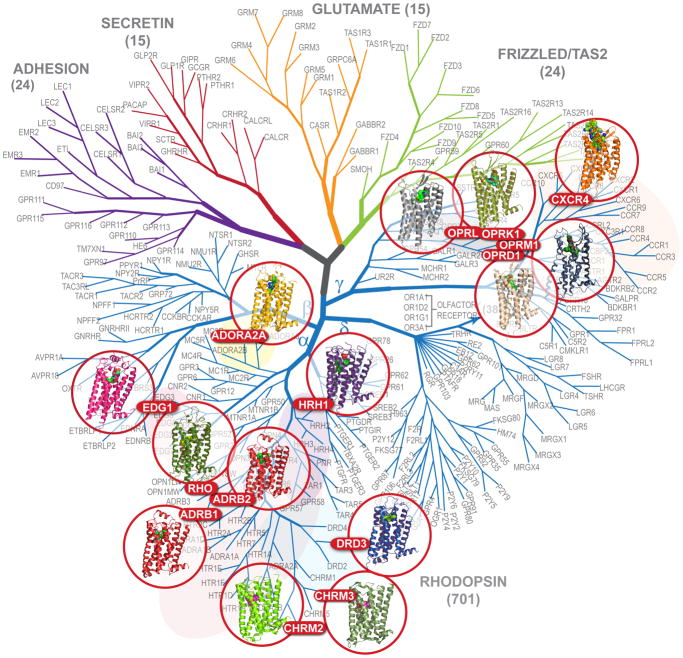
Prior to 2007, the structure and function of GPCRs, which had been extensively probed with biophysical and biochemical methods, was interpreted via ab initio or rhodopsin-based (9) modeling (e.g., reviewed in (10, 11)). Breakthrough developments in protein engineering (12, 13) and crystallography (14, 15) galvanized exponential growth in GPCR structure determination, which have thus far (as of May, 2012) resulted in structures of 14 receptors (Figure 1 and Table 1). Each of these GPCRs were initially captured in inactive states stabilized by antagonists or inverse agonists while the elevated conformational plasticity and heterogeneity of the activated states posed additional challenges for crystallization.
Figure 1.
Dendrogram of the human GPCR superfamily with the crystal structures solved. The tree based on sequence similarity in the seven-transmembrane domain is redrawn from (2). According to this notation, human GPCRs include Class A (Rhodpsin family), Class B (Secreting and Adhesion families), Class C (Glutamate family) and Frizzled/TAS2 Family. The Rhodopsin family is divided into Groups (α-γ). GPCRs can be further provisionally divided into clusters (e.g., aminergic), subfamilies (e.g., adrenergic or opioid) and individual GPCR subtypes (e.g., dopamine receptor subtype D3). Olfactory receptors comprise the largest distinct cluster of 388 receptors (only 4 subtypes shown) in δ-group of the Class A (Rhodopsin family) of GPCRs.
Table 1.
Crystal Structures of GPCRs
| Receptor | Gene Name | Species | Number of structures | References | Years | Resolution range (Å) |
|---|---|---|---|---|---|---|
| A2A adenosine | ADORA2A | Human | 12 | (19, 20, 44, 45, 111, 112, 57) | 2008—2012 | 1.80–3.34 |
| β1 adrenergic | ADRB1 | Turkey | 12 | (13, 42, 43,43a) | 2008—2012 | 2.30–3.65 |
| β2 adrenergic | ADRB2 | Human | 11 | (12, 21, 22, 27, 41, 109,113) | 2007—2011 | 2.40–3.50 |
| Chemokine CXCR4 | CXCR4 | Human | 5 | (33) | 2010 | 2.50–3.20 |
| Dopamine D3 | DRD3 | Human | 1 | (29) | 2010 | 2.89 |
| Histamine H1 | HRH1 | Human | 1 | (28) | 2011 | 3.10 |
| κ-opioid | OPRK1 | Human | 1 | (36) | 2012 | 2.90 |
| μ-opioid | OPRM1 | Mouse | 1 | (34) | 2012 | 2.90 |
| δ-opioid | OPRD1 | Mouse | 1 | (37) | 2012 | 3.40 |
| Nociceptin/orpha nin FQ (NOP) | OPRL1 | Human | 1 | (35) | 2012 | 3.00 |
| M2 muscarinic | CHRM2 | Human | 1 | (30) | 2012 | 3.00 |
| M3 muscarinic | CHRM3 | Rat | 1 | (31) | 2012 | 3.40 |
| S1P1 sphingolipid | EDG1 | Human | 1 | (32) | 2012 | 2.80–3.35 |
| Rhodopsin | RHO | Bovine/S quid | 23/4 | (9, 39, 15)a | 2000—2012 | 2.20–4.00 |
Rhodopsin structures citations shown for the first, the highest resolution and the first active-state structure
The first insights into active-state crystal structures were obtained in 2008 for ligand-free rhodopsin (opsin) (14, 16). Selective stabilization of the active-state resulted in active—state structures of rhodopsin (17, 18), A2AAR (19, 20), as well β2AR bound to an agonist and stabilized by a heterotrimeric Gs protein (21) or nanobody (22). These crystallographic insights are being greatly complemented by studies of GPCR conformational changes and dynamics obtained using biochemical methods and spectroscopy.
This review will discuss how the recent structural and biophysical insights contribute to our understanding of molecular interactions, subtype and functional selectivity of ligands, activation mechanisms and conformational dynamics, and other aspects of GPCR biological function. We will also outline emerging and future lines of inquiry for studying GPCR signaling mechanisms and harnessing them for drug discovery (23, 24).
Structural coverage of GPCR superfamily
Recent progress in GPCR structural characterization
Until 2007, structural information on the highly diverse repertoire of GPCRs (2) was limited to crystal structures of bovine rhodopsin (Class A) and structures of extracellular (EC) domains of Secretin (Class B) (25) and Glutamate (Class C) (26) families. By May of 2012, structures of 14 different Class A GPCRs, have been determined (Figure 1 and Table 1), seven of them published in the year 2012 alone, suggesting at this point an exponential growth trend. Half of the solved structures belong to aminergic receptors, a cluster of the Class A α-group GPCRs that binds monoamine neurotransmitters and acetylcholine and has traditionally played a key role in pharmacology and drug development. These receptors include β-adrenergic receptors, β2AR (27) and β1AR (13), histamine H1 receptor (28), dopamine D3 receptor (29), and two muscarinic acetylcholine receptors, M2 and M3 (30, 31). In addition to aminergic receptors and rhodopsin, the α-group structures include A2AAR (28), as well as sphingosine-1-phosphate1 (S1P1) (32) (the first example of a lipid-activated GPCR). Beyond the α-group, the structures of several peptide-binding receptors from the γ-group of Class A GPCRs have been solved, including a chemokine receptor CXCR4 (33), and the opioid receptors: κ-opioid, μ-opioid, δ-opioid and the nociceptin/orphanin FQ peptide receptor (NOP) (34–37).
Opioid receptors are thus far the most comprehensively covered subfamily, with crystal structures of all four closely related opioid subtypes solved, followed by β-adrenergic receptors (2 out of 3 subtypes) and muscarinic acetylcholine receptors (2 out of 5 subtypes). The other solved structures are single representatives of their subfamilies. At the group level, substantial structural coverage has been achieved for the α-group, where solved structures of 9 GPCRs represent 7 distinct subfamilies, about half of the subfamilies in the group. At the same time, structural coverage of the γ-group is still very sparse, while the structures of other two major groups (β and δ) of Class A and of other GPCR classes are all, as yet, not known.
Several GPCRs, such as rhodopsin (9, 14, 15, 17, 18, 38, 39), β2AR (21, 22, 27, 40, 41),β1AR (13, 42, 43), A2AAR (19, 20, 44, 45) and CXCR4 (33), have been co-crystallized in complexes with different ligands, in different crystal forms or using different approaches to receptor stabilization and crystallization. Importantly, the conformational differences between the multiple inactive structures of the same receptor were found to be minor (all atom root mean squared deviations (RMSD) < 0.8 Å), leaving most key receptor-specific structural features well preserved. This reproducibility establishes an important baseline for structural comparisons between different GPCRs and between different functional states of these receptors. Moderate levels of induced fit in the GPCR binding pockets (41) is also a key for effective applications of structures to rational drug discovery, where hit identification rates as high as 20 to 70% have been shown in virtual screening for novel ligand chemotypes for β2AR (46), A2AAR (47, 48), histamine H1 (49), dopamine D3 (50) and chemokine CXCR4 (50a) receptors, and for rational optimization of established scaffolds in adenosine receptors (51).
Structural diversity between GPCRs subfamilies and subtypes
Although all GPCRs are characterized by a similar seven transmembrane (7TM) topology, the five major families of human GPCRs (2, 52) (Figure 1) share very little sequence identity (SI, < 20% in the TM domain) and possess different extracellular N-terminal domains. The largest and most diverse Rhodopsin-like family (also known as Class A) consists of about 700 GPCRs in humans, and can be further classified into four subgroups (α-δ) (2) (SI ≥ 25%). Each subgroup contains numerous subfamilies that share higher sequence similarity (SI ≥ 30%) and often common ligand selectivity.
