Abstract
The non-enzymatic free radical generation of reactive aldehydes is known to contribute to diseases of sustained oxidative stress including rheumatoid arthritis, atherosclerosis, neurodegenerative and a number of liver diseases. At the same time, the accumulation of lipid electrophiles has been demonstrated to play a role in cell signaling events through modification of proteins critical for cellular homeostasis. Given the broad scope of reactivity profiles and the ability to modify numerous proteomic and genomic processes, new emphasis is being placed on a systems-based analysis of the consequences of electrophilic adduction. This review focuses on the generation and chemical reactivity of lipid-derived aldehydes with a special focus on the homeostatic responses to electrophilic stress.
Keywords: Lipid Peroxidation, Reactive Aldehyde, 4-hydroxynonenal, 4-HNE
Introduction
Unsaturated fatty acids are abundant constituents sequestered in sn-1 and sn-2 esterfied forms in glycerol phospholipids within cellular membranes. In addition, unsaturated free fatty acids are abundant within various cellular compartments. The fact that lipids constitute a major portion of the plasma, mitochondrial and endoplasmic membranes establishes the presence of massive concentrations of unsaturated fatty acids within membranous structures. Likewise, the concentration of unsaturated fatty acids, in the free form or bound to specific transport proteins, in cells is noteworthy. It is also well recognized that the polyunsaturated fatty acids are bioactive mediators of diverse pathways involved in cellular homeostasis or, in some cases, interact with cellular macromolecules resulting in cell death. These cellular responses may be a consequence of the vulnerability of unsaturated fatty acids to diverse oxidation reactions or radical reactions both of which result in formation of electrophilic lipid products. Certain of the oxidation reactions involving unsaturated fatty acids are enzymatically mediated by families of non-heme-containing metallo-enzymes including the lipoxygenases (LOX), cyclooxygenases (COX), and cytochrome P450. Because these reactions are integral for an organisms response to a range of stimuli, they are generally well-controlled and generate a spectrum of bioactive products which are ligands for highly specific, receptor-mediated responses including vasodilation, vasoconstriction as well as pro-inflammatory or anti-inflammatory cascades (for informative comprehensive reviews of the production and actions of enzymatically generated bioactive lipids see the entire volume of Chem Rev. 111:2011) [1].
The electrophilic lipid products generated by free radical mediated lipid oxidation are different in many respects than those generated by enzyme-mediated oxidation. First, free radical mediated production of electrophilic products of polyunsaturated fatty acids proceeds by autocatalysis and is, as a result, not well regulated. Thus, free radical mediated lipid peroxidation is more commonly associated with diseases of sustained oxidative stress including rheumatoid arthritis, atherosclerosis, neurodegenerative and a number of liver diseases. Further, the overproduction of electrophilic lipids resulting from free radical mediated peroxidation of polyunsaturated fatty acids has been demonstrated to initiate cell death through modification of proteins critical for cellular homeostasis. A series of recent reviews highlight the advances in identification of lipid substrates, mechanisms of lipid peroxidation and protein targets of electrophilic lipid modification [2, 3]. At the same time, there is emerging data demonstrating that, like the lipid products produced by the LOX, COX, and P450-mediated oxidation of unsaturated fatty acids, the electrophilic products of radical-generated lipid peroxidation also initiate responses that are cytoprotective and thus integral for cellular homeostasis and survival. This review will provide an overview of free radical mediated lipid peroxidation involved in cellular injury followed by a more comprehensive examination of recent studies describing the potential of electrophilic products of lipid peroxidation to modulate signaling pathways involved in antioxidant responses. The final section of this perspective will assess the potential of antioxidant therapy to enhance or abrogate antioxidant responses initiated by electrophilic lipid signaling.
Generation of Reactive Aldehydes
The generation of oxygen-derived free radicals and oxidants is a consequence of cellular metabolism and bioenergetic processes throughout the animal kingdom [4]. It is evident that a host of biochemical mechanisms mediate persistent free radical generation and damage. An insightful review by Hermann Esterbauer details the integration of oxidative processes in human physiology [5]. In it, he states that the adult human with a daily energy 0requirement of 10,000 kJ consumes approximately 660 g of oxygen per day, with roughly 90–95% of that oxygen converted to harmless water through mitochondrial respiration. The remaining 5–10% of that oxygen undergoes univalent and divalent reduction, yielding reactive oxygen species, such as superoxide radicals. When taken into context of the average human lifespan, the human body consumes a massive 17,000 kg of oxygen which results in the concomitant production of 800–1700 kg of oxygen radicals. These figures provide a fascinating perspective on the detoxifying mechanisms aerobic organisms have integrated into biology and the critical role they play in diffusing oxygen radicals and maintaining life.
The endogenous generation of reactive aldehydes has been studied for decades and is known to contribute to numerous disease pathologies by altering proteomic, genomic, cell signaling, and metabolic processes [6, 7]. Reactive aldehydes compose a class of highly reactive organic chemical compounds obtained by oxidation of primary alcohols, characterized by the common group R-CHO consisting of a carbonyl center bonded to hydrogen and an R group [8]. These compounds arise predominantly as a consequence of oxidative stress within the cellular microenvironment, where prooxidant forces overcome natural antioxidant capacities. Numerous studies demonstrate that at low levels, these compounds contribute to regulating cell proliferation among many other processes; however, a delicate balance exists between basal levels of these aldehydes and cytotoxic concentrations [5, 6, 9, 10].
