Abstract
Brucella is an animal and human pathogen that expresses several virulence factors required for host cell invasion and intracellular survival. It produces LPS with unusually low toxicity, which hamper the detection of bacteria by the host immune system and thus provides resistance against intracellular antimicrobial mechanisms of the host. By chemical and spectroscopic methods we determined the structure of LPS core and of a non-repetitive oligosaccharide fragment at the reducing end of the O-specific polysaccharide. These data should be useful for understanding biological role of the Brucella LPS.
Keywords: Brucella, LPS, structure, NMR, MS
1. Introduction
Brucellae are Gram-negative coccobacilli and facultative intracellular pathogens. They cause animal diseases, transmitted to humans by contact with animals infected with B. abortus, B. melitensis or B. suis or their products, resulting in acute or chronic disease with non-specific symptoms, difficult to diagnose and treat1. Brucella is considered a potential bioterrorism agents. The bacteria are highly contagious and it was suggested that 10–100 sprayed bacteria could cause disease in humans.
The lipopolysaccharide (LPS) produced by Brucella posses unusual immunological properties such as low toxicity, intensively studied during last years (reviewed in 2). It is considered as a major virulence factor of Brucella3. Structure of the polysaccharide chain of the LPS was studied for B. abortus, B. melitensis, and B. suis4–6. In all cases it was a homopolymer of 4-formamido-4,6-dideoxy-D-mannose (N-formylperosamine, Rha4NFo), mostly 1-2-linked, comprising two antigenic determinants named A (from abortus) and M (from melitensis). Antigenic differences between strains were ascribed to the presence of some amount of 1-3-linkages between perosamine residues, leading to the appearance of M-epitopes. The highest concentration of M-epitope was found in the O-chain of B. melitensis strain 16M7. It was proposed that in this strain alternating residues of 1-2- and 1-3-linked Rha4NFo form a pentasaccharide repeating units containing one α-1,3-linked and four α-1,2-linked monosaccharides4. The structure of Brucella lipid A has been published8, the structure of the core part of the LPS has not been reported. Since biological studies require structural data for meaningful explanation of observations, we undertook the investigation of detailed structure of B, abortus, B. melitensis and B. suis LPSs.
2. Results
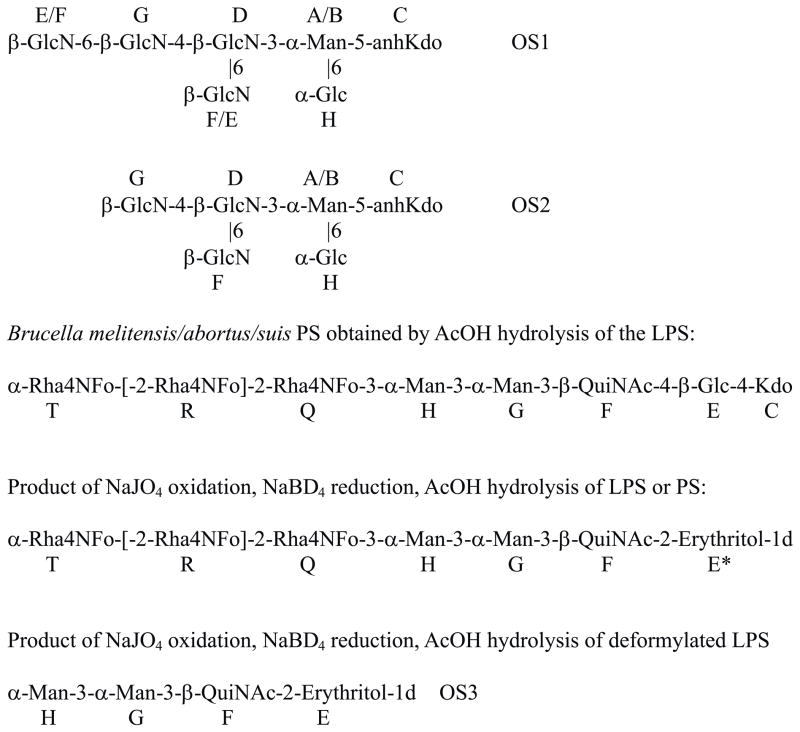