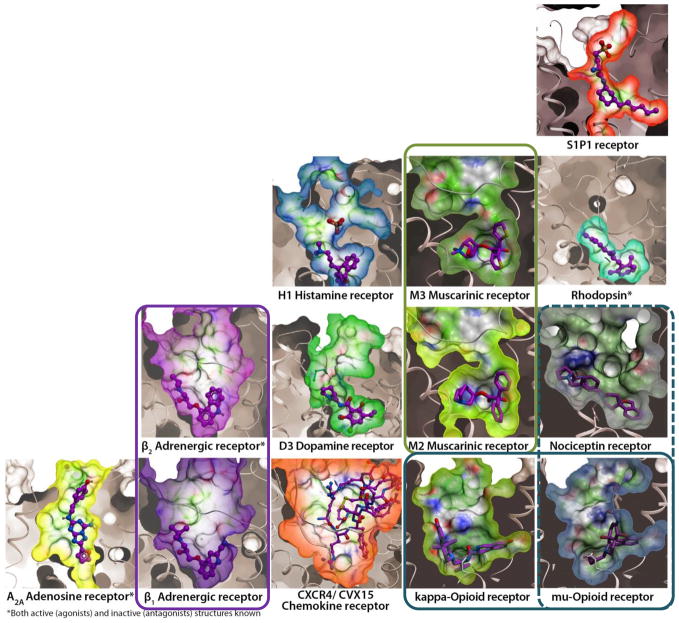
Analysis of structural variations at different levels of homology provides important insights into the scope of structural diversity in GPCRs, as has been reviewed (53). Within subfamilies, sequence and structural similarity can be high enough to allow for accurate predictions by homology modeling, which is useful in such practical applications as ligand docking (54), or virtual screening for dopamine D3 antagonists (50), as well as profiling of ligand selectivity within the adenosine receptor subfamily (55). However, structural differences between different GPCR subfamilies and groups are much more dramatic. The most striking variations revealed by crystallography are in the extracellular loop region, which presents a diverse repertoire of secondary structures and disulfide crosslinking patterns (56). Key variations are also found in the 7TM helical bundle itself, including both proline and nonproline kinks and “bulges,” π-helices and other local variations, resulting in as much as ~7 Å deviations in the extracellular tips of transmembrane helices. Most importantly, such variations in extracellular loops, transmembrane helices and side chains create a remarkable variety of sizes, shapes and electrostatic properties of the ligand-binding pockets in different GPCR subfamilies, reflecting the diversity of their corresponding ligands (Figure 2).
Figure 2.
Diversity of ligand binding pockets in GPCRs. Pockets are shown as molecular surfaces for available inactive-state GPCR structures in complex with corresponding antagonists. Receptor orientations and the surface clipping planes are the same for all receptors. Pairs of closely related GPCR subtypes with similar pockets are highlighted by colored frames.
With the structure determination of S1P1 (32), muscarinic acetylcholine (30, 31), and opioid (34–37) receptors, further examples of structural diversity are added to this repertoire. For example, the structure of a lipid binding GPCR, S1P1, reveals a unique configuration of the extracellular loop 2 and N-terminus, which completely seals the extracellular entrance into the binding pocket. Although rhodopsin also has a tight extracellular “lid” covering the pocket, the backbone structure of these elements in S1P1 is different, extracellular loop 2 is lacking in secondary structure, a disulfide bond to helix III that is highly conserved in other GPCRs is missing, and the N-terminus forms an α-helix on top of the binding pocket. Consistent with the highly lipophilic nature of S1P1 ligands, the structure reveals a “side” opening between helices I and VII, which provides direct access of the ligand into the pocket from the lipid bilayer. In contrast, a prominent structural feature of the M2 and M3 muscarinic acetylcholine receptors (30, 31) includes an apparent constriction site in the orthosteric binding pocket controlled by several large aromatic residues. This narrowing creates a separate allosteric pocket in the extracellular loops of the receptor, which potentially plays a role in the ligand binding kinetics and can accommodate an allosteric modulator or an extended bitopic ligand (31).
Comprehensive structural coverage of the opioid subfamily
Comprehensive structural coverage of the opioid receptor subfamily (34–37) is especially insightful for appreciation of structural conservation and diversity in GPCRs. Opioid receptor structures exemplify the second subfamily in the peptide binding γ-branch of the GPCR tree, previously represented only by the chemokine receptor CXCR4 (33). Despite rather low homology between opioid and chemokine receptors (sequence identity <27%), both subfamilies have a very similar β-hairpin conformation of their extracellular loop 2, which emerges as a common peptide-binding motif in the large and diverse γ-group of GPCRs. In contrast, the 7TM bundle region of opioid receptors appears structurally closer to aminergic receptors from α-group, with some key features in common between them. For example, all opioid receptors have the same aspartate residue (using Ballesteros-Weinstein numbering (115) this residue is identified as Asp3.32), which is also fully conserved in aminergic receptors; in both families the acidic side chain of Asp3.32 serves as a major salt bridge anchor critical for ligand binding and receptor activation.
Intriguingly, the “opioid-like” receptor NOP (35), despite very high overall sequence identity (60%) with the “classical” μ-opioid (34), κ-opioid (36), and δ-opioid (37) receptors, displays very different ligand selectivity profiles to both endogenous peptides and small molecule morphine derivatives. These changes in ligand selectivity are consistent with dramatic structural rearrangements between classical opioid receptors and the NOP , where replacements of only a few key residues in the ligand binding pocket region results in conformational changes in the backbone structure, including shifts in helices V and VI (35). Such deviations in both ligand selectivity and binding pocket conformations, in spite of the overall high sequence identity, are in stark contrast with the tight conservation of pockets previously observed in other closely related GPCR subtypes. For example, between β2AR and β1AR, as well as between M2 and M3 muscarinic receptors, the side chains of the core orthosteric binding pockets are all identical and have very similar conformations. This observation suggests a divergence of NOP from classical opioid receptors driven by the switch in ligand selectivity, which emphasizes the highly dynamic nature of GPCR evolution in mammalian genomes (58).
Structural framework for understanding GPCR activation
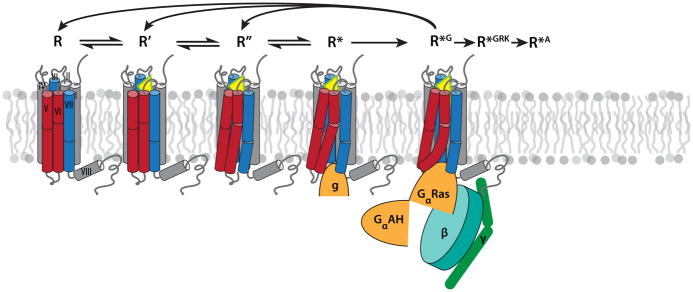
The multitude of biochemical, biophysical and structural data suggests that most GPCRs exist in a dynamic equilibrium between inactive (R, R’) and active states (R’’, R*), which can be further converted to the signaling state (R*G) in the presence of heterotrimeric G protein, as illustrated in Figure 3. Distribution between states in ligand-free receptors can vary drastically, reflecting their different levels of basal activities (59, 60). Inverse agonists shift the equilibrium towards inactive states, decreasing the level of basal activity, while neutral antagonists do not affect the basal equilibrium. Binding of agonists shifts the equilibrium towards the activated states characterized by large-scale conformational changes at the receptor’s intracellular side (61). These five conformationally distinct functional states have now been characterized crystallographically in rhodopsin, A2AAR and β2AR receptors as listed in Table 2.
Figure 3.
Key intermediates in GPCR activation mechanism, characterized crystallographically (see Table 1 for PDB codes and references): R represents inactive (ground) states, which can be stabilized by binding of inverse agonists or antagonists. R’ represents inactive low affinity agonist-bound states, which differ from R by only small local changes in the receptor binding pocket. R’’ represents activated state(s), characterized by substantial global rearrangement of helices and side chain microswitches on the intracellular side that expose, at least partially, the G protein binding crevice. R* represents activated substates with initial insertion of G protein C-terminal α-helix (or its surrogate mimic g) into the intracellular (IC) crevice. Finally, R*G is a distinct G protein signaling conformation of a receptor, which can be achieved upon full engagement and activation of the GPCR-Gαβγ-complex. Other conformationally distinct active states (not depicted) also likely exist, for example for GPCR binding to G protein receptor kinases (R*GRK) and to β-arrestin (R*A). Note that transition from initial G protein binding (R*) to full signaling state R*G is accompanied by release of GDP and, therefore, proceeds unidirectionally; subsequent return to pre-signaling states requires dissociation of the protein complex and binding of a new Gαβγ-GDP unit to the receptor.
Table 2.
Five distinct activation states represented by crystal structures of GPCRs (PDB codes and corresponding publications shown).
| Receptor | R Inactive State | R’ Inactive Agonist Bound | R’’ Active State | R* Active, with G& Mimic | R*G G protein Signaling |
|---|---|---|---|---|---|
| A2AAR | 3EML (45), 3REY, 3RFM, 3PWH (44), 3VGA, 3VG9 (111), 3UZA, 3UZC (112), 4EIY (57) | n/a | 3QAKa (19), 2YDOb, 2YDVb (20) | n/a | N/a |
| β 1 AR | 2VT4 (13), 2YCW, 2YCX, 2YCY, 2YCZ (42), 4AMI, 4AMJ (43a) | 2Y00, 2Y01, 2Y02, 2Y03, 2Y04 (43) | n/a | n/a | N/a |
| β 2 AR | 2RH1 (12, 27), 2R4R, 2R4S (113), 3D4S (40), 3KJ6 (103), 3NY8, 3NY9, 3NYA (41) | 3PDS (109) | n/a | 3P0Gc (22) | 3SN6d (21) |
| Rhodopsin | 1F88 (9), 1U19 (39)e | 2G87, 2HPY (38) | 3CAPf (15) | 3DQBfg (14), 2X72g (17), 3PQRg, 3PXOg (18) 4A4Mgb (18a) | N/a |
Stabilized by a conformationally selective agonist
Stabilized in activated state by point mutations
Complex with a nanobody mimicking Gα interactions
Complex with Gαβγ, additionally stabilized by a nanobody
Out of 15 dark state rhodopsin structures, only the first and the highest resolution structures are shown
Retinal-free opsin
Complex with C-terminal peptide Gα
Large-scale rearrangements of transmembrane helices
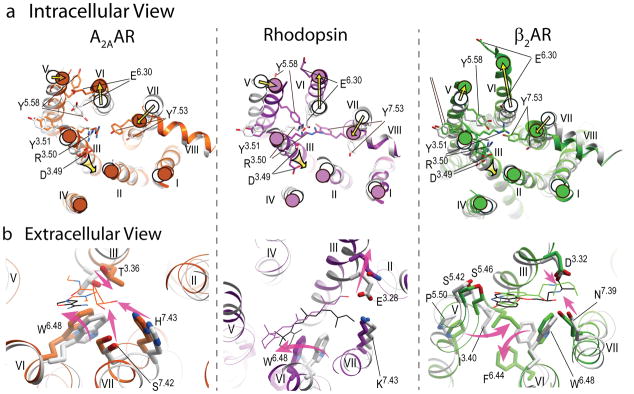
Quite remarkably, despite the differences in the identity of receptors, comparisons of inactive and active state structures reveal common activation-related features with respect to conformational changes on the intracellular side of the receptors (Figure 4a). The most pronounced common rearrangement of helices on the intracellular side includes an outward “swinging” motion of helix VI in concert with a movement of helix V, as a part of the previously proposed “global toggle switch” mechanism (10, 61). The magnitude of this motion varies between different GPCRs and different active states, for example, the intracellular tip of helix VI moves ~3.5 Å in A2AAR (R’’), ~6 Å in opsin (R’’ and R*) and as much as 11 Å and 14 Å in β2AR complex with nanobody (22) and G protein (R*G) (21), respectively. Moreover, the final positions of the intracellular tips of helices V and VI in the signaling R*G states seem to be largely controlled by dramatic rearrangements in the G protein itself and are accompanied by GDP release (see Sidebar and Figure 5b).