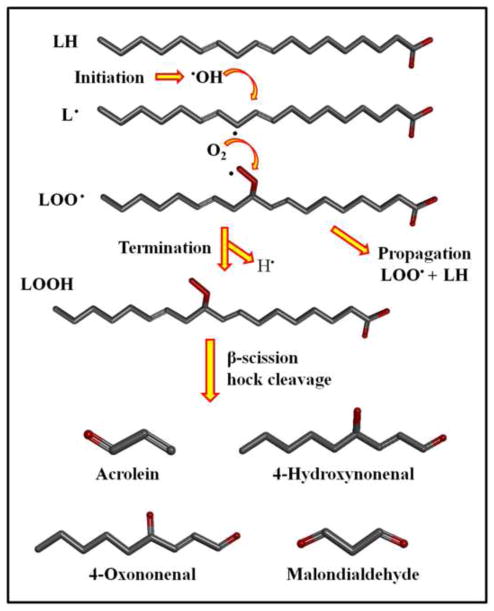
The production of reactive aldehydes occurs through a number of pathways, including enzymatic and non-enzymatic processes. One of the most prominent sources for the generation of reactive aldehydes is through non-enzymatic free radical mechanisms [11]. Specifically, a profusion of reactive nitrogen species (RNS) and reactive oxygen species (ROS), including peroxynitrite, superoxide radicals, and hydroxyl radicals provide an overabundance of initiating chemicals. A major source of these aldehydes is the autoxidation of polyunsaturated fatty acids (PUFA), including arachidonic and linoleic acid. Lipid peroxidation occurs in three major phases, comprised of an initiation event, chain propagation, and termination (Figure 1.) [3]. Typical initiating chemical species are hydroxyl and superoxide radicals. Both are present under normal physiologic conditions and are produced at much higher concentrations in situations of oxidative stress. Initiation occurs through the abstraction of a bis-allylic hydrogen from a lipid chain (LH) to yield a lipid radical (L·). Propagation proceeds when oxygen is added to the carbon-centered radical, where L· is rapidly converted to an oxygen-centered peroxyl radical (LOO·). The LOO· reacts with another LH to generate L· and an unstable lipid hydroperoxide (LOOH), which in turn yields new peroxyl and alkoxyl radicals, degrading further to secondary products through β-scission and hock cleavage, among others. A single initiation reaction is postulated to result in 200 to 400 propagation cycles, rapidly amplifying free radical damage under highly oxidizing PUFA-rich environments [12]. Chain breaking events are known to occur, as α-tocopherol (vitamin E) radicals form while converting LOO· to LOOH [13]. Termination of the propagation cycle occurs when two free radical species combine to yield non-radical species. The degradation of peroxyl and alkoxyl radicals into secondary products is thought to generate well over a hundred different reactive species, each with a wide range of reactivity, size, and specificity [5, 6]. This overabundance of chemical species greatly increases the complexity of studies attempting to characterize lipid peroxidation derived electrophilic damage. The more prominent products 4-hydroxynonenal (4-HNE), malondialdehyde (MDA), and acrolein (ACR) have been examined under various disease models through in vitro assays, immunohistochemical staining, and analytical HPLC methods [7, 14, 15].
Figure 1.
The generation of reactive aldehydes through the initiation, propagation, and termination stages of lipid peroxidation. For simplicity, Figure 1 shows the generation of reactive aldehydes from a C18 fatty acid; however, in cells, the fatty acids are largely esterified within phospholipids. Furthermore, reactive aldehydes can also occur in esterified form, which may also form adducts with proteins and other macromolecules, although this is a less well studied area.
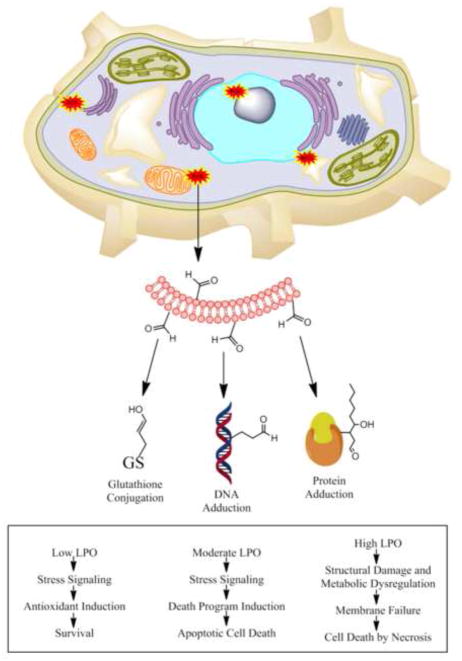
Cellular location is an important consideration in determining the extent of free radical mediated damage resulting in the generation of reactive aldehyde species. As illustrated in Figure 2, lipid rich membranes provide an optimal environment for producing a high abundance of these cytotoxic compounds. Microsomal and mitochondrial intra-membrane concentrations of these lipid peroxidation derived electrophilic compounds have been estimated to accumulate to as high as 10 mM in vivo [9, 16]. The regulatory function and cytotoxicity of these aldehydes hinges on abundance, reactivity and longevity. Longevity varies greatly, with compounds such as 4-oxononenal (4-ONE) and 4-HNE displaying half-lives of roughly 1 second and 2 minutes, respectively [9, 17]. Persisting on a scale of minutes, some aldehydes preferentially retain the ability to transiently modify distant proteins, membranes, and DNA. These factors may contribute to a localized bias for molecular adduction by reactive aldehydes, particularly those with short half-lives, as nucleophiles located within the vicinity of lipid peroxidation remain more susceptible to higher concentrations of electrophiles. However, long-lived species may have a more pronounced global impact by altering a broader array of proteomic and genomic targets due to their transient nature.