Mild acid hydrolysis of purified B. suis LPS produced a polysaccharide and minor amounts of oligosaccharide material. Oligosaccharides, presumably originating from LPS core, were contaminated with agar fragments and cyclic glucan. They were positively charged and purified by anion (eluted with solvent front) and then cation-exchange chromatography on Hitrap Q and S columns, respectively, in NaCl gradient, to give two fractions, OS1 and OS2. NMR study of the product OS1 (Table 1) revealed an unusual structure containing four β-GlcN residues with (Fig. 1). Determination of the configuration of the monosaccharides in pyranose form was based on vicinal proton coupling constants, as shown on Fig. 1. Presence of aminogroups was deduced from 13C signal position (Table 1). Kdo was tentatively identified based on the presence of several variants of reducing-end component with a CH2-group, which is characteristic for anhydro-Kdo forms produced by mild acid hydrolysis of LPS, and confirmed by mass spectral data. Substitution of Kdo at O-5 is given tentatively as the most common in bacterial LPS. All GlcN residues had unsubstituted aminogroups, resulting in high-field position of their H-2 signals (Table 1), which agreed with the MS data. OS1 was a mixture of two compounds, differing by the presence of Glc H. Due to the overlap of H/C-6 signals of GlcN residues G and D it was not possible to distinguish between residues E and F (Table 1), which however did not influence established structure. OS2 had the same structure as OS1 but without GlcN at G6. ESI mass spectral data were in agreement with the proposed structure: negative mode spectrum of the OS1 showed signals at m/z 1026.0 (no Glc H), 1188.0 (with Glc H), positive mode m/z 1028.7, 1191.0; calculated Hex1HexN4anhKdo1 = 1026.8 Da; OS2 produced ions 161 Da lower due to the loss of GlcN.
Table 1.
NMR data for OS1.
| 1 | 2 | 3 | 4 | 5 | 6 | ||
|---|---|---|---|---|---|---|---|
| α-Man A | H | 5.40 | 4.20 | 4.00 | 3.94 | 3.84 | 3.74; 4.03 |
| C | 101.4 | 69.1 | 79.8 | 65.6 | 73.0 | 66.5 | |
| α-Man B | H | 5.19 | 4.16 | 4.04 | 3.94 | 3.78 | 3.74; 4.03 |
| C | 99.0 | 69.1 | 80.0 | 65.6 | 72.9 | 66.5 | |
| α-Man A | H | 5.40 | 4.20 | 4.00 | 3.82 | 3.68 | 3.77; 3.88 |
| no H | C | 101.4 | 69.1 | 79.8 | 65.8 | 74.6 | 61.7 |
| α-Man B | H | 5.19 | 4.16 | 4.04 | 3.64 | 3.63 | 3.77; 3.88 |
| no H | C | 99.0 | 69.1 | 80.0 | 74.6 | 74.6 | 61.7 |
| α-Glc H | H | 4.98 | 3.57 | 3.74 | 3.43 | 3.73 | 3.76; 3.87 |
| C | 98.9 | 72.5 | 74.2 | 70.6 | 72.9 | 61.8 | |
| α-GlcN D | H | 4.88 | 3.18 | 3.87 | 3.87 | 3.95 | 3.98; 4.22 |
| C | 98.3 | 56.4 | 72.1 | 79.6 | 74.7 | 69.8 | |
| α-GlcN E | H | 4.82 | 3.11 | 3.67 | 3.48 | 3.52 | 3.77; 3.95 |
| C | 100.6 | 56.9 | 73.2 | 70.7 | 77.3 | 61.5 | |
| α-GlcN F | H | 4.78 | 3.08 | 3.69 | 3.48 | 3.52 | 3.78; 3.94 |
| C | 100.4 | 56.9 | 73.2 | 70.7 | 77.3 | 61.5 | |
| α-GlcN G | H | 4.76 | 3.07 | 3.65 | 3.55 | 3.74 | 4.27; 3.96 |
| C | 100.4 | 56.9 | 73.3 | 70.7 | 75.8 | 69.7 |
Fig. 1.
Structure of the compounds prepared from Brucella LPS. Polysaccharide includes additionally mannose residue M at undefined position. 3-Substituted Rha4NFo residues in the O-chain are not shown.
To confirm the presence of GlcN with free aminogroups LPSs were deaminated with NaNO2-AcOH, reduced and acetylated. GC-MS analysis of the products identified 2,5-anhydromannitol, expected product of this treatment, in all LPSs. GC analysis of alditol acetates, after acid hydrolysis of LPS, showed small amounts of GlcN because it did not undergo hydrolysis without an N-acyl substituent.
The presence of Kdo indicated that OS1 and OS2 were part of LPS-like molecule, presumably representing LPS core. Mass spectra of the oligosaccharides isolated from B. abortus and B. melitensis contained the same ions as described for B. suis OS1, suggesting NMR analysis of B. abortus and B. melitensis oligosaccharides was not performed because of insufficient amounts of their LPSs.
GC analyses of alditol acetates of B. abortus, B. melitensis and B. suis O-chain polysaccharides detected of mannose, glucose and quinovosamine in similar ratios, and perosamine. Quantification of Rha4N by GC produces inadequately low results due to its decomposition during acid hydrolysis.
Methylation analysis (Ciucanu-Kerek procedure9 of the polysaccharide led to the identification of 3-substituted mannose, 3-substituted quinovosamine and 4-substituted glucose.
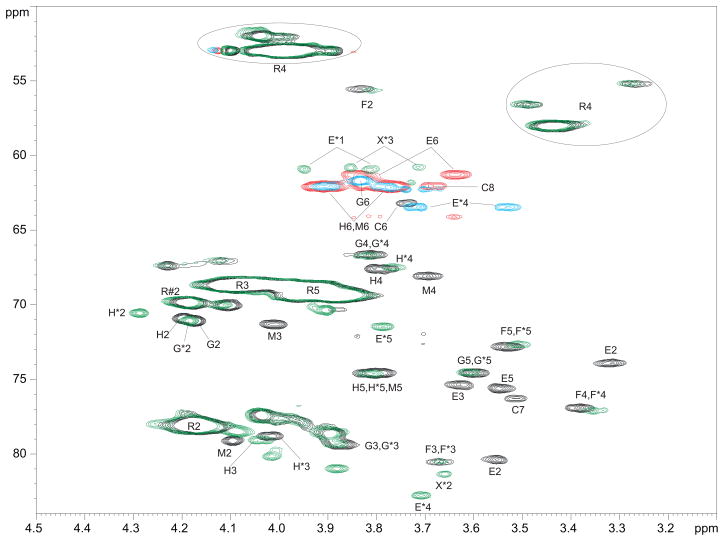
2D NMR spectra of the polysaccharides were complicated and similar in all strains. Assignment of the spectral signals was performed by standard methotds and is shown on Fig. 2 and in Tables 2 and 3. Main difference between strains was in the intensity of the signals of perosamine (Rha4N), which reflected differences in O-chain length. Minor signals were the same in all studied strains, suggesting that they represented components of the linkage region between polysaccharide and other parts of LPS or the non-reducing end of the polysaccharide chain.
Fig. 2.
Fragment of 1H-13C HSQC NMR spectra of B. suis polysaccharide (CH - black, CH2 - red) and NaIO4 oxidized, NaBD4-reduced, AcOH hydrolyzed B. suis polysaccharide (CH - green, CH2 - blue). Oxidized product signals are marked with *; R is 2- or 3-substituted Rha4NFo; R#2 is the signal of H/C-2 of 3-substituted Rha4NFo. Note the disappearance of units C, E, and M signals after the oxidation. Signals of mannose H shift probably due to the removal of the residue M, which was close to H. Hydroxymethyl groups E*1 and X*3 appear as CH because of deuterium substitution. R4 appears at ~3.4 and 4.0 ppm due to formyl group E/Z tautomerism.
Table 2.
NMR data for 2,7-anhydro-Kdo-furanoside from Brucella LPS O-chain and model compound10.
| H/C-1 | H/C-2 | H/C-3 | H/C-4 | H/C-5 | H/C-6 | H/C-7 | H/C-8 | |
|---|---|---|---|---|---|---|---|---|
| anh-Kdo | 2.27; 2.69 | 4.76 | 4.60 | 3.74 | 3.51 | 3.68; 3.81 | ||
| 42.3 | 77.7 | 84.7 | 63.2 | 76.2 | 62.0 | |||
| model | 2.11; 2.67 | 4.61 | 4.35 | 3.71 | 3.52 | 3.66; 3.80 | ||
| 176.0 | 106.1 | 44.2 | 69.9 | 86.4 | 63.3 | 76.3 | 62.1 |
Table 3.
NMR data for non-Rha4N components of Brucella polysaccharides and OS3.
| Unit | H/C 1 | H/C 2 | H/C 3 | H/C 4 | H/C 5 | H/C 6 |
|---|---|---|---|---|---|---|
| PS | ||||||
| Glc E | 4.54–4.62 | 3.31 | 3.63 | 3.55 | 3.54 | 3.64; 3.84 |
| 102.4 | 74.0 | 75.3 | 80.5 | 75.6 | 61.4 | |
| QuiNAc F | 4.62 | 3.80 | 3.65 | 3.35 | 3.53 | 1.31 |
| 102.4 | 55.7 | 80.6 | 77.2 | 72.8 | 17.7 | |
| Man G | 5.25 | 4.18 | 3.88 | 3.81 | 3.61 | 3.83; 3.83 |
| 101.9 | 71.2 | 79.4 | 66.7 | 74.6 | 61.7 | |
| Man H | 5.11 | 4.28 | 4.02 | 3.77 | 3.82 | 3.77; 3.89 |
| 103.6 | 70.8 | 78.8 | 67.6 | 74.6 | 62.0 | |
| Man M | 5.25 | 4.12 | 4.03 | 3.70 | 3.79 | 3.78; 3.92 |
| 101.9 | 79.6 | 71.3 | 68.1 | 74.5 | 62.0 | |
| OS3 | ||||||
| QuiNAc F | 4.61 | 3.80 | 3.66 | 3.34 | 3.50 | 1.31 |
| 102.6 | 55.6 | 80.7 | 77.1 | 72.7 | 17.7 | |
| Man G | 5.25 | 4.17 | 3.88 | 3.81 | 3.60 | 3.83; 3.83 |
| 102.1 | 71.1 | 79.2 | 66.7 | 74.6 | 61.8 | |
| Man H | 5.11 | 4.07 | 3.88 | 3.65 | 3.77 | 3.76; 3.90 |
| 103.4 | 71.2 | 71.5 | 68.0 | 74.5 | 62.3 | |
| Ery-ol | 3.72 | 3.70 | 3.78 | 3.52; 3.71 | ||
| 62.0 | 82.9 | 71.5 | 63.5 |
Reducing-end sugar of the polysaccharide chain was identified as Kdo, presented in several anhydro-variants. No umnodified Kdo-pyranose was detected. Major form of Kdo was 2,7-anhydro-α-furanose (2,7-anhydro-Kdo), a product of acid induced Kdo trasformation10. Kdo was substituted at O-4 with β-Glc E. Substitution position of Kdo was found on the variant with 2,7-anhydro-Kdo, where NOE from Glc H-1 to Kdo H-3,4 were observed. Substitution at O-4 caused large downfield shift of 2,7-anhydro-Kdo C-4 signal (Table 2). Several variants of Glc E were present due to its attachment to different Kdo variants. All other sugars were not sensitive to multiple Kdo forms and showed only one spin system each. Signals of QuiNAc usually appear at very low intensity, in some samples no correlations from its H-1 to H-2,3,4 was visible. However, H-1:H-6 correlation was always strong, and the whole QuiNAc spin-system could be identified by strong TOCSY from H-6 to all ring protons. β-Glc E was substituted at O-4 with β-QuiNAc F, which carried two mannose sequence (Fig. 1). Mannose H was substituted at position 3, as followed from downfield shift of C-3 signal and results of periodate oxidation (see below).
O-Chain polysaccharide contained another mannose residue M substituted at position 2, as deduced from the observation of a NOE correlation from Rha4NFo to its H-2, and position of 13C NMR signals with downfield shift of C-2 (Table 3, Fig. 2). Methylation analysis shows very small amount of 2-substituted mannose as compared to 3-substituted one. A NOE correlation from H-1 of Man M to H-3 of the Man H was always observed, which remained without explanation, since periodate oxidation removed Man M but Man H remained substituted at O-3 as follows from the position of its 13C signals.
LPS and AcOH-released polysaccharide were oxidized with NaIO4, reduced with NaBD4, hydrolyzed with 1% AcOH, and purified by gel-chromatography on Sephadex G50. This procedure resulted in oxidized polysaccharides, but no oligosaccharides was isolated. The structure of oxidized polymers was analyzed by NMR (Table 2, Fig. 1,2). It was found that Man M, Glc E and Kdo disappeared. Glc E upon oxidation-reduction was converted into 1d-erythritol (E). Signals of Rha4NFo have not changed after oxidation (Fig. 1). Several low-field signals appeared in HSQC spectrum of the oxidized polymer (Fig. 2) and remained without explanation. Spectra of different samples of oxidized polymers contained low intensity signals of 3d-glyceraldehyde (marked by X on Fig. 2), possibly produced by oxidation of PerN with lost formyl group on N-4. These signals were absent in the spectra of some samples. Formyl group is not completely stable to acid hydrolysis and might be partially lost during LPS hydrolysis or hydrolysis of oxidized material. The fact that mannose M was lost without influence to the presence of other components showed that it was a part of a side-chain or non-reducing end fragment at the PS chain, which was oxidized so that no oligosaccharides was produced.
Oxidation of deformylated PS or LPS produced an oligosaccharide OS3, consisting of two mannose residues, QuiNAc and erythritol (NMR data in Table 3). Structure was confirmed by ESI MS: observed peaks in positive mode m/z 635.4, negative mode m/z 633.6, calculated mass 634.3 Da. Formation of this compound proved that Man H was substituted by 2-substituted Rha4NFo residue, which became susceptible to oxidation after the removal of N-formyl group.
Polysaccharide chain could be linked to lipid through OS1/2. If it was linked to GlcN with free aminogroup, polysaccharide should be released by deamination of the LPS. This experiment was performed but produced no water-soluble polysaccharide (LPS was removed by ultracentrifugation, supernate liquid contained no polysaccharide). Deamination of N-deformylated LPS also gave no oligosaccharides, only small molecular mass compounds could be isolated by Sephadex G-15 chromatography. H-G-F-E-C fragment was not found among soluble compounds. Thus GlcN residues of the OS1 and OS2 were not the attachment site of the O-chain.
Complete deacylation of the LPS with 4M KOH gave polysaccharide which had no visible signals of chain ends in NMR spectra. This experiment did not produce any information.
3. Discussion
Our results bring important new data regarding the structure of the Brucella LPS. Brucella is related to Ochrobactrum, and it was suggested that they should have similar LPS architecture. Core structure was published for O. anthropi11, which indeed contained a fragment β-GlcN-3-α-Man-5-α-Kdo, also identified in the core of Brucella described here (OS1 and 2).
The presence of the core with several positively charged aminogroups may be responsible for preventing of killing of Brucella by cationic peptides and recognition by components of the innate immune system. Mutation affecting core structure was shown to make bacteria sensitive to polymixin B and colistin12. Mutation was done on the gene encoding mannosyltransferase and did not affect O-chain, however it was not known which mannose was removed. Based on the results described here, one may conclude that it was the mannose residue in the OS1/2. Such mutation would abolish the attachment of all GlcN and remove positive charge from the core, changing bacterial susceptibility to cationic peptides, and not affecting the attachment of the O-chain.
Position of attachment of the polysaccharide to the core has not been determined directly, but the number of possible attachment sites was reduced. We have shown that it was not linked to glucosamine components of the core. Mutation studies12 indicate that it is also not linked to core glucose and mannose, thus leaving Kdo as the only possible attachment site. Another possibility however exists that the polysaccharide is linked to a different lipid, which could be the reason of detection of larger than expected amount of straight chain fatty acids in the LPS preparation, and a low yield of lipid A8.
Polysaccharides with monosaccharide repeating units are usually polymerized by a processive glycosyl transfer mechanism, i.e. sequential addition of monosaccharides to the non-reducing end of the nascent chain. Structures of this type may contain specific non-repetitive fragments at the reducing end, biosynthetically different from the core or the O-chain fragments, like for example in Bordetella 13. The sequence -3-α-Man-3-α-Man-3-β-QuiNAc-4-β-Glc-4-Kdo, identified in this work, represents specific reducing end fragment of the O-chain.
4. Experimental part
4.1. Source of LPS
In this work LPS of B. abortus biotype 4, B. suis strain 4, and B. melitensis strain 3, used by Dr. Perry for his study of the O-chain structure6, were analyzed. All LPS contained some irrelevant lipid, which was removed by MeOH wash.
4.2. NMR spectroscopy
NMR experiments were carried out on a Varian INOVA 500 and 600 MHz (1H) spectrometer with 3 mm gradient probe at 25 °C with acetone internal reference (2.23 ppm for 1H and 31.45 ppm for 13C) using standard pulse sequences gCOSY, TOCSY (mixing time 120 ms), NOESY (for polymers) or ROESY (for oligosaccharides) (mixing time 300 ms), gHSQC, gHMBC (100 ms long range transfer delay), and gHSQCTOCSY (mixing time 80 ms, suppress direct correlations). AQ time was kept at 0.8–1 sec for H-H correlations and 0.25 sec for HSQC, 512 increments was acquired for t1.
Assignment of spectra was performed using Topspin 2 (Bruker Biospin) program for spectra visualization and overlap. Monosaccharides were identified by COSY, TOCSY and NOESY cross peak patterns and 13C NMR chemical shifts. Aminogroup location was concluded from high field signal position of aminated carbons (CH at 45–60 ppm). Connections between monosaccharides were determined from transglycosidic NOE and HMBC correlations.
4.3. Mass spectrometry
Capillary electrophoresis-mass spectrometry (CE-MS) analysis was performed using a 4000 Q-Trap mass spectrometer (Applied Biosystems/Sciex, Concord, Ontario, Canada) via a CE-MS interface with 90 cm length of bare fused-silica capillary using 15 mM ammonium acetate in deionized water, pH 7.0. A sheath solution (isopropanol-methanol, 2:1) was delivered at a flow rate of 1.5 μL/min. The orifice voltage was set at −110 V. For MS/MS analysis, fragment ions formed by collision activation of selected precursor ions with nitrogen in the RF-only quadrupole collision cell and recorded using time-of-fight mass analyser. For pseudo MS/MS/MS analysis, the precursor ions were generated with an orifice voltage of +120 V and mass spectra were acquired with nitrogen in the RF-only quadrupole collision cell.
GC-MS was carried out on Varian Saturn 2000 electron impact ion-trap instrument.
4.4. N-Deacylation
Preparation of the completely deacylated LPS. LPS samples (80 mg each) in polypropylene vials were dissolved in 4 M NaOH (4 ml each), kept overnight at 120° C, and neutralized with 2 M HCl. Precipitated material was removed by centrifugation and deacylated material isolated by gel chromatography on Biogel P10 column (2.5×60 cm). The fractions were analysed by NMR.
For removal of formyl groups 0.5 M NaOH (100 °C, 1 h) was used with the same work-up procedure.
4.5. Periodate oxidation
Polysaccharide or LPS (10–50 mg) was dissolved in 2–5 ml of water, NaIO4 was added to 0.1 M concentration, solution kept for 2 days, treated with 0.2 mL of ethyleneglycol, reduced with excess of NaBD4, desalted on Sephadex G-15 column, hydrolyzed with 2% AcOH (100 °C, 2 h), products separated on Sephadex G-50 column, analyzed by NMR.
4.6. Purification of the core oligosaccharides
Oligosaccharide fraction isolated by Sephadex G50 column chromatography from LPS hydrolyzate (2% AcOH, 100 °C, 3 h) was separated by anion exchange chromatography on Hitrap Q column (5 mL size, Amersham) in a linear gradient of NaCl - (0–1 M, 1 h). All core oligosaccharides were eluted with solvent front and reseparated on cation-exchange Hitrap S column (5 mL size, Amersham) in a linear gradient of NaCl - (0–1 M, 1 h). Carbohydrate containing fractions were detected by charring the spots from each fraction on TLC plate after dipping in 5% H2SO4 in EtOH. Desalting was performed on Sephadex G15 prior to analysis by NMR.
Brucella LPS protects bacteria from the host immune system and has unusual biological properties
We have determined previously unknown structure of the core of Brucella LPS.
Polysaccharide has specific sequence at the reducing end:
[-2-Rha4NFo]n-3-α-Man-3-α-Man-3-β-QuiNAc-4-β-Glc-4-Kdo
Acknowledgments
Authors are greatly indebted to Dr. M.B. Perry (NRC Canada) for Brucella LPS samples. This work was partially supported by the intramural programs of NICHD, National Institutes of Health, Bethesda, MD.
Abbreviations
- Rha4NFo
4-formamido-4,6-dideoxy-D-mannose (N-formylperosamine)
- Rha4N
4-amino-4,6-dideoxy-D-mannose (perosamine)
- QuiN
4-amino-4,6-dideoxy-D-glucose (quinovosamine)
- LPS
lipopolysaccharide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Franco MP, Mulder M, Smits HL. Lancet Infect Dis. 2007;7:775–786. doi: 10.1016/S1473-3099(07)70286-4. [DOI] [PubMed] [Google Scholar]
- 2.Cardoso PG, Macedo GC, Azevedo V, Oliveira SC. Microb Cell Fact. 2006;5:13. doi: 10.1186/1475-2859-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lapaque N, Moriyon I, Moreno E, Gorvel JP. Curr Opin Microbiol. 2005;8:60–66. doi: 10.1016/j.mib.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Bundle DR, Cherwonogrodzky JW, Perry MB. Biochemistry. 1987;26:8717–8726. doi: 10.1021/bi00400a034. [DOI] [PubMed] [Google Scholar]
- 5.Bundle DR, Cherwonogrodzky JW, Perry MB. FEBS Lett. 1987;216:261–264. doi: 10.1016/0014-5793(87)80702-0. [DOI] [PubMed] [Google Scholar]
- 6.Meikle PJ, Perry MB, Cherwonogrodzky JW, Bundle DR. Infect Immun. 1989;57:2820–2828. doi: 10.1128/iai.57.9.2820-2828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cherwonogrodzky JW, Perry MB, Bundle DR. Can J Microbiol. 1987;33:979–981. doi: 10.1139/m87-172. [DOI] [PubMed] [Google Scholar]
- 8.Qureshi N, Takayama K, Seydel U, Wang R, Cotter RJ, Agrawal PK, Bush CA, Kurtz R, Berman DT. J Endotoxin Res. 1994;1:137–148. [Google Scholar]
- 9.Ciucanu I, Kerek F. Carbohydr Res. 1984;131:209–217. [Google Scholar]
- 10.McNicholas PA, Batley M, Redmond JW. Carbohydr Res. 1987;165:17–22. doi: 10.1016/0008-6215(86)85041-8. [DOI] [PubMed] [Google Scholar]
- 11.Velasco J, Moll H, Knirel YA, Sinnwell V, Moriyon I, Zahringer U. Carbohydr Res. 1998;306:283–290. doi: 10.1016/s0008-6215(97)10029-5. [DOI] [PubMed] [Google Scholar]
- 12.Conde-Alvarez R, rce-Gorvel V, Iriarte M, Mancek-Keber M, Barquero-Calvo E, Palacios-Chaves L, Chacon-Diaz C, Chaves-Olarte E, Martirosyan A, von BK, Grillo MJ, Jerala R, Brandenburg K, Llobet E, Bengoechea JA, Moreno E, Moriyon I, Gorvel JP. PLoS Pathog. 2012;8:e1002675. doi: 10.1371/journal.ppat.1002675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Preston A, Petersen BO, Duus JO, Kubler-Kielb J, Ben-Menachem G, Li J, Vinogradov E. J Biol Chem. 2006;281:18135–18144. doi: 10.1074/jbc.M513904200. [DOI] [PubMed] [Google Scholar]