Figure 4.
Major conformational rearrangements and ligand-dependent triggers in the three available structural models of GPCR activation. a) Intracellular view: common shifts of the intracellular tips of transmembrane helices (yellow arrows), include outward swinging of helix VI, accompanied by movement of helix V, as well as inward shift of helix VII and axial shift of helix III. The established conserved microswitches, shown by stick presentations and labeled, undergo rotamer changes upon activation. b) Extracellular view: the key ligand-dependent “triggers” of GPCR activation (highlighted by the shaped magenta arrows) found in the orthosteric site. Note that trigger residues and their Ballesteros-Weinstein positions are not conserved between these receptors, and the directions of the helical shifts are different. Ligands are shown by thin lines with black carbons for all antagonists (ZM241385, 11-cis retinal and carazolol, respectively) and colored carbons for agonists (NECA, all-trans retinal and BI-167107 colored orange, purple and green, respectively). In all panels, inactive conformations are shown in gray, and corresponding activated conformations are colored orange for A2AAR, PDB codes 3EML (45) and 3QAK (19), purple for Rhodopsin, PDB codes 1GZM (114) and 2X72 (17), and green for β2AR, PDB codes 2RH1 (27) and 3SN6 (21).
SIDEBAR. Movements of TM helices: rigid body or not?
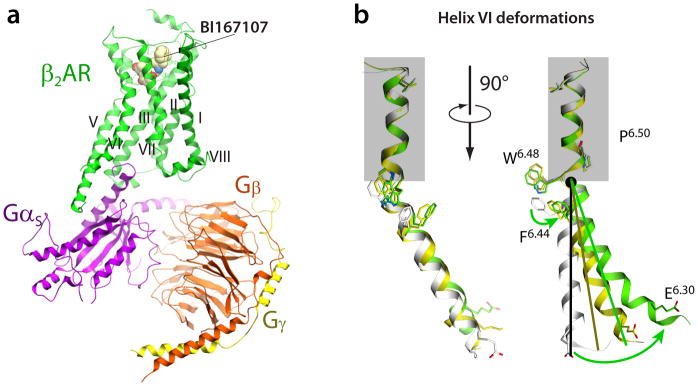
The global rearrangements of transmembrane helices during activation are often approximated as rigid body motions (18). Comparison of individual helices in crystal structures of inactive and active GPCRs shows examples of local deformations in helices during activation. One of the most pronounced is a kink around the highly conserved Pro6.50 in helix VI. A superimposition of the extracellular part of helix VI in inactive and activated structures (see of β2AR example in Figure 5) reveals an activation-related “swinging” motion of the intracellular part of helix VI, which involves some unwinding of the helix at the Pro6.50 kink. Additional elastic bending of intracellular tip of helix VI in the contact area with G protein or G protein-mimicking nanobody is apparent from the structures, suggesting a force applied in this region by G protein or nanobody insertion. In contrast, in A2AAR/agonist complex, or in rhodopsin/opsin bound to Gα C-terminal peptide, the intracellular tips of helices V and VI are straight or slightly bent in the direction opposite to the overall helical movement (19).
Figure 5.
a) Structure of β2AR complex with agonist BI167107 and the G protein heterotrimer (PDB code 3SN6) (21). The receptor and G protein are shown by colored ribbons, while the agonist is illustrated by spheres with carbon atoms colored yellow. Stabilizing nanobody and T4 lysozyme used for crystallization are not shown for clarity. b) Conformational changes in helix VI upon β2AR activation. Structures of inactive R (PDB code 2RH1, gray) (27), nanobody-bound R* (PDB code 3P0G, yellow) (22) and G protein-bound R*G (PDB code 3SN6, orange) (21) states are superimposed at the extracellular part of the helix above Pro2886.50, framed by blue rectangle. While the bend angle of the Pro6.50-induced kink is maintained, the ligand-stabilized movement of Phe2826.44 unwinds the Pro6.50 kink, resulting in a swinging motion (combined tilt and rotation) of the intracellular portion of helix VI. Note also that the motion of the intracellular part cannot be described entirely in the rigid body terms, but shows substantial elastic behavior. This additional bend and displacement of the helix VI tip is apparently induced by insertion of G-protein or the nanobody mimic.
Helices III and VII also undergo substantial rearrangements during activation in all three receptors, with the most pronounced changes in this region observed in A2AAR agonist-bound structures (19, 20). The intracellular part of helix VII moves inward towards the middle axis of 7TM helical bundle and undergoes a marked backbone distortion in the region of the conserved NPxxY motif. Motion of helix III is comprised of an upward shift along its axis and some lateral movement. In β2AR the overall shifts of helices III and VII are less pronounced than in A2AAR and rhodopsin, though there is still a pronounced distortion in the helix VII NPxxY motif.
These observations suggest that movements of helices V and VI are absolutely essential for G protein binding and activation, and are likely conserved in Class A GPCRs. Movements of helices III and VII more likely depend on the particular receptor and ligand, and although their role in G protein activation is not clear, may contribute to G protein-independent signaling pathways (104) as described later.
Conserved microswitches in GPCR activation
The global movements of helices during activation are accompanied by a common set of local “microswitches” in the intracellular part of GPCRs. The microswitches are characterized by rotamer changes in highly conserved side chains (61), which stabilize the global movements of helices and help to prime the intracellular side of GPCR for G protein binding (Figure 4a). The D[E]RY sequence in helix III represents one of the most conserved motifs of Class A GPCRs, in which residue Arg3.50 (96% conservation among Class A GPCRs) forms a salt bridge to the neighboring acidic side chain Asp(Glu)3.49 (Asp 68%, Glu 20%) (62), as found in all inactive state GPCRs structures to date. Interestingly, the Arg3.50-Asp3.49 salt bridge remains intact in the active β2AR -nanobody complex (22), as well as in the active state A2AAR structures (R’’) (19, 20). Only in the active state rhodopsin (R*) and β2AR (R*G) structures is the salt bridge broken, and the Arg3.50 guanidine switches rotamer to interact with the C-terminal helix of the Gα subunit (14, 17, 18, 21), thus suggesting that the switch in Arg3.50 requires the presence of a G protein (Figure 4a).
The Arg3.50 side chain can also form an interhelical salt bridge to Asp6.30, known as the “ionic lock,” which connects the intracellular ends of helices III and VI. The ionic lock was first observed in the structures of dark-adapted bovine rhodopsin, stabilizing this receptor in a fully inactive state (62). The stabilizing role of the ionic lock may be less pronounced in other GPCRs, as it is absent in many GPCRs structures, including those with an intact intracellular loop 3 (42), and computer simulations suggest a highly dynamic nature for this interaction (63). Moreover, an acidic residue in position 6.30 is conserved in only about 30% of GPCRs, for example it is missing in chemokine receptors. Instead, some crystal structures reveal hydrogen bonding interactions between the Arg3.50 side chain and other polar residues in helix VI, for example Thr6.34 in κ-opioid and μ-opioid receptors, which may also play a role in the regulation of receptor signaling.
The “NPxxY” motif is located near the intracellular end of helix VII and contains a highly (92%) conserved Tyr7.53 serving as a major activation microswitch in GPCRs. In inactive GPCR structures the side chain of Tyr7.53 points towards helices I, II or VIII. In contrast, in all active state GPCR crystal structures, the Tyr7.53 side chain changes its rotamer conformation and points towards the middle axis of the 7TM bundle, forming interactions with side chains of helices VI and III. In the active state structures of β2AR and rhodopsin, the Tyr7.53 hydroxyl may also form a tentative water-mediated hydrogen bond with another putative microswitch, Tyr5.58 (89% conserved). Interestingly, the Tyr5.58 side chain behaves very differently in all three activation models: in rhodopsin it switches from outside-to-inside of the helical bundles, in A2AAR it makes an opposite switch from inside-to-outside, while in all β2AR complexes this residue remains in the interior of the 7TM bundle. Note also, that mutation of Tyr5.58 to alanine contributes to the stabilization of β1AR in its inactive state (64), and reduction of basal activity in muscarinic M3 (65) and thyrotropin receptors (65a), though the precise role of this residue in various GPCRs needs further study
Ligand–dependent “triggers” of GPCR activation
One of the most intriguing questions of GPCR activation is: How does binding of different ligands in the highly diverse extracellular pockets translate into common large scale rearrangements on the intracellular side of different GPCRs? Identification of specific ligand-receptor interactions, or “triggers,” that control equilibrium between receptor functional states is critical for understanding and potentially designing efficient agonists, including agonists with desired functional selectivity or “bias” (66–69). Comparison of multiple active and inactive structures shows modest, but well defined, changes in the binding pockets residues in each receptor (all atom RMSD ~1.3 Å for the ligand-contacting residues in β2AR and A2AAR, and ~2.0 Å for rhodopsin), as illustrated in Figure 4b. These ligand-dependent changes in the binding pockets are also consistent with specific ligand interactions, and their repertoires vary among the receptors. Some examples of triggers are briefly discussed below; other triggers are likely to be discovered, especially for GPCRs modulated by peptides and/or allosteric ligands.
