Abstract
Proteins of the AAA (ATPases Associated with various cellular Activities) often have complex modes of regulation due to their central position in important cellular processes. P60 katanin, a AAA protein that severs and depolymerizes microtubules, is subject to multiple modes of regulation including a phosphorylation in the N-terminal domain involved in mitotic control of severing. Phosphorylation decreases severing activity in Xenopus egg extracts and is involved in controlling spindle length. Here we show that the evolutionarily divergent N-terminal domains of p60 have maintained hotspots of mitotic kinase regulation. By reconstituting in vitro severing reactions, we show that phosphomimetic modification at amino acid position 131 in X. laevis p60 decreases severing and microtubule stimulated ATPase activity without affecting the binding affinity of p60 for microtubules. At high concentrations of the phosphomimetic mutant p60, near wild-type levels of activity could be observed, indicating a more switch-like threshold of activity that is controlled by regulating oligomerization on the microtubule. This provides a cellular mechanism for high local concentrations of p60, like that found on spindle poles, to maintain severing activity while most of the protein is inhibited. Overall, we have shown that the modular domain architecture of AAA proteins allows for precise control of cellular activities with simple modifications.
Keywords: AAA ATPase, microtubule severing, phosphorylation, local concentration, conservation of regulation
Introduction
Members of the AAA ATPase (ATPases Associated with various cellular Activities) family of proteins function in a diverse array of cellular processes including protein degradation, vesicle trafficking, protein quality control and cell division1,2,3,4. Typically, the highly conserved C-terminal catalytic domain is regulated by the N-terminal region, which is highly divergent and also mediates interactions with adaptor proteins that facilitate sub-cellular targeting or otherwise directly regulate AAA protein function3,4. For example, two distinct cellular activities of the AAA protein p97/VCP are achieved by competitive binding of two different sets of adaptor proteins as well as a host of phosphorylations and acetylations at its N-terminus5,6,7. However, how the majority of AAA proteins are mechanistically regulated remains unknown.
Katanin is a heterodimeric microtubule severing AAA ATPase composed of a catalytic p60 subunit containing the AAA domain and a targeting subunit p808. Like many AAA proteins, active katanin functions as a hexamer, though the exact mechanism of its microtubule severing activity is unclear9. In vivo, katanin plays critical roles in meiosis10, cilia and flagella assembly11,12,13,14,15, plant cell division16,17,18, and has also been suggested to generate non-centrosomal microtubule arrays in neurons19.
Recently, we demonstrated another interesting role for katanin in governing meiotic spindle length differences between two closely related species of frogs, Xenopus laevis and Xenopus tropicalis20. We found that suppression of microtubule severing by katanin is responsible in large part for the longer spindles assembled in X. laevis egg extracts relative to X. tropicalis. This difference in activity was not caused by differences in protein levels, but rather by the presence of an inhibitory Aurora B phosphorylation site at Serine 131 in the N-terminus of X. laevis p60 katanin, which is absent from X. tropicalis p6020. Addition of a recombinant mutant of X. laevis p60 katanin with Serine 131 changed to Alanine led to rapid microtubule severing in metaphase-arrested egg extracts, while a phosphomimetic mutant displayed reduced activity similar to the wild-type p6020. Here we investigate the biochemical basis of inhibitory phosphorylation at S131 by comparing the activities of wild-type and phosphomimetic mutants of p60 in vitro with pure microtubules. We show that phosphorylation of the N-terminal domain represents a novel mode of AAA ATPase regulation that may provide general insights into the mechanism and regulation of AAA proteins and microtubule severing proteins in particular. Our results suggest a means for the cell to tune the activity of a AAA protein as a function of its local concentration.