Figure 2. Cellular effects of oxidative stress and lipid peroxidation.
The non-enzymatic free radical generation of reactive aldehydes by lipid peroxidation occurs near lipid-rich cellular membranes and produces both membrane-bound and free electrophiles. These electrophilic species go on to modify proteomic and genomic processes. Antioxidant responses such as glutathione conjugation play a role in defending against the damaging effects of these reactive aldehydes. (LPO, lipid peroxidation)
Chemical Mechanisms
The chemical specificity of reactive aldehydes is impacted through direct and indirect forces. Direct factors include characteristics of the electrophile and nucleophile species, such as pKa, chemical potential, and electrophilic index. Indirect forces include solvent pH and adjacent peptide sequence, which can play an important role through steric hindrance and charge stabilization. Both factors play an integrated role in determining the specificity for any given set of electrophile-nucleophile pairs. To better understand the stability and reactivity of these adducts, a number of researchers have employed computational models coupled to in vitro studies [18–22]. Importantly, these studies provide key mechanistic insight into the reactivity and specificity of reactive aldehydes toward cellular targets. A detailed analysis of the selectivity of electrophile-nucleophile interactions applying the hard, soft, acids, and bases (HSAB) theory is covered extensively in a recent perspective [23]. Basically, electrophiles and nucleophiles are classified as either soft (polarizable) or hard (nonpolarizable). Based on these parameters, among others, the specificity of an electrophile for any given nucleophile is determined with a certain degree of confidence. Therefore, these factors can be applied to chemical toxicology to aid in the identification of sites of modification and mechanisms of toxicant action. Further, through the application of quantum mechanical parameters to in vitro experiments, it was confirmed that soft electrophiles bind more favorably with soft nucleophiles and hard electrophiles bind more favorably with hard nucleophiles [23, 24]. An in vitro study using purified human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) intricately demonstrates that a weak electrophile (acrylamide) was able to discriminate among highly reactive cysteines from less reactive residues [19]. Furthermore, a quantitative relationship was determined correlating adduct formation and protein dysfunction. Certainly, in vivo and in vitro model systems contain highly complex matrices of biomolecules which may confound the predictive nature of these experiments employing purified protein and theoretical computational models. However, these examples provide support for the combined application of computational analysis with physiologically applied models to better understand the chemical reactivity of these reactive aldehydes.
The covalent modification of biomolecules by reactive aldehydes is a well-documented phenomenon [25, 26]; however, underlying chemical mechanisms of specificity and reactivity remain somewhat unclear. Basic mechanisms of aldehyde modification include Michael addition and Schiff base reactions with the thiol group of cysteine and secondary amines of lysine, histidine, and arginine [3, 27]. DNA adducts arise through direct base adduction, for example, the Michael addition of deoxyguanosine by 4-HNE [21]. In general, these electrophilic adducts are thought to generate an array of stable, covalent modifications, yet numerous reports present a contrasting account of adduct stability [20, 28–30]. Instability among theses electrophilic adducts provides a likely explanation for the lack of reported in vivo adducts by mass spectrometry techniques. While continued research is providing an ever-expanding list of proteins modified in vivo by lipid electrophiles like 4-HNE [25, 31], the modified amino acid is rarely identified. To circumvent potential instability of adducts, sodium borohydride treatment is used to stabilize these adducts through reduction, increasing the chances for identifying modified proteins [32]. Recently, a number of novel methods have been developed to capture these carbonylated proteins and provide a large toolset for identifying and characterizing these PTMs [33–36]. These strategies were recently reviewed and include a click chemistry approach as well as those based primarily on hydrazide reactivity with protein carbonyls, such as biotin hydrazide, aldehyde reactive probes, and Girard’s P reagent [3].
The majority of research studying chemical mechanisms of aldehyde adduction has occurred under controlled experimental conditions. In these studies, protein targets previously identified through western blotting or immunoprecipitation techniques are treated with known quantities of aldehyde in a test tube [18, 37–39]. While these studies have revealed important information relating to amino acid targets and kinetic parameters for these reactive aldehydes, little insight is gained in establishing a substantial connection between the electrophilic modification of a specific protein and a related physiologic response. Importantly, a number of studies have demonstrated that these electrophilic adducts may be used as biomarkers for a range of disease pathologies; however, what role these adducts play in the initiation and/or progression of these diseases remain unclear [7]. Only recently has a global analysis of protein damage by a lipid electrophile been accomplished [25]. In this study, human colon carcinoma RKO cells were treated with increasing concentrations of 4-HNE followed by the isolation of modified proteins for MS/MS analysis. Proteomic surveys of protein carbonylation typically generate large lists of proteins (~500), where highly abundant proteins are found modified with greater frequency, which leads to the question of 4-HNE specificity versus concentration-type interactions. An important consideration when examining these adducts is whether they form through concentration-dependent mechanisms. It is anticipated that true protein targets of 4-HNE display an electrophile concentration-dependent relationship. As expected, this study confirmed previous conclusions regarding pseudo-first order kinetics of 4-HNE protein adduction [40]. A key analysis is to examine if protein damage reflects specificity for cell components and processes. To address this particular issue, Codreanu et al. performed a networks and communities analysis of modified proteins [25]. By mapping the identified 4-HNE-adducted proteins to a human protein interaction network they were able to provide systems-level insight into the cellular impact of 4-HNE adduction. Importantly, the authors were able to utilize protein clustering coefficients to characterize protein communities targeted by 4-HNE and elucidate cellular processes that may be impacted.