One of the prominent examples of ligand-dependent rearrangements in the binding pocket involves a shift of the Trp6.48 residue, which belongs to the conserved CWxP motif on helix VI. Although this side chain was previously classified as a microswitch (or “rotamer toggle switch”) (70), crystal structures of active state GPCRs did not show rotamer changes in this residue upon activation. Moreover, a modified rotamer state of the Trp6.48 side chain was observed in inactive M2 and M3 muscarinic receptor structures (30, 31), showing the that rotamer state of Trp6.48 does not correlate with the functional state of the receptor. Nevertheless, the crystal structures suggest a prominent role for Trp6.48 as a major ligand-dependent “trigger” in some GPCRs. Thus, A2AAR and rhodopsin active state structures reveal direct steric interactions of Trp6.48 side chain with agonists (19, 20), which stabilize a pronounced shift of Trp6.48 and the corresponding swinging movement of helix VI (Figure 5). On the other hand, the inactive position of Trp6.48 can be stabilized by direct interactions with inverse agonists, as observed in rhodopsin/11-cis-retinal (9), A2AAR/ZM241385 (45), and histamine H1/doxepin(28) complexes.
Unlike A2AAR and rhodopsin, the β2AR and β1AR structures lack any direct contacts between the Trp6.48 side chain and agonists or inverse agonists. Instead, conformational changes that promote helix VI motion in β2AR (22) depend largely on agonists engaging in polar interactions with Ser2035.42 and Ser2075.46 and stabilizing an ~2 Å inward shift of the extracellular part of helix V, as initially predicted from modeling results obtained with the inactive β2AR structure (71, 72). Further details of this conformational change, resolved in the crystal structures of active β2AR (21, 22), show that this movement in helix V results in a rotamer switching in Ile1213.40, which is in turn coupled with a 4 Å movement of the Phe2826.44 side chain and a corresponding swing of helix VI. Thus, in the case of β2AR, Trp6.48 seems to have only an indirect role in activation, as also apparent from its much smaller (<1 Å) movement than in other receptors. Therefore, the role of Trp6.48 is not universal even among the three currently available models of GPCR activation; moreover about 30% of Class A GPCRs have different side chains in the 6.48 position.
Another common site of conformational changes in the binding pocket involves helices III and VII, which are bridged by strong ligand-mediated interactions in all three GPCR activation models. The specifics of the changes in this site, however, vary dramatically for different receptors (Figure 4b). In rhodopsin, light activation results in disruption of a salt bridge between Glu1133.28 and the Lys2967.43 Schiff base linked to retinal, and corresponds to an increase of the distance between helices III and VII by about 2–3 Å. Conversely, in A2AAR, of the ribose rings of agonists participate in a strong hydrogen bonding network with Thr883.36 and Ser2777.42/His2787.43, bringing helices III and VII closer by about 2 Å, as compared to the structure with inverse agonist. In β2AR, Asp1133.32 and Asn3127.39 are bridged by an ethanolamine tail in both agonists and antagonist complexes, so the role of this bridge in β2AR activation is not clear from the crystal structures. Further insights into how engagement of this and other triggers can affect signaling (including biased) can be gained by biophysical methods sensitive to receptor dynamics, as discussed below.
G protein binding and signaling
The structure of β2AR/Gαβγ signaling complex (21) (Figure 5a) reveals a series of additional conformational changes at the receptor that are largely controlled by the full Gαβγ engagement and activation. As suggested by Rasmussen et al. (21), initial insertion of the Gαs C-terminal α5-helix into the transiently accessible site between β2AR helices is likely to resemble insertion of a Gα C-terminal peptide into the rhodopsin structure (14). Engagement of other receptor-Gαβγ interactions leads to a crowbar-like rotation of Gαs α5-helix by about 30°, remodeling the β6-α5 loop region, followed by a dramatic rotation of the GαAH domain and GDP release (21). All these rearrangements apparently force further displacement of β2AR helices V and VI, so that helix VI is dramatically curved outward (see Figure 5b) and its tip is displaced by as much as 14 Å (21) as compared to inactive states.
The G protein-bound crystal structure also reveals the large β2AR/Gα protein interface formed by intracellular loop 2 and helices V/VI of the receptor, and by α4/α5-helices, the αN–β1 junction and β3-strand of GαsRas. Notably, the crystallographically resolved parts of the receptor lack direct interactions with the Gαβγ subunits. Additional structures of GPCR/G protein complexes may be needed to explain selectivity among Gα subtypes, which in some cases may be differentially regulated by biased ligands (66–69). The structure of the complex also, surprisingly, does not reveal any G-protein contacts with β2AR helices VII and VIII, suggesting that activation-related changes in these helices may not be necessary for G protein signaling, but possibly may mediate other GPCR interactions, for example, with β-arrestins.
Structural insights into allosterism and oligomerization
Structural basis for allosteric modulation of GPCRs
In addition to orthosteric ligands, GPCR signaling can be affected by a variety of endogenous allosteric modulators, such as regulatory proteins (64, 65), lipids and sterols (59, 73) and ions (66). Moreover, synthetic allosteric and bitopic (7) modulators have been identified for many GPCRs, showing promise as drug candidates with improved subtype selectivity and pharmacological profiles. Several potential allosteric sites in GPCRs revealed by crystal structures have been reviewed previously (53), including a phosphate ion binding site in the extracellular subpocket of the histamine H1 structure (28), a cholesterol binding site in β2AR (40) and a ‘druggable’ extracellular extension of the dopamine D3 orthosteric subpocket (29).
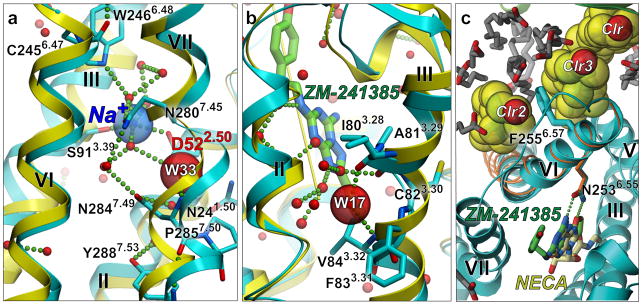
New insights into GPCR allosteric regulation come from the 1.8 Å resolution structure of inactive A2AAR (57) that revealed a tight water-filled channel connecting extracellular and intracellular sides of receptor. A small (ca. 200 Å 3) allosteric pocket in the middle part of the channel was found to bind a Na+ ion surrounded by a cluster of 10 structured waters (Figure 6a). The pocket is bounded by Trp2466.48 (conserved in 71% of Class A GPCRs) on the extracellular side and Tyr2887.53 (92%) on the intracellular side, as well as residues Asn241.50 (100%), Asp522.50 (94%), Ser913.39 (Ser:53%, Asp:26%), Asn2807.45 (67%) and Asn2847.49 (75%), previously predicted to participate in a water-mediated hydrogen-bonding network (61). The negative allosteric modulation effect of Na+ on agonist binding and activation has been observed in many GPCRs (74), and was tentatively linked to the Asp2.50 residue by site-directed mutagenesis (75–77); however, the ion remained undetected in all previous medium resolution structures. The high resolution A2AAR structure revealed the location of the Na+ ion coordinated by conserved Asp522.50 and Ser913.39 residues and structured water, rationalizing the stabilizing effect of the Na+ ion on the inactive receptor state (57). Moreover, analysis of A2AAR crystal structures shows that in the active-state A2AAR (19, 20) the Na+/water pocket collapses from ca. 200 to 70 Å 3 due to movements of transmembrane helices. The collapsed pocket does not provide adequate coordination for Na+, suggesting that the ion abandons the pocket and potentially leaves the receptor upon activation (57).
Figure 6.
Allosteric sites in the high-resolution inactive A2AAR structure (blue ribbon and side chains) (57), as compared to the active state A2AAR complex (yellow ribbon) (19). (a) Highly conserved in Class A GPCR is an allosteric site in the middle of the transmembrane bundle. A Na+/water cluster is observed in the inactive state, but the collapsed pocket in the active state precludes Na+ binding. (b) Tight binding of a structured water in a non-proline kink in helix III of A2AAR, is abolished in the active state, where helix III is straightened (yellow ribbon). (c) Two cholesterol molecules (Clr2 and Clr3) sandwich the phenol ring of Phe2556.57 in close proximity of the binding pocket and stabilize the conformation of the extracellular part of helix VI.
The key amino acids in the Na+/water pocket represent the most conserved spatial cluster of residues in Class A GPCRs (61), which suggests they have a key common role in receptor function. At the same time, any variations in these and surrounding residues that affect the water cluster and Na+ ion may also alter the dynamics and activation profiles in different GPCRs. Interestingly, while the extracellular entrance to the Na+/water site is somewhat restricted by the Trp6.48 side chain in most GPCR structures that have been solved, this passage is more open in the muscarinic M2 and M3 receptors (30, 31). Further understanding of the structural and functional details of the pocket create an opportunity to target this conserved site by allosteric molecules (e.g., amiloride analogs (57)), or bitopic compounds, some of which would have an enhanced ability to stabilize the receptor in the inactive form.