Results
A phosphomimetic mutant p60S131E displays concentration dependent inhibition of severing in vitro
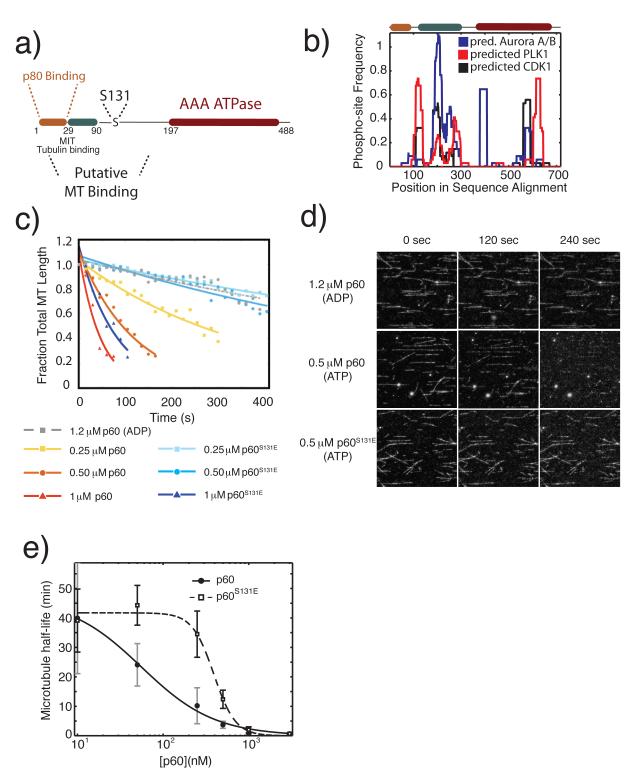
We previously identified Serine 131 as a site of negative regulation in X. laevis p60 katanin. Missing from X. tropicalis, this site is conserved among many vertebrate species but absent from D. melanogaster, C. elegans and S. purpuratus (sea urchin), indicating that regulatory control at this site may only be necessary under certain physiological contexts or that other selective pressures have cause the regulatory sites in this domain to be shifted within the primary sequence20. Serine 131 lies outside any well-defined protein interaction site, but within a large putative microtubule-binding domain predicted to be natively unstructured (Fig. 1a). To determine whether this domain has maintained regulatory potential despite only partial conservation of the mapped phosphorylation site and whether the phosphorylation of Serine 131 is part of larger regulatory network within the N-terminus of p60 we developed a multiple sequence alignment based metric called the domain phospho-regulatory potential (Fig. 1b).
Figure 1. A phosphomimetic mutation in an N-terminal regulatory domain of p60 katanin directly inhibits microtubule severing.
a) Schematic domain structure of p60 katanin and the position of Serine 13124,25. b) The predicted domain regulatory potential for three mitotic kinases. Frequency of phospho-sites is calculated as the fraction of predicted phospho-sites within a sliding window of 30 amino acids across all sequences in the multiple sequence alignment. Predicted phospho-sites for CDK1, PLK1 and Aurora A and B combined were determined using the Group-based prediction system 2.1 online server26. Multiple sequence alignments were generated in Matlab (Mathworks) using the multialign function in the bioinformatics toolbox. c) Quantification of the time course of severing of taxol stabilized microtubules by purified X. laevis p60 katanin or p60S131E at indicated concentrations, measured in fraction of total microtubule length, and fit to an exponential decay model. Representative data from 3 independent experiments except 1.2 μM ADP, which was a single experiment to indicate the requirement for ATP. d) Images from the time lapse data quantified in c. Protein expression, purification, data collection, and processing were as previously described20. e) Plot of microtubule half-life relative to p60 concentration. Data were fitted to the 4 parameter logistic equation to indicate the variation in point of 50% maximal activity and identify the relative differences in Hill slope.
Using 34 p60 katanin and katanin-like protein sequences assembled from across the phylogenetic tree, frequency of predicted phospho-regulation was calculated as the integration of predicted phosphorylation sites across all sequences within a sliding window of 30 amino acids divided by the number of sequences. This method allows the identification of regions of regulation even if individual sites are poorly conserved. Analysis of three mitotic kinases, cyclin dependent kinase 1, polo-like kinase 1, and aurora kinases (predictions of Aurora A and Aurora B sites combined) suggests the N-terminal domain of p60 katanin is enriched in predicted regulatory sites. Serine 131 falls within a region where all three mitotic kinases have increased frequency of predicted phosphorylation sites suggesting complex phospho-regulation. While Serine 131 is weakly conserved across all phyla, essentially all katanin related proteins have some form of mitotic regulation within a 30 amino acid window around Serine 131 and most have a predicted aurora kinase site in this region. Other regions of highly predicted regulation occur near positions 100 and 600 in the multiple sequence alignment (Fig. 1b) just after the p80 binding region and within the ATPase domain, respectively. These results suggest that investigating the regulation within the N-terminal domain of p60 katanin should yield significant mechanistic insight.