Representing a paradigm shift in studying cellular responses to electrophiles, Jacobs and Marnett provide a novel approach to define the chemistry of protein modification using lipid electrophiles as a model system [33]. Employing a 4-HNE-derived click chemistry treatment to RKO cells, the authors identified 4-HNE activated transcriptional pathways though microarray, real-time PCR, Western blot, and luciferase reporter assays. Their global approach incorporated data analyses for electrophile-protein modification as well as alterations in gene expression. Informatics tools provided a theoretical connection by linking electrophile-induced proteomic and genomic effects through related transcription factors. Interestingly, pathways that were found to be activated by 4-HNE treatment include DNA damage, antioxidant, ER stress, and heat shock responses. This strategy correlates proteome-wide electrophile modification to cellular responses, bridging our understanding of the chemical reactivity of aldehydes to human health and disease. Indeed, electrophile-induced protein modification and downstream effects reveal an extremely complex cascade of cellular responses to oxidative stress and lipid peroxidation in particular.
Homeostatic Responses to Electrophilic Stress
The proposition that 4-HNE and other lipid electrophiles produced from the radical oxidation of lipids mediate cellular toxicity through adduction of proteins has been investigated extensively and is the subject of many recent reviews. However, investigators have observed that cellular responses are divergent depending on the dose or concentration of 4-HNE that the cells are exposed to. For instance, in the classic review presented by Esterbauer [9] it is noted that cells exposed to sub-cytotoxic concentrations (0.5–2μM) of 4-HNE displayed activation of phospholipase c activity or, in the case of smooth muscle vascular cells, proliferation. These observations were followed by a report [41] that sub-cytotoxic concentrations of 4-HNE increase expression of the glutathione s-transferases (GST) in a hepatoma cell line through activation of the antioxidant response element (ARE). Because GST-mediated conjugation of 4-HNE with glutathione (GSH) is a major pathway for detoxification of this lipid electrophile, the results of this study suggested that the exposure of cells to sub-cytotoxic concentrations of 4-HNE result in induction of pathways which facilitate its elimination. This notion was subsequently confirmed by a report that exposure of HBE1 cells to physiologically relevant concentrations of 4-HNE resulted in induction of GSH synthesis through induction of glutamate cysteine ligase (GCL) which is the rate limiting step in glutathione synthesis [42]. The fact that GCL induction was attenuated by a JNK inhibitor demonstrated that 4-HNE signaled this response through the JNK pathway. Taken together, these early studies suggested that when present at low concentrations, 4-HNE has the ability to activate a spectrum of cytoprotective signaling cascades in order to maintain cellular homeostasis.
There is general agreement that the cellular accumulation of 4-HNE leads to apoptosis and the specific mechanisms by which 4-HNE modulates signaling stress-induced programmed cell death has received considerable attention and is detailed in a recent review [43]. The involvement of 4-HNE in modulation of programmed cell death is multifactorial in that this lipid electrophile modulates signaling pathways involved in extrinsic and intrinsic pathways of apoptosis. In terms of the extrinsic pathway, reports have appeared describing the potential of 4-HNE initiate binding of the death-associated protein Daxx with the intracellular surface of the Fas receptor [44–46]. Once Daxx binding to Fas occurs, apoptosis is modulated through the down-stream signaling proteins, ASK1, JNK and Caspase 3. Interestingly, the silencing of Daxx results in increased sensitivity of Jurkat cells to 4-HNE-induced apoptosis suggesting that this lipid electrophile, when present at low concentrations, can play a self-regulatory or self-limiting role in apoptosis [46]. Activation of the intrinsic pathway of apoptosis involves the p53 tumor-suppressor protein [47]. The ability of 4-HNE generated by oxidative stress to initiate apoptosis has been attributed to the ability of this lipid electrophile to increase p53 expression followed by activation of p21, Bax, JNK and caspase 3 [45, 48]. The involvement of 4-HNE in apoptosis through extrinsic and intrinsic pathways is interesting in terms of self-regulating the extent of programed cell death in response to oxidative stress through a common down-stream signaling protein JNK. As this phenomenon continues to be investigated, it will be important to determine if programmed cell death by 4-HNE is regulated through the same mechanisms in all cell types.
The concept that cellular responses to 4-HNE and oxidant stress is supported by a decade old study documenting modulation of 35 genes in human retinal pigment epithelial cells exposed to 4-HNE or the oxidants H2O2 or tert-butylhydroperoxide [49]. The mechanisms of gene modulation by 4-HNE were revealed in a more recent comprehensive report of gene profiling expression in RKO cells exposed to sub-cytotoxic concentrations of 4-HNE for 6 or 24 hours [50]. Among the prominent responsive genes identified by these investigators were those involved in oxidative damage or ER stress. Of particular interest was the experimental approach which facilitated the observation that gene responsiveness was mechanistically linked to signaling responses associated with adduction of relevant proteins.
As noted earlier in this review, protein thiols are preferential targets for modification by 4-HNE. It is not surprising that the transcription factor Keap-1-Nrf2, a chief regulator of cellular antioxidant responses through redox sensitive cysteine residues, is a sensitive target protein for 4-HNE adduction resulting in activation of cellular antioxidant systems. The specific mechanisms of 4-HNE- Keap-1-Nrf2 interactions and cellular responses are detailed in a recent review [51]. Therefore, we will present only an overview of reports describing the interaction of 4-HNE with the thiol sensitive systems involving Keap-1-Nrf-2 which, in turn, impact protective cellular responses to stress.