Other, receptor-specific allosteric sites have also been revealed by recent crystal structures, including a “vestibule” pocket in the extracellular loops of the muscarinic receptors (30, 31), which may bind some of the known allosteric ligands (68). The high-resolution A2AAR structure (57) also suggests a second cholesterol binding site (Figure 6c), distinct from a previously described site in β2AR (40). Another intriguing observation in this A2AAR structure is a water molecule intercalated at the nonproline kink between residues Ile803.28 and Val843.32, which apparently stabilizes the kinked conformation of helix III (Figure 6b). Importantly, in all agonist- bound active state structures of A2AAR, the kink in this region of helix III is completely straightened, thus precluding water binding. The rearrangement in this water binding site therefore might play an important role in the activation mechanism of the adenosine receptor subfamily, and can be exploited for allosteric modulation by extending a ligand scaffold to interact with this site.
GPCR dimerization and oligomerization
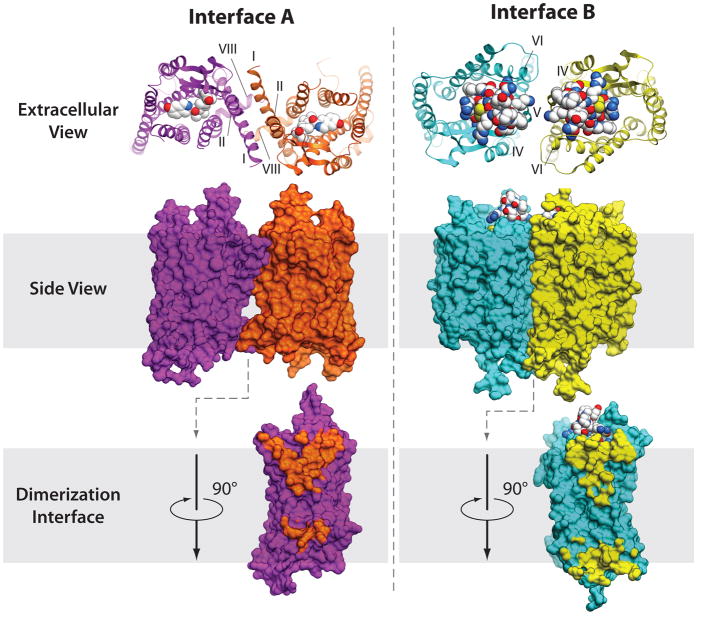
Although Class A GPCRs can effectively signal as monomers, numerous studies have suggested the existence of homodimers, heterodimers and higher oligomers in many GPCRs and proposed their role in the regulation of GPCR function and crosstalk (reviewed in (78, 79)). GPCR dimerization and oligomerization has been extensively studied by atomic microscopy, FRET, BRET, cross-linking, time-resolved spectroscopy and molecular modeling (reviewed in (80, 81)), with results systematically collected in the GPCR-OKB database (82). High-resolution structural information on GPCR-GPCR interactions has just started to emerge from crystallographic studies. Parallel dimers with substantial protein-protein interface (>800 Å 2) have been found in crystal structures of 4 different GPCRs, including rhodopsin (83), CXCR4 (33), κ-opioid (36) and μ-opioid (34) receptors (see Table 3). Overall, there are two consensus clusters of symmetric interfaces suggested by these structures, which concur with biochemical data for rhodopsin (84, 85), serotonin 5HT2C (86), dopamine D2 (87, 88), adrenergic α1bAR (89, 90), chemotactic C5a (91), chemokine CCR5 (92), and other GPCRs. The first symmetric interface A (Figure 7a), found in rhodopsin (15, 83), κ-opioid (36) and μ-opioid (34) structures is formed by helices I, II and VIII, comprising two separate interaction patches on the extracellular and intracellular sides. Biophysical studies revealed that this dimerization mode is insensitive to receptor activation (86), in agreement with minimal activation-related conformational changes in these interface helices. The second symmetric interface B (Figure 7b) involves helices V and VI, and in the case of CXCR4/peptide complexes can also involve helices III and IV on the intracellular side (33). Similar overall orientation of the dimer subunits, although with a different contact interface formed by the transmembrane bundle of helices V and VI, has been observed in μ-opioid receptor (34). Interface B is likely to interfere with the activation state, because activation requires large conformational movements of helices V and VI. Combined in a repeated pattern, the A and B interfaces are compatible with a high-level oligomerization found in rhodopsin arrays (87, 93), although it is not clear if such a pattern is relevant for the oligomerization observed in some other GPCRs (e.g., (89, 90). Further crystallographic, biochemical and biophysical studies will help to better understand the structural basis and functional role of dimerization interfaces and their variability between different GPCRs.
Table 3.
GPCR transmembrane dimerization interfaces found in crystal structures
| Interacting helices | Receptor | Interface (Å2) | PDB code | Other evidence | Potential Implication |
|---|---|---|---|---|---|
| Interface A I, II and VIII | Rhodopsin | ~800 | 2I36, 2I37, 2I35, 3CAP, 4A4M | Crosslinking (84, 85), Atomic force Microscopy (93) | functional oligomer |
| κ-OR | ~1080 | 4DJH | Crosslinking in 5HT2C, dopamine D2 (87) | activation insensitive | |
| μ-OR | ~600 | 4DKL | |||
| Interface B V, VI (EC); III,IV (IC) | CXCR4 | ~1250 850 (EC)+ 400 (IC) |
3ODU, 3OE0 | Crosslinking in Rhodopsin (84, 85) 5HT2C (86), dopamine D2 (88) | activation sensitive |
| V, VI bundle | μ-OR | ~1400 | 4DKL |
Figure 7.
Two major types of symmetric dimer interfaces observed in GPCR structures. A representative structure of dimer interface A with contacts via helices I, II, VIII is shown here for κ-opioid, PDB code 3DJH (36) (orange and magenta show receptor, the JDTic ligand is shown by spheres with white carbons). Interface A has been also observed within μ-opioid, rhodopsin and opsins structures. Another cluster of dimer interfaces B involves contacts via helices IV, V, VI (cyan and yellow) and is shown here for the CXCR4 complex with peptide antagonist PDB code 3OE0 (33). Similar orientation of subunits has also been observed in μ-opioid structure, PDB code 3DKL (34), with an extensive interface formed via helices V and VI.
Complementary biophysical studies of conformational dynamics in GPCRs
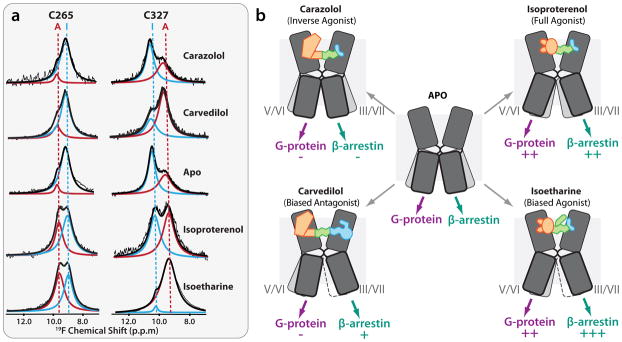
As mentioned above, GPCR activation dynamics are characterized by a complex equilibrium between multiple conformational states (94, 95), manifesting in significant basal activity, existence of allosteric modulators (68, 96) and a whole spectrum of functional responses including biased signaling (69). Understanding of these phenomena requires an ability to probe the equilibrium between different conformations, which is beyond the capabilities of crystallographic methods that give a highly detailed, but “frozen picture” of the lowest energy receptor state. Various biophysical approaches have shown utility in assessing local and global conformational changes in GPCRs, including fluorescent spectroscopy (94, 97, 98), double electron–electron resonance (DEER) (99), hydrogen-deuterim exchange coupled with mass spectrometry (HDX-MS) (100–102) and nuclear magnetic resonance (NMR) spectroscopy (103, 104) . Methods are also being developed to assess kinetics of changes in GPCRs using time-resolved single-molecule studies (105, 106). The availability of 3D structures now allows for the design of optimized labeling sites and provides a structural framework for the analysis of obtained data. Thus, 13C NMR spectroscopy has been employed to assess ligand-dependent conformational changes in the extracellular region by measuring formation of a salt bridge that connects extracellular loops 2 and 3 (103), previously found in the β2AR crystal structure (27). Another NMR technique that has been used to probe dynamics of the intracellular changes in β2AR exploits spectra of sensitive 19F labels (104). Most notably, the 19F NMR spectra suggested that helices VI and VII can each adopt at least two major conformational states that have a slow (>100 ms) exchange rate (Figure 8). The equilibrium distribution between these states is controlled by ligand binding: while antagonists and inverse agonists keep β2AR in a predominantly inactive state, binding of partial, and especially full, agonists shifts the conformational equilibrium towards an active state in both helix VI and helix VII. The most striking differential effect was observed for the arrestin-biased βAR ligands carvedilol and isoetharine, which both showed dramatic (>40%) increase in the occupancy of the helix VII activated state as compared to their unbiased analogs carazolol and isoproterenol. These results demonstrate at least a partial decoupling between conformational changes in helices VI and VII, and a specific association of helix VII changes with arrestin-biased signaling of carvedilol and isoetharine. As described here, association of helix VII with arrestin-biased signaling is also supported by recent FRET studies (107), key interactions of arrestin with the C-terminal part of GPCRs adjacent to helix VII (108), as well as lack of G protein contacts with helix VII in the β2AR-Gαβγ complex structure (21).
Figure 8.