The combination of sequence analysis and our previous work on phospho-regulation of p60 katanin in Xenopus egg extracts indicates the importance of phosphorylation at Serine 131 in the context of evolution and cellular function, but did not address the biochemical mechanism by which phosphorylation exerts control over p60 katanin’s activity. To determine whether phosphorylation directly affects the microtubule severing activity of p60 in vitro, we quantified microtubule severing rates in flow cells containing taxol-stabilized microtubules. At the lower concentrations used, 250 and 500 nM, p60S131E showed severely attenuated microtubule severing activity relative to wild-type p60, indicating that the reduced severing activity of p60S131E previously observed in Xenopus egg extracts reflects an intrinsic biochemical alteration in the protein itself and is not a secondary effect due to interaction with other proteins found in the egg extract. Whereas microtubules persisted with a fitted half life of 12.1±3.1 min (mean±SEM) in flow cells containing 500 nM p60 S131E, rapid severing occurred in the presence of the same concentration of wildtype p60, giving a half life of 3.72±1.7 min (Fig. 1c). Interestingly, however, at 1 μM p60WT and p60S131E both showed rapid rates of severing, with microtubule half-lives of 0.82±0.20 min and 2.04±1.04 min, respectively (Fig. 1c) and at 3 μM the severing rates were indistinguishable (0.66±0.06 and 0.71±0.24 for wild-type and p60 S131E, respectively). This restoration of wild-type levels of severing activity at higher concentrations suggests that the mutant is not misfolded, and that phosphorylation at Serine 131 does not block p60’s catalytic activity outright, but instead modulates a concentration-dependent activity of p60. A plot of fitted microtubule half-lives for both wild-type and p60 S131E over a range of concentrations explicitly displays this concentration dependent modulation (Fig. 1e). Whereas wild-type p60 severing activity increases with increasing concentration, the p60 S131E mutant shows virtually no severing activity below a threshold of 0.25 μM. Above this threshold, however, the mutant shows an abrupt, switch-like activation of severing activity that converges on wild-type levels at and above 1 μM.
P60 microtubule binding is unaffected by Ser131 phosphomimetic mutation
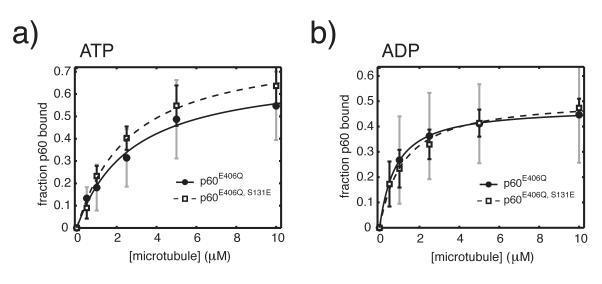
Because S131 falls within p60’s putative microtubule binding domain, modulation of p60’s affinity for microtubule binding was an obvious possible explanation for phospho-regulated inhibition of severing and could explain the concentration-dependent activity observed in severing assays. A decreased binding affinity would also predict that normal severing rates can be restored at sufficiently high p60 concentrations. To begin to address the specific underlying biochemical cause for the concentration-dependent severing behavior of p60S131E seen in flow cells, we introduced the well-characterized Walker B point mutation, E406Q, to generate ATP hydrolysis deficient wild type and phosphomimetic p60. We then compared the microtubule-binding ability of wild-type p60E406Q versus p60S131E, E406Q in defined nucleotide states. In both the ATP and ADP state, we observed that wild-type p60 E406Q and p60S131E, E406Q possessed similar microtubule binding affinities (Fig. 2). Estimated dissociation constants (KD) were 2.83±0.92 μM for wild-type p60E406Q (maximum likelihood estimation of mean±SD of fitted parameters based on Monte Carlo sampling and fitting 250 values within the experimental error) and 2.63±0.53 μM for p60S131E, E406Q in the ATP state, and 0.81±0.37 μM and 1.23±0.43 μM in the ADP state, respectively (Fig. 2). Monte Carlo sampling and fitting a large number of possible datasets within the experimental error allowed an examination of the distribution of possible fitted KD‘s, leading to the conclusion that a difference in microtubule binding affinity is not responsible for the large difference in activity between p60 and p60S131.