As detailed in two recent reviews [51, 52] the involvement of Keap-1-Nrf2 complex in antioxidant responses, through activation of the antioxidant-response elements (AREs) has received considerable attention. The mechanistic aspects of this system are intriguing in that electrophiles such as 4-HNE target cysteine thiols in Keap-1 resulting in release of Nrf2 which translocates to the nucleus where it dimerizes with nuclear factors forming a complex activating expression of ARE genes for the phase II antioxidant proteins NADH-dependent quinone reductase, heme oxygenase-1, glutathione-S-transferases and glutamate-cysteine ligase. The release of Nrf2 from Keap-1 is mediated by modification of specific thiol sensors in Keap-1 by electrophiles such as 4-HNE. Although Keap-1 contains multiple thiols, those at locations C151 and C288 appear to be specific for 4-HNE and other alkenals [53]. Interestingly, these specific cysteine residues are separate from those targeted by other stress associated endogenous agents such as nitric oxide or Zn+ suggesting a high degree of specificity for the signaling capabilities of Keap-1 to endogenous signals.
The thiol sensitivity of the nuclear factor kappa b (NFκB) system was first proposed in a study describing what appeared to be inhibition of its activation in RKO cells and H1299 cells exposed to 4-HNE [54]. NFκB is a transcription factor commonly associated with proinflammatory signaling and is regulated by binding of the inhibitor kappa B (IκBα) protein. Thus, under homeostatic cellular conditions, IκBα binds to NFκB sequestering it in an inactive form. Under conditions of cell stress, the enzyme IκB kinase phosphorylates IκBα facilitating its dissociation from NFκB allowing translocation to the nucleus for transcription of Bcl-2 family members which mediate antiapoptotic effects. Similarly, the expression of proinflammatory cytokines such as IL-6 is also mediated by NFκB. Interestingly, systematic evaluation of the NFκB inhibition revealed that 4-HNE inhibited IkB kinase (IKK), thus compromising IkBα degradation [54]. The sensitivity of IKK to 4-HNE inhibition is marked, as the studies with kupffer cells, which are the resident macrophage in liver, have demonstrated inhibition of IL-6 release is severely impaired by sub-micromolar concentrations of 4-HNE [55]. Thus, 4-HNE produced during controlled lipid peroxidation such as that occurring during homeostatic cell metabolism, may play an important role in regulating inflammatory responses. Taken together, the effects of 4-HNE on activation of phase II antioxidant enzymes through Keap-1 and repression of pro-inflammatory cytokine release by inhibition of IKK suggest that basal concentrations of this and other lipid electrophiles may play very important homeostatic roles in a variety of cell types.
Considerations of Pharmacologic Strategies for Mitigating Lipoxidation Derived Reactive Aldehydes
There is substantial evidence that the overproduction of electrophilic lipid aldehydes such as 4-HNE result in protein carbonylation associated with a spectrum of disorders including diabetes, neurodegenerative diseases and liver disease. Therefore, it is logical that a number of strategies have been evaluated to arrest oxidative stress through the use of antioxidants or attempts to chemically trap the reactive lipid electrophiles. To date, there is little evidence that prophylactic antioxidant therapy is effective in preventing or slowing progression of diseases involving oxidative stress. The approach of using specific agents to trap reactive aldehydes is based on the chemical properties of the thiol- and histidine-containing compounds that form Michael adducts with the lipid electrophiles. The agents evaluated to date are described in detail in a comprehensive review [56]. Briefly, one strategy includes utilizing endogenous pathways of detoxification such as spontaneous and GST-catalyzed glutathionylation, where irrigation with γ-glutamyl-cysteine may increase GSH levels to normalize pathophysiological 4-HNE concentrations to pre-insult levels. Other interventions include ascorbate (vitamin C) and carnosine (B-alanyl-L-histidine), each used as a quencher of cytotoxic carbonyl species via Michael addition [7, 56]. Whereas these compounds are effective in trapping reactive aldehydes in isolated cells subjected to significant oxidative stress, their efficacy in animal models or humans remains to be established. As with any potential therapeutic agent, a thorough assessment must be performed to identify off-target effects and resulting toxicities. This is complicated by the potential role of 4-HNE in regulating homeostatic responses to oxidative stress through ARE activation [53] or inhibition of the release of pro-inflammatory cytokines through modulation of NFκB [54, 55]. Likewise, especially challenging is the selectivity of targeting specific protein thiols, such as those identified in keap-1, which selectively interact with 4-HNE rather than Zn+ or nitric oxide [53]. Clearly, caution is needed when prophylactic treatment with antioxidant therapies such as vitamin E supplementation is employed. One must assess the overall impact to signaling cascades as well as oxidative stress using a system-wide approach.
Conclusions
The generation of reactive aldehydes through non-enzymatic free radical mechanisms is a complex process resulting in the formation of over a hundred different species of electrophilic compounds containing a broad spectrum of reactivity profiles. Given that oxidative stress is integrated throughout the cell, most notably through cellular respiration in the mitochondria, it is not surprising that multiple antioxidant responses exist. Indeed, a systems biology approach investigating both protective and deleterious effects of reactive aldehydes is essential to understanding the chemico-biological nature of aldehyde-regulated cellular homeostasis. While reactive aldehydes are known to alter proteomic, lipidomic, and genomic processes, very little is known about the role these compounds play in the initiation and progression of disease pathologies. Further studies are required to elucidate the physiological impact these electrophiles have on biological processes.