NMR experiments shed light on major signaling pathways in β2AR (104). (a) 19F NMR spectra suggest existence of at least two distinct states in both Cys265 (Helix VI) and Cys327 (Helix VII). Equilibrium in each helix is differentially regulated by an agonist (isoproterenol), inverse agonist (carazolol), arrestin-biased agonist (isoetharine) and arrestin-biased antagonist (carvedilol). (b) Suggested differential effect of unbiased (top row) and biased (bottom row) ligands of G-protein-mediated and β-arrestin-mediated signaling. Helices V/VI and III/VII of β2AR are shown as two sets of bent boxes, of which the most highly occupied states are shown in dark gray, less occupied states in light gray, and minimally occupied states framed with a dotted line. The arrows at the bottom indicate the flow of signals through each helix to the corresponding downstream effector, with an increasing number of plus-signs (+) indicating higher levels of signaling, and a minus-sign (–) indicating reduced signaling, as compared to the basal level.
Complementary insights into GPCR function can be obtained by molecular modeling approaches, including conformational sampling and flexible ligand-receptor docking (71, 72). Also, thanks to a rapid growth of computer power and parallelization, unbiased molecular dynamics simulations at ~10 microsecond scale has allowed for observations of large conformational changes, i.e., active-to-inactive transitions in β2AR (109) as well as prediction of ligand binding paths and kinetics (110).
Further structural, biophysical and computational studies of receptor dynamics, including single molecule time-resolved studies will help to characterize the relationship between various activation states and substates in different GPCRs, and to study the possibilities of selectively modulating these states by “biased” and/or allosteric ligands.
Summary points
With the recent breakthroughs in GPCR crystallography, structural coverage of GPCRs has experienced an exponential growth trend, suggesting that receptors from a majority of subfamilies will be solved within the next decade.
Crystal structures of inactive and active state GPCRs provide an atomic level 3D framework for biochemical, biophysical and computational inquiries into GPCR function and dynamics.
All GPCRs have a common 7TM topology, however they present a great variety of features in their structure, dynamics, selectivity to ligands, modulators and downstream signaling effectors. While the largest structural differences can be found among GPCR classes and subfamilies, structures of the opioid receptors reveal substantial structural deviations within the subfamily, even at 60% sequence identity.
Activation mechanisms in GPCRs share a number of common features, including movements of helices, side chain microswitches, and potential rearrangements in the Na+/water cluster. Changes on the intracellular side are softly coupled to ligand binding through a number of anchor ligand-receptor contacts (or “triggers”). A set of specific triggers can vary between ligands and between different GPCRs. Preferential engagement of specific “triggers” by structurally distinct ligands may explain the variety of conformational and functional responses known as “biased signaling.”
Homodimerization, heterodimerization and oligomerization have been suggested in GPCR regulation and crosstalk. While crystallography reveals several distinct homodimer interfaces, additional biochemical studies are needed to understand their functional roles in different GPCRs.
Crystal structures provide new insights into allosteric regulation of GPCRs by endogenous ions (e.g. sodium), lipids, cholesterol and polypeptides, as well as by synthetic molecules, which can modulate GPCR function and pharmacological response. These discoveries highlight that GPCRs are complex allosteric machines, controlled by more than just their pharmacological ligand.
Computer modeling has an important role in creating a comprehensive picture of GPCR structure-function by filling remaining gaps in the superfamily coverage and molecular interactions, and providing a platform for rational drug discovery.
Future Directions
Determine representative structures for β and δ groups of the Rhodopsin family, as well as for the other GPCR families (secretin, adhesion, glutamate, frizzled/TAS2), in complex with antagonists and agonists.
Increase structural coverage within specific GPCR subfamilies via structure determination and homology modeling.
Further explore GPCR dynamics via combination of structural and biophysical tools (e.g., HDX, NMR, EPR) including time-resolved single molecule approaches, as well as molecular dynamics.
Explore the structural basis of G protein and β-arrestin type selectivity.
Obtain structures of GPCR complexes with allosteric modulators.
Improve our understanding of biased signaling via structural and biophysical studies.
Apply structural knowledge to successful structure-based drug discovery.
Acknowledgments
We would like to thank Enrique Abola, Jeff Liu, Irina Kufareva and Hugo Gutiérrez de Terán for helpful discussions, Katya Kadyshevskaya for help with figure preparation and Angela Walker for help with manuscript preparation. The work is funded by NIH/NIGMS PSI:Biology grant U54 GM094618.
Acronyms/Terms (up to 20, each 20 words)
- TM
transmembrane
- EC
extracellular
- IC
intracellular
- SI
sequence identity
- Ballesteros-Weinstein (BW) number
enumeration of GPCR transmembrane residues in X.YY format, where X is the helix number, and YY is the residue position relative to the most conserved residue in the helix, designated X.50)(115)
- bRho
bovine rhodopsin
- β2AR, β1AR
β1 and β2 adrenergic receptors
- A2AAR
adenosine A2A receptor
- D3R
dopamine D3 receptor
- CXCR4
chemokine CXCR4 receptor
- H1R
histamine H1 receptor
- M2, M3
muscarinic M2 and M3 receptors
- κ-OR, μ-OR, δ-OR, NOP
opioid receptors κ, m, δ and nociceptin/orphanin FQ receptor
- S1P1 (or EDGE1))
sphingosine 1-phosphate receptor 1
- Microswitches
rotamer changes in side chains highly conserved between GPCRs in the IC part of the receptors
- Triggers
direct ligand-receptor interactions, that modulate equilibrium between GPCR functional states
- Allosteric site
receptor binding site distinct from the endogenous ligand (orthosteric) site
- Bitopic ligand
chemical compound targeting both ortho- and allosteric sites
Literature Cited
- 1.Lagerstrom MC, Schioth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–57. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 2.Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–72. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 3.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat Rev Drug Discov. 2006;5:993–6. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 4.Tyndall JD, Sandilya R. GPCR agonists and antagonists in the clinic. Med Chem. 2005;1:405–21. doi: 10.2174/1573406054368675. [DOI] [PubMed] [Google Scholar]
- 5.Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat Rev Drug Discov. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- 6.Allen JA, Roth BL. Strategies to discover unexpected targets for drugs active at G protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2011;51:117–44. doi: 10.1146/annurev-pharmtox-010510-100553. [DOI] [PubMed] [Google Scholar]
- 7.Valant C, Robert Lane J, Sexton PM, Christopoulos A. The best of both worlds? Bitopic orthosteric/allosteric ligands of g protein-coupled receptors. Annu Rev Pharmacol Toxicol. 2012;52:153–78. doi: 10.1146/annurev-pharmtox-010611-134514. [DOI] [PubMed] [Google Scholar]
- 8.Reiter E, Ahn S, Shukla AK, Lefkowitz RJ. Molecular Mechanism of beta-Arrestin-Biased Agonism at Seven-Transmembrane Receptors. Annu Rev Pharmacol Toxicol. 2012;52:179–97. doi: 10.1146/annurev.pharmtox.010909.105800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–45. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 10.Schwartz TW, Frimurer TM, Holst B, Rosenkilde MM, Elling CE. Molecular mechanism of 7TM receptor activation–a global toggle switch model. Annu Rev Pharmacol Toxicol. 2006;46:481–519. doi: 10.1146/annurev.pharmtox.46.120604.141218. [DOI] [PubMed] [Google Scholar]
- 11.Kobilka BK. G protein coupled receptor structure and activation. Biochim Biophys Acta. 2007;1768:794–807. doi: 10.1016/j.bbamem.2006.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–73. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 13.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, et al. Structure of a beta(1)-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–91. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 15.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–7. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 16.Park PS, Lodowski DT, Palczewski K. Activation of G protein-coupled receptors: beyond two-state models and tertiary conformational changes. Annu Rev Pharmacol Toxicol. 2008;48:107–41. doi: 10.1146/annurev.pharmtox.48.113006.094630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Standfuss J, Edwards PC, D’Antona A, Fransen M, Xie G, et al. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2011;471:656–60. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choe HW, Park JH, Kim YJ, Ernst OP. Transmembrane signaling by GPCRs: insight from rhodopsin and opsin structures. Neuropharmacology. 2011;60:52–7. doi: 10.1016/j.neuropharm.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 18a.Deupi X, Edwards P, Singhal A, Nickle B, Oprian D, et al. Stabilized G protein binding site in the structure of constitutively active metarhodopsin-II. Proc Natl Acad Sci USA. 2012;109:119–24. doi: 10.1073/pnas.1114089108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu F, Wu H, Katritch V, Han GW, Jacobson KA, et al. Structure of an Agonist-Bound Human A2A Adenosine Receptor. Science. 2011;332:322–7. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lebon G, Warne T, Edwards PC, Bennett K, Langmead CJ, et al. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature. 2011;474:521–5. doi: 10.1038/nature10136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–55. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasmussen SG, Choi HJ, Fung JJ, Pardon E, Casarosa P, et al. Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature. 2011;469:175–80. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynolds K, Abagyan R, Katritch V. Structure and Modeling of GPCRs: Implications for Drug Discovery. In: Gilchrist A, editor. GPCR Molecular Pharmacology and Drug Targeting: Shifting Paradigms and New Directions. Hoboken, NJ: Wiley & Sons, Inc; 2010. pp. 385–433. [Google Scholar]