Figure 2. The affinity of p60 for microtubules is unchanged by a phosphomimetic mutation.
250 nM X. laevis p60 or p60S131E with either ATP (a) or ADP (b) was added to increasing concentrations of taxol-stabilized MTs from 0-10 μM in BRB80 with 10μM Taxol. Reactions were incubated at room temperature for 15 min, pelleted by centifugation and analyzed by SDS-PAGE. Binding data was fit to the quadratic binding equation as previously described20. Error in fitted parameters was estimated by monte carlo sampling within the experimentally determined error and fitting the resulting datasets27. This procedure generates hundreds of theoretical datasets that fit the measured data by randomly sampling a single value at each protein concentration that falls within the experimental error of that data point. The theoretical datasets are individually fitted normally. The maximum likelihood mean±SD was determined from 250 individual fits and provides the reported KD value and error. Plotted data is from two independent experiments with two separate preparations of each protein.
Microtubule-stimulated ATPase activity of p60S131E is decreased at low concentration
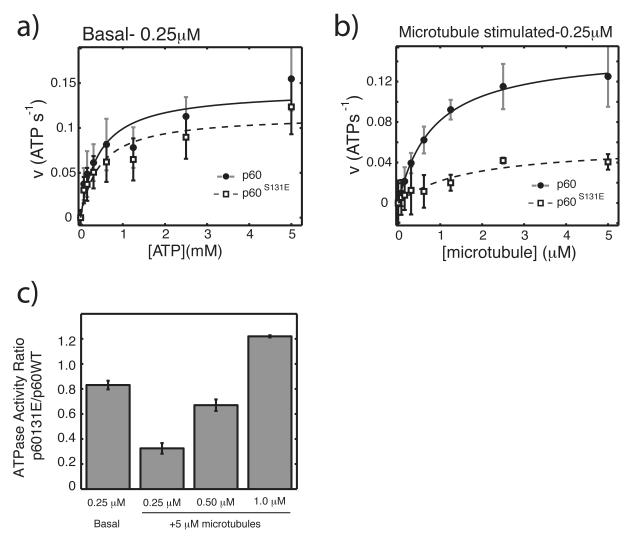
To determine whether the site of phospho-regulation affected the ATPase domain of p60, we analyzed both the basal and microtubule-stimulated ATPase activity of p60 and p60S131E under steady-state conditions using an enzyme coupled reaction (Fig. 3). Basal rates of ATP hydrolysis were determined for 250 nM wild type p60 and p60S131E with concentrations of ATP ranging from 0.075-5 mM. We found very similar rates of ATP hydrolysis with no significant difference in fitted kinetic parameters under these conditions. For wild-type p60, we measured a Vmax of 0.15±0.03 s−1 and a Km-ATP of 0.57±0.39 mM (maximum likelihood estimated mean±SD of fitted parameters is based on Monte Carlo sampling and fitting 200 values within the experimental error). Steady-state ATP hydrolysis parameters for p60S131E were very similar, with a Vmax of 0.12±0.03 s−1 and a Km-ATP of 0.62±0.56 mM (Fig. 3a) and in a similar range of other AAA ATPases 21,22. These nearly identical kinetic parameters confirm that the phospho-mimetic mutant is not severely disrupted at a structural level and suggest that the enzymatic machinery necessary to hydrolyze ATP is not the point of regulation.
Figure 3. Microtubule stimulated ATPase activity is inhibited by a phosphomimetic mutation.
a) Measurement of basal ATPase activity of 250 nM p60 and p60S131E with increasing ATP concentration. b) Measurement of ATPase activity of 250 nM p60 and p60S131E in response to increasing microtubule concentration and 1mM ATP, corrected for basal activity. ATPase activity was determined using an enzyme-coupled assay as previously described except that reactions were in 20 mM HEPES pH 7.7, 25 mM potassium glutamate, 10% glycerol, 0.02% Triton-X100, 10 mM MgCl228. Data was fit to the Michaelis-Menton equation and estimates of the error in fitted parameters was determined by monte carlo sampling of data within the experimental error and fitting each resulting dataset27. The maximum likelihood mean±SD