Highlights.
An overview of the chemistry and biology of reactive aldehydes is presented.
Non-enzymatic generation of reactive aldehydes contributes to numerous diseases.
Electrophiles mediate cellular toxicity via protein and small molecule adduction.
New emphasis is placed on systems-based analysis of aldehyde reactivity.
Acknowledgments
Funding
This work was supported by, in part, by Grants NIH/NIAAA5R37AA009300–16 (to D.R.P.) and NIH/NIDDK 5R01DK074487–05 (to D.R.P.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brown HA, Marnett LJ. Introduction to lipid biochemistry, metabolism, and signaling. Chem Rev. 2011;111:5817–5820. doi: 10.1021/cr200363s. [DOI] [PubMed] [Google Scholar]
- 2.Catala A. Lipid peroxidation of membrane phospholipids generates hydroxy-alkenals and oxidized phospholipids active in physiological and/or pathological conditions. Chem Phys Lipids. 2009;157:1–11. doi: 10.1016/j.chemphyslip.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Fritz KS, Petersen DR. Exploring the biology of lipid peroxidation-derived protein carbonylation. Chem Res Toxicol. 2011;24:1411–1419. doi: 10.1021/tx200169n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol. 2007;39:44–84. doi: 10.1016/j.biocel.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Esterbauer H. Cytotoxicity and genotoxicity of lipid-oxidation products. Am J Clin Nutr. 1993;57:779S–785S. doi: 10.1093/ajcn/57.5.779S. discussion 785S–786S. [DOI] [PubMed] [Google Scholar]
- 6.Gueraud F, Atalay M, Bresgen N, Cipak A, Eckl PM, Huc L, Jouanin I, Siems W, Uchida K. Chemistry and biochemistry of lipid peroxidation products. Free Radic Res. 2010;44:1098–1124. doi: 10.3109/10715762.2010.498477. [DOI] [PubMed] [Google Scholar]
- 7.Poli G, Schaur RJ, Siems WG, Leonarduzzi G. 4-hydroxynonenal: a membrane lipid oxidation product of medicinal interest. Med Res Rev. 2008;28:569–631. doi: 10.1002/med.20117. [DOI] [PubMed] [Google Scholar]
- 8.McNaught AD, Wilkinson A. Compendium of chemical terminology: IUPAC recommendations. Oxford [England]; Malden, MA, USA: Blackwell Science; 1997. International Union of Pure and Applied Chemistry. [Google Scholar]
- 9.Esterbauer H, Schaur RJ, Zollner H. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med. 1991;11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 10.Pizzimenti S, Barrera G, Dianzani MU, Brusselbach S. Inhibition of D1, D2, and A-cyclin expression in HL-60 cells by the lipid peroxydation product 4-hydroxynonenal. Free Radic Biol Med. 1999;26:1578–1586. doi: 10.1016/s0891-5849(99)00022-2. [DOI] [PubMed] [Google Scholar]
- 11.Niki E, Yoshida Y, Saito Y, Noguchi N. Lipid peroxidation: mechanisms, inhibition, and biological effects. Biochem Biophys Res Commun. 2005;338:668–676. doi: 10.1016/j.bbrc.2005.08.072. [DOI] [PubMed] [Google Scholar]
- 12.Jones DP. Radical-free biology of oxidative stress. Am J Physiol Cell Physiol. 2008;295:C849–868. doi: 10.1152/ajpcell.00283.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reed TT. Lipid peroxidation and neurodegenerative disease. Free Radic Biol Med. 2011;51:1302–1319. doi: 10.1016/j.freeradbiomed.2011.06.027. [DOI] [PubMed] [Google Scholar]
- 14.Wakita C, Honda K, Shibata T, Akagawa M, Uchida K. A method for detection of 4-hydroxy-2-nonenal adducts in proteins. Free Radic Biol Med. 2011;51:1–4. doi: 10.1016/j.freeradbiomed.2011.02.037. [DOI] [PubMed] [Google Scholar]
- 15.Sampey BP, Korourian S, Ronis MJ, Badger TM, Petersen DR. Immunohistochemical characterization of hepatic malondialdehyde and 4-hydroxynonenal modified proteins during early stages of ethanol-induced liver injury. Alcohol Clin Exp Res. 2003;27:1015–1022. doi: 10.1097/01.ALC.0000071928.16732.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benedetti A, Comporti M, Fulceri R, Esterbauer H. Cytotoxic aldehydes originating from the peroxidation of liver microsomal lipids. Identification of 4,5-dihydroxydecenal. Biochim Biophys Acta. 1984;792:172–181. doi: 10.1016/0005-2760(84)90219-4. [DOI] [PubMed] [Google Scholar]
- 17.Doorn JA, Petersen DR. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem Res Toxicol. 2002;15:1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 18.Wakita C, Maeshima T, Yamazaki A, Shibata T, Ito S, Akagawa M, Ojika M, Yodoi J, Uchida K. Stereochemical configuration of 4-hydroxy-2-nonenal-cysteine adducts and their stereoselective formation in a redox-regulated protein. J Biol Chem. 2009;284:28810–28822. doi: 10.1074/jbc.M109.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martyniuk CJ, Fang B, Koomen JM, Gavin T, Zhang L, Barber DS, Lopachin RM. Molecular mechanism of glyceraldehyde-3-phosphate dehydrogenase inactivation by alpha,beta-unsaturated carbonyl derivatives. Chem Res Toxicol. 2011;24:2302–2311. doi: 10.1021/tx200437y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fritz KS, Kellersberger KA, Gomez JD, Petersen DR. 4-HNE Adduct Stability Characterized by Collision-Induced Dissociation and Electron Transfer Dissociation Mass Spectrometry. Chem Res Toxicol. 2012 doi: 10.1021/tx300100w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Banerjee S, Christov PP, Kozekova A, Rizzo CJ, Egli M, Stone MP. Replication Bypass of the trans-4-Hydroxynonenal-Derived (6S,8R,11S)-1,N(2)-Deoxyguanosine DNA Adduct by the Sulfolobus solfataricus DNA Polymerase IV. Chem Res Toxicol. 2012;25:422–435. doi: 10.1021/tx200460j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han B, Stevens JF, Maier CS. Design, synthesis, and application of a hydrazide-functionalized isotope-coded affinity tag for the quantification of oxylipid-protein conjugates. Anal Chem. 2007;79:3342–3354. doi: 10.1021/ac062262a. [DOI] [PubMed] [Google Scholar]
- 23.Lopachin RM, Gavin T, Decaprio A, Barber DS. Application of the Hard and Soft, Acids and Bases (HSAB) Theory to Toxicant-Target Interactions. Chem Res Toxicol. 2012;25:239–251. doi: 10.1021/tx2003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LoPachin RM, Gavin T, Petersen DR, Barber DS. Molecular mechanisms of 4-hydroxy-2-nonenal and acrolein toxicity: nucleophilic targets and adduct formation. Chem Res Toxicol. 2009;22:1499–1508. doi: 10.1021/tx900147g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Codreanu SG, Zhang B, Sobecki SM, Billheimer DD, Liebler DC. Global analysis of protein damage by the lipid electrophile 4-hydroxy-2-nonenal. Mol Cell Proteomics. 2009;8:670–680. doi: 10.1074/mcp.M800070-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petersen DR, Doorn JA. Reactions of 4-hydroxynonenal with proteins and cellular targets. Free Radic Biol Med. 2004;37:937–945. doi: 10.1016/j.freeradbiomed.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 27.Liebler DC. Protein damage by reactive electrophiles: targets and consequences. Chem Res Toxicol. 2008;21:117–128. doi: 10.1021/tx700235t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rauniyar N, Stevens SM, Prokai-Tatrai K, Prokai L. Characterization of 4-hydroxy-2-nonenal-modified peptides by liquid chromatography-tandem mass spectrometry using data-dependent acquisition: neutral loss-driven MS3 versus neutral loss-driven electron capture dissociation. Anal Chem. 2009;81:782–789. doi: 10.1021/ac802015m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo J, Prokai L. To tag or not to tag: a comparative evaluation of immunoaffinity-labeling and tandem mass spectrometry for the identification and localization of posttranslational protein carbonylation by 4-hydroxy-2-nonenal, an end-product of lipid peroxidation. J Proteomics. 2011;74:2360–2369. doi: 10.1016/j.jprot.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin D, Saleh S, Liebler DC. Reversibility of Covalent Electrophile-Protein Adducts and Chemical Toxicity. Chem Res Toxicol. 2008 doi: 10.1021/tx800248x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shin NY, Liu Q, Stamer SL, Liebler DC. Protein targets of reactive electrophiles in human liver microsomes. Chem Res Toxicol. 2007;20:859–867. doi: 10.1021/tx700031r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uchida K, Stadtman ER. Selective cleavage of thioether linkage in proteins modified with 4-hydroxynonenal. Proc Natl Acad Sci U S A. 1992;89:5611–5615. doi: 10.1073/pnas.89.12.5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs AT, Marnett LJ. Systems analysis of protein modification and cellular responses induced by electrophile stress. Acc Chem Res. 2010;43:673–683. doi: 10.1021/ar900286y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim HY, Tallman KA, Liebler DC, Porter NA. An azido-biotin reagent for use in the isolation of protein adducts of lipid-derived electrophiles by streptavidin catch and photorelease. Mol Cell Proteomics. 2009;8:2080–2089. doi: 10.1074/mcp.M900121-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirzaei H, Regnier F. Enrichment of carbonylated peptides using Girard P reagent and strong cation exchange chromatography. Anal Chem. 2006;78:770–778. doi: 10.1021/ac0514220. [DOI] [PubMed] [Google Scholar]
- 36.Chung WG, Miranda CL, Maier CS. Detection of carbonyl-modified proteins in interfibrillar rat mitochondria using N′-aminooxymethylcarbonylhydrazino-D-biotin as an aldehyde/keto-reactive probe in combination with Western blot analysis and tandem mass spectrometry. Electrophoresis. 2008;29:1317–1324. doi: 10.1002/elps.200700606. [DOI] [PubMed] [Google Scholar]
- 37.Roede JR, Carbone DL, Doorn JA, Kirichenko OV, Reigan P, Petersen DR. In vitro and in silico characterization of peroxiredoxin 6 modified by 4-hydroxynonenal and 4-oxononenal. Chem Res Toxicol. 2008;21:2289–2299. doi: 10.1021/tx800244u. [DOI] [PubMed] [Google Scholar]
- 38.Ishii T, Tatsuda E, Kumazawa S, Nakayama T, Uchida K. Molecular basis of enzyme inactivation by an endogenous electrophile 4-hydroxy-2-nonenal: identification of modification sites in glyceraldehyde-3-phosphate dehydrogenase. Biochemistry (Mosc) 2003;42:3474–3480. doi: 10.1021/bi027172o. [DOI] [PubMed] [Google Scholar]
- 39.Fritz KS, Galligan JJ, Smathers RL, Roede JR, Shearn CT, Reigan P, Petersen DR. 4-Hydroxynonenal inhibits SIRT3 via thiol-specific modification. Chem Res Toxicol. 2011;24:651–662. doi: 10.1021/tx100355a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szapacs ME, Riggins JN, Zimmerman LJ, Liebler DC. Covalent adduction of human serum albumin by 4-hydroxy-2-nonenal: kinetic analysis of competing alkylation reactions. Biochemistry (Mosc) 2006;45:10521–10528. doi: 10.1021/bi060535q. [DOI] [PubMed] [Google Scholar]
- 41.Tjalkens RB, Luckey SW, Kroll DJ, Petersen DR. Alpha,beta-unsaturated aldehydes increase glutathione S-transferase mRNA and protein: correlation with activation of the antioxidant response element. Arch Biochem Biophys. 1998;359:42–50. doi: 10.1006/abbi.1998.0895. [DOI] [PubMed] [Google Scholar]
- 42.Dickinson DA, Iles KE, Watanabe N, Iwamoto T, Zhang H, Krzywanski DM, Forman HJ. 4-hydroxynonenal induces glutamate cysteine ligase through JNK in HBE1 cells. Free Radic Biol Med. 2002;33:974. doi: 10.1016/s0891-5849(02)00991-7. [DOI] [PubMed] [Google Scholar]
- 43.Awasthi YC, Sharma R, Sharma A, Yadav S, Singhal SS, Chaudhary P, Awasthi S. Self-regulatory role of 4-hydroxynonenal in signaling for stress-induced programmed cell death. Free Radic Biol Med. 2008;45:111–118. doi: 10.1016/j.freeradbiomed.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X, Khosravi-Far R, Chang HY, Baltimore D. Daxx, a novel Fas-binding protein that activates JNK and apoptosis. Cell. 1997;89:1067–1076. doi: 10.1016/s0092-8674(00)80294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Sharma R, Patrick B, Sharma A, Jeyabal PV, Reddy PM, Saini MK, Dwivedi S, Dhanani S, Ansari NH, Zimniak P, Awasthi S, Awasthi YC. Regulation of CD95 (Fas) expression and Fas-mediated apoptotic signaling in HLE B-3 cells by 4-hydroxynonenal. Biochemistry (Mosc) 2006;45:12253–12264. doi: 10.1021/bi060780+. [DOI] [PubMed] [Google Scholar]
- 46.Sharma R, Sharma A, Dwivedi S, Zimniak P, Awasthi S, Awasthi YC. 4-Hydroxynonenal self-limits fas-mediated DISC-independent apoptosis by promoting export of Daxx from the nucleus to the cytosol and its binding to Fas. Biochemistry (Mosc) 2008;47:143–156. doi: 10.1021/bi701559f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 48.Laurora S, Tamagno E, Briatore F, Bardini P, Pizzimenti S, Toaldo C, Reffo P, Costelli P, Dianzani MU, Danni O, Barrera G. 4-Hydroxynonenal modulation of p53 family gene expression in the SK-N-BE neuroblastoma cell line. Free Radic Biol Med. 2005;38:215–225. doi: 10.1016/j.freeradbiomed.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 49.Weigel AL, Handa JT, Hjelmeland LM. Microarray analysis of H2O2-, HNE-, or tBH-treated ARPE-19 cells. Free Radic Biol Med. 2002;33:1419–1432. doi: 10.1016/s0891-5849(02)01082-1. [DOI] [PubMed] [Google Scholar]
- 50.West JD, Marnett LJ. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chem Res Toxicol. 2006;19:173–194. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- 51.Kansanen E, Jyrkkanen HK, Levonen AL. Activation of stress signaling pathways by electrophilic oxidized and nitrated lipids. Free Radic Biol Med. 2012;52:973–982. doi: 10.1016/j.freeradbiomed.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 52.Schopfer FJ, Cipollina C, Freeman BA. Formation and signaling actions of electrophilic lipids. Chem Rev. 2011;111:5997–6021. doi: 10.1021/cr200131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McMahon M, Lamont DJ, Beattie KA, Hayes JD. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc Natl Acad Sci U S A. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ji C, Kozak KR, Marnett LJ. IkappaB kinase, a molecular target for inhibition by 4-hydroxy-2-nonenal. J Biol Chem. 2001;276:18223–18228. doi: 10.1074/jbc.M101266200. [DOI] [PubMed] [Google Scholar]
- 55.Luckey SW, Taylor M, Sampey BP, Scheinman RI, Petersen DR. 4-hydroxynonenal decreases interleukin-6 expression and protein production in primary rat Kupffer cells by inhibiting nuclear factor-kappaB activation. J Pharmacol Exp Ther. 2002;302:296–303. doi: 10.1124/jpet.102.033522. [DOI] [PubMed] [Google Scholar]
- 56.Aldini G, Dalle-Donne I, Facino RM, Milzani A, Carini M. Intervention strategies to inhibit protein carbonylation by lipoxidation-derived reactive carbonyls. Med Res Rev. 2007;27:817–868. doi: 10.1002/med.20073. [DOI] [PubMed] [Google Scholar]