- 24.Congreve M, Langmead CJ, Mason JS, Marshall FH. Progress in structure based drug design for G protein-coupled receptors. J Med Chem. 2011;54:4283–311. doi: 10.1021/jm200371q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Archbold JK, Flanagan JU, Watkins HA, Gingell JJ, Hay DL. Structural insights into RAMP modification of secretin family G protein-coupled receptors: implications for drug development. Trends Pharmacol Sci. 2011;32:591–600. doi: 10.1016/j.tips.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 26.Kniazeff J, Prezeau L, Rondard P, Pin JP, Goudet C. Dimers and beyond: The functional puzzles of class C GPCRs. Pharmacol Ther. 2011;130:9–25. doi: 10.1016/j.pharmthera.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 27.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, et al. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 2007;318:1258–65. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011;475:65–70. doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chien EY, Liu W, Zhao Q, Katritch V, Han GW, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–5. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–51. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kruse AC, Hu J, Pan AC, Arlow DH, Rosenbaum DM, et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–6. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson MA, Roth CB, Jo E, Griffith MT, Scott FL, et al. Crystal structure of a lipid G protein-coupled receptor. Science. 2012;335:851–5. doi: 10.1126/science.1215904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu B, Chien EY, Mol CD, Fenalti G, Liu W, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330:1066–71. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, et al. Crystal structure of the mu-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–6. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson AA, Liu W, Chun E, Katritch V, Wu H, et al. Structure of the Nociceptin/Orphanin FQ Receptor in Complex with a Peptide Mimetic. Nature. 2012;485:395–9. doi: 10.1038/nature11085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu H, Wacker D, Mileni M, Katritch V, Han GW, et al. Structure of the human kappa-opioid receptor in complex with JDTic. Nature. 2012;485:327–32. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, Weis WI, Kobilka BK. Structure of the delta opioid receptor bound to naltrindole. Nature. 2012;485:400–4. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamichi H, Okada T. Local peptide movement in the photoreaction intermediate of rhodopsin. Proc Natl Acad Sci USA. 2006;103:12729–34. doi: 10.1073/pnas.0601765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342:571–83. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 40.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, et al. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wacker D, Fenalti G, Brown MA, Katritch V, Abagyan R, et al. Conserved binding mode of human beta2 adrenergic receptor inverse agonists and antagonist revealed by X-ray crystallography. J Am Chem Soc. 2010;132:11443–5. doi: 10.1021/ja105108q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moukhametzianov R, Warne T, Edwards PC, Serrano-Vega MJ, Leslie AG, et al. Two distinct conformations of helix 6 observed in antagonist-bound structures of a {beta}1-adrenergic receptor. Proc Natl Acad Sci USA. 2011;108:8228–32. doi: 10.1073/pnas.1100185108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warne T, Moukhametzianov R, Baker JG, Nehme R, Edwards PC, et al. The structural basis for agonist and partial agonist action on a beta(1)-adrenergic receptor. Nature. 2011;469:241–4. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Warne T, Edwards PC, Leslie AG, Tate CG. Crystal structures of a stabilized beta1-adrenoceptor bound to the biased agonists bucindolol and carvedilol. Structure. 2012;20:841–9. doi: 10.1016/j.str.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dore AS, Robertson N, Errey JC, Ng I, Hollenstein K, et al. Structure of the adenosine A(2A) receptor in complex with ZM241385 and the xanthines XAC and caffeine. Structure. 2011;19:1283–93. doi: 10.1016/j.str.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaakola VP, Griffith MT, Hanson MA, Cherezov V, Chien EY, et al. The 2.6 angstrom crystal structure of a human A2A adenosine receptor bound to an antagonist. Science. 2008;322:1211–7. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kolb P, Rosenbaum DM, Irwin JJ, Fung JJ, Kobilka BK, Shoichet BK. Structure-based discovery of beta2-adrenergic receptor ligands. Proc Natl Acad Sci USA. 2009;106:6843–8. doi: 10.1073/pnas.0812657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katritch V, Jaakola VP, Lane JR, Lin J, Ijzerman AP, et al. Structure-based discovery of novel chemotypes for adenosine A(2A) receptor antagonists. J Med Chem. 2010;53:1799–809. doi: 10.1021/jm901647p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carlsson J, Yoo L, Gao ZG, Irwin JJ, Shoichet BK, Jacobson KA. Structure-based discovery of A2A adenosine receptor ligands. J Med Chem. 2010;53:3748–55. doi: 10.1021/jm100240h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Graaf C, Kooistra AJ, Vischer HF, Katritch V, Kuijer M, et al. Crystal structure-based virtual screening for fragment-like ligands of the human histamine H(1) receptor. J Med Chem. 2011;54:8195–206. doi: 10.1021/jm2011589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carlsson J, Coleman RG, Setola V, Irwin JJ, Fan H, et al. Ligand discovery from a dopamine D(3) receptor homology model and crystal structure. Nat Chem Biol. 2011;7:769–78. doi: 10.1038/nchembio.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a.Mysinger MM, Weiss DR, Ziarek JJ, Gravel S, Doak AK, et al. Structure-based ligand discovery for the protein-protein interface of chemokine receptor CXCR4. Proc Natl Acad Sci USA. 2012;109:5517–22. doi: 10.1073/pnas.1120431109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tosh DK, Phan K, Gao ZG, Gakh AA, Xu F, et al. Optimization of Adenosine 5’-Carboxamide Derivatives as Adenosine Receptor Agonists Using Structure-Based Ligand Design and Fragment-Based Searching. J Med Chem. 2012;55:4297–308. doi: 10.1021/jm300095s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fredriksson R, Schioth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol Pharmacol. 2005;67:1414–25. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- 53.Katritch V, Cherezov V, Stevens RC. Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci. 2012;33:17–27. doi: 10.1016/j.tips.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kufareva I, Rueda M, Katritch V, Stevens RC, Abagyan R. Status of GPCR modeling and docking as reflected by community-wide GPCR Dock 2010 assessment. Structure. 2011;19:1108–26. doi: 10.1016/j.str.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katritch V, Kufareva I, Abagyan R. Structure based prediction of subtype-selectivity for adenosine receptor antagonists. Neuropharmacology. 2011;60:108–15. doi: 10.1016/j.neuropharm.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wheatley M, Wootten D, Conner M, Simms J, Kendrick R, et al. Lifting the lid on G-protein-coupled receptors: The role of extracellular loops. Br J Pharmacol. 2011;165:1688–703. doi: 10.1111/j.1476-5381.2011.01629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu W, Chun E, Thompson A, Chubukov P, Xu F, et al. Structural basis for allosteric regulation of GPCRS by sodium ions. Science. 2012;337:232–6. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strotmann R, Schrock K, Boselt I, Staubert C, Russ A, Schoneberg T. Evolution of GPCR: change and continuity. Mol Cell Endocrinol. 2011;331:170–8. doi: 10.1016/j.mce.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 59.Seifert R, Wenzel-Seifert K. Constitutive activity of G-protein-coupled receptors: cause of disease and common property of wild-type receptors. Naunyn-Schmiedeberg’s Arch Pharmacol. 2002;366:381–416. doi: 10.1007/s00210-002-0588-0. [DOI] [PubMed] [Google Scholar]
- 60.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 61.Nygaard R, Frimurer TM, Holst B, Rosenkilde MM, Schwartz TW. Ligand binding and micro-switches in 7TM receptor structures. Trends Pharmacol Sci. 2009;30:249–59. doi: 10.1016/j.tips.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 62.Vogel R, Mahalingam M, Ludeke S, Huber T, Siebert F, Sakmar TP. Functional role of the “ionic lock”–an interhelical hydrogen-bond network in family A heptahelical receptors. J Mol Biol. 2008;380:648–55. doi: 10.1016/j.jmb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 63.Vanni S, Neri M, Tavernelli I, Rothlisberger U. Observation of “ionic lock” formation in molecular dynamics simulations of wild-type beta 1 and beta 2 adrenergic receptors. Biochemistry. 2009;48:4789–97. doi: 10.1021/bi900299f. [DOI] [PubMed] [Google Scholar]
- 64.Warne T, Serrano-Vega MJ, Tate CG, Schertler GF. Development and crystallization of a minimal thermostabilised G protein-coupled receptor. Protein Expr Purif. 2009;65:204–13. doi: 10.1016/j.pep.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 65.Li B, Nowak NM, Kim SK, Jacobson KA, Bagheri A, et al. Random mutagenesis of the M3 muscarinic acetylcholine receptor expressed in yeast: identification of second-site mutations that restore function to a coupling-deficient mutant M3 receptor. J Biol Chem. 2005;280:5664–75. doi: 10.1074/jbc.M411623200. [DOI] [PubMed] [Google Scholar]
- 65a.Biebermann H, Schöneberg T, Schulz A, Krause G, Grüters A, et al. A conserved tyrosine residue (Y601) in transmembrane domain 5 of the human thyrotropin receptor serves as a molecular switch to determine G-protein coupling. FASEB J. 1998;12:1461–71. doi: 10.1096/fasebj.12.14.1461. [DOI] [PubMed] [Google Scholar]
- 66.Gao ZG, Jacobson KA. Keynote review: allosterism in membrane receptors. Drug Discov Today. 2006;11:191–202. doi: 10.1016/S1359-6446(05)03689-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kenakin T, Miller LJ. Seven transmembrane receptors as shapeshifting proteins: the impact of allosteric modulation and functional selectivity on new drug discovery. Pharmacol Rev. 2010;62:265–304. doi: 10.1124/pr.108.000992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Keov P, Sexton PM, Christopoulos A. Allosteric modulation of G protein-coupled receptors: a pharmacological perspective. Neuropharmacology. 2011;60:24–35. doi: 10.1016/j.neuropharm.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 69.Whalen EJ, Rajagopal S, Lefkowitz RJ. Therapeutic potential of beta-arrestin- and G protein-biased agonists. Trends Mol Med. 2011;17:126–39. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi L, Liapakis G, Xu R, Guarnieri F, Ballesteros JA, Javitch JA. Beta2 adrenergic receptor activation. Modulation of the proline kink in transmembrane 6 by a rotamer toggle switch. J Biol Chem. 2002;277:40989–96. doi: 10.1074/jbc.M206801200. [DOI] [PubMed] [Google Scholar]
- 71.Katritch V, Reynolds KA, Cherezov V, Hanson MA, Roth CB, et al. Analysis of full and partial agonists binding to beta2-adrenergic receptor suggests a role of transmembrane helix V in agonist-specific conformational changes. J Mol Recognit. 2009;22:307–18. doi: 10.1002/jmr.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vilar S, Karpiak J, Berk B, Costanzi S. In silico analysis of the binding of agonists and blockers to the beta2-adrenergic receptor. J Mol Graph Model. 2011;29:809–17. doi: 10.1016/j.jmgm.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi L, Javitch JA. The binding site of aminergic G protein-coupled receptors: the transmembrane segments and second extracellular loop. Annu Rev Pharmacol Toxicol. 2002;42:437–67. doi: 10.1146/annurev.pharmtox.42.091101.144224. [DOI] [PubMed] [Google Scholar]
- 74.Pert CB, Pasternak G, Snyder SH. Opiate agonists and antagonists discriminated by receptor binding in brain. Science. 1973;182:1359–61. doi: 10.1126/science.182.4119.1359. [DOI] [PubMed] [Google Scholar]
- 75.Jiang Q, Lee BX, Glashofer M, van Rhee AM, Jacobson KA. Mutagenesis reveals structure-activity parallels between human A2A adenosine receptors and biogenic amine G protein-coupled receptors. J Med Chem. 1997;40:2588–95. doi: 10.1021/jm970084v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parker MS, Wong YY, Parker SL. An ion-responsive motif in the second transmembrane segment of rhodopsin-like receptors. Amino Acids. 2008;35:1–15. doi: 10.1007/s00726-008-0637-6. [DOI] [PubMed] [Google Scholar]
- 77.Selent J, Sanz F, Pastor M, De Fabritiis G. Induced effects of sodium ions on dopaminergic G-protein coupled receptors. PLoS Comput Biol. 2010;6:e1000884. doi: 10.1371/journal.pcbi.1000884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Milligan G. G protein-coupled receptor hetero-dimerization: contribution to pharmacology and function. Br J Pharmacol. 2009;158:5–14. doi: 10.1111/j.1476-5381.2009.00169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maurice P, Kamal M, Jockers R. Asymmetry of GPCR oligomers supports their functional relevance. Trends Pharmacol Sci. 2011;32:514–20. doi: 10.1016/j.tips.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 80.Kaczor AA, Selent J. Oligomerization of G protein-coupled receptors: biochemical and biophysical methods. Curr Med Chem. 2011;18:4606–34. doi: 10.2174/092986711797379285. [DOI] [PubMed] [Google Scholar]
- 81.Selent J, Kaczor AA. Oligomerization of G protein-coupled receptors: computational methods. Curr Med Chem. 2011;18:4588–605. doi: 10.2174/092986711797379320. [DOI] [PubMed] [Google Scholar]
- 82.Khelashvili G, Dorff K, Shan J, Camacho-Artacho M, Skrabanek L, et al. GPCR-OKB: the G protein coupled receptor oligomer knowledge base. Bioinformatics. 2010;26:1804–5. doi: 10.1093/bioinformatics/btq264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, et al. Crystal structure of a photoactivated deprotonated intermediate of rhodopsin. Proc Natl Acad Sci USA. 2006;103:16123–8. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Taylor MS, Fung HK, Rajgaria R, Filizola M, Weinstein H, Floudas CA. Mutations affecting the oligomerization interface of G-protein-coupled receptors revealed by a novel de novo protein design framework. Biophys J. 2008;94:2470–81. doi: 10.1529/biophysj.107.117622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suda K, Filipek S, Palczewski K, Engel A, Fotiadis D. The supramolecular structure of the GPCR rhodopsin in solution and native disc membranes. Mol Membr Biol. 2004;21:435–46. doi: 10.1080/09687860400020291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mancia F, Assur Z, Herman AG, Siegel R, Hendrickson WA. Ligand sensitivity in dimeric associations of the serotonin 5HT2c receptor. EMBO Rep. 2008;9:363–9. doi: 10.1038/embor.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guo W, Urizar E, Kralikova M, Mobarec JC, Shi L, et al. Dopamine D2 receptors form higher order oligomers at physiological expression levels. EMBO J. 2008;27:2293–304. doi: 10.1038/emboj.2008.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo W, Shi L, Filizola M, Weinstein H, Javitch JA. Crosstalk in G protein-coupled receptors: changes at the transmembrane homodimer interface determine activation. Proc Natl Acad Sci USA. 2005;102:17495–500. doi: 10.1073/pnas.0508950102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lopez-Gimenez JF, Canals M, Pediani JD, Milligan G. The alpha1b-adrenoceptor exists as a higher-order oligomer: effective oligomerization is required for receptor maturation, surface delivery, and function. Mol Pharmacol. 2007;71:1015–29. doi: 10.1124/mol.106.033035. [DOI] [PubMed] [Google Scholar]
- 90.Milligan G, Pediani JD, Canals M, Lopez-Gimenez JF. Oligomeric structure of the alpha1b-adrenoceptor: comparisons with rhodopsin. Vision Res. 2006;46:4434–41. doi: 10.1016/j.visres.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 91.Klco JM, Lassere TB, Baranski TJ. C5a receptor oligomerization. I Disulfide trapping reveals oligomers and potential contact surfaces in a G protein-coupled receptor. J Biol Chem. 2003;278:35345–53. doi: 10.1074/jbc.M305606200. [DOI] [PubMed] [Google Scholar]
- 92.Hernanz-Falcon P, Rodriguez-Frade JM, Serrano A, Juan D, del Sol A, et al. Identification of amino acid residues crucial for chemokine receptor dimerization. Nat Immunol. 2004;5:216–23. doi: 10.1038/ni1027. [DOI] [PubMed] [Google Scholar]
- 93.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. The G protein-coupled receptor rhodopsin in the native membrane. FEBS Lett. 2004;564:281–8. doi: 10.1016/S0014-5793(04)00194-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yao XJ, Velez Ruiz G, Whorton MR, Rasmussen SG, DeVree BT, et al. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci USA. 2009;106:9501–6. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Deupi X, Kobilka BK. Energy landscapes as a tool to integrate GPCR structure, dynamics, and function. Physiology (Bethesda) 2010;25:293–303. doi: 10.1152/physiol.00002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goblyos A, Ijzerman AP. Allosteric modulation of adenosine receptors. Biochim Biophys Acta. 2011;1808:1309–18. doi: 10.1016/j.bbamem.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 97.Gether U, Lin S, Kobilka BK. Fluorescent labeling of purified beta 2 adrenergic receptor. Evidence for ligand-specific conformational changes. J Biol Chem. 1995;270:28268–75. doi: 10.1074/jbc.270.47.28268. [DOI] [PubMed] [Google Scholar]
- 98.Ghanouni P, Gryczynski Z, Steenhuis JJ, Lee TW, Farrens DL, et al. Functionally different agonists induce distinct conformations in the G protein coupling domain of the beta 2 adrenergic receptor. J Biol Chem. 2001;276:24433–6. doi: 10.1074/jbc.C100162200. [DOI] [PubMed] [Google Scholar]
- 99.Altenbach C, Kusnetzow AK, Ernst OP, Hofmann KP, Hubbell WL. High-resolution distance mapping in rhodopsin reveals the pattern of helix movement due to activation. Proc Natl Acad Sci USA. 2008;105:7439–44. doi: 10.1073/pnas.0802515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lodowski DT, Palczewski K, Miyagi M. Conformational changes in the G protein-coupled receptor rhodopsin revealed by histidine hydrogen-deuterium exchange. Biochemistry. 2010;49:9425–7. doi: 10.1021/bi101502v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.West GM, Chien EY, Katritch V, Gatchalian J, Chalmers MJ, et al. Ligand-dependent perturbation of the conformational ensemble for the GPCR beta(2) adrenergic receptor revealed by HDX. Structure. 2011;19:1424–32. doi: 10.1016/j.str.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chung KY, Rasmussen SG, Liu T, Li S, DeVree BT, et al. Conformational changes in the G protein Gs induced by the beta2 adrenergic receptor. Nature. 2011;477:611–5. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bokoch MP, Zou Y, Rasmussen SG, Liu CW, Nygaard R, et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature. 2010;463:108–12. doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Liu JJ, Horst R, Katritch V, Stevens RC, Wuthrich K. biased signaling pathways in beta2-adrenergic receptor characterized by 19F-NMR. Science. 2012;335:1106–10. doi: 10.1126/science.1215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim TY, Uji-i H, Moller M, Muls B, Hofkens J, Alexiev U. Monitoring the interaction of a single G-protein key binding site with rhodopsin disk membranes upon light activation. Biochemistry. 2009;48:3801–3. doi: 10.1021/bi900308c. [DOI] [PubMed] [Google Scholar]
- 106.Bockenhauer S, Furstenberg A, Yao XJ, Kobilka B, Moerner WE. Conformational dynamics of single G protein-coupled receptors in solution. J Phys Chem B. 2011;115:13328–88. doi: 10.1021/jp204843r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rahmeh R, Damian M, Cottet M, Orcel H, Mendre C, et al. Structural insights into biased G protein-coupled receptor signaling revealed by fluorescence spectroscopy. Proc Natl Acad Sci USA. 2012;109:6733–8. doi: 10.1073/pnas.1201093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nobles KN, Xiao K, Ahn S, Shukla AK, Lam CM, et al. Distinct phosphorylation sites on the {beta}2-adrenergic receptor establish a barcode that encodes differential functions of {beta}-arrestin. Sci Signal. 2011;4:ra51. doi: 10.1126/scisignal.2001707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, et al. Structure and function of an irreversible agonist-beta(2) adrenoceptor complex. Nature. 2011;469:236–40. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dror RO, Pan AC, Arlow DH, Borhani DW, Maragakis P, et al. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc Natl Acad Sci USA. 2011;108:13118–23. doi: 10.1073/pnas.1104614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hino T, Arakawa T, Iwanari H, Yurugi-Kobayashi T, Ikeda-Suno C, et al. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 2012;482:237–40. doi: 10.1038/nature10750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Congreve M, Andrews SP, Dore AS, Hollenstein K, Hurrell E, et al. Discovery of 1,2,4-triazine derivatives as adenosine A(2A) antagonists using structure based drug design. J Med Chem. 2012;55:1898–903. doi: 10.1021/jm201376w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–7. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 114.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–38. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 115.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure–function relations in G-protein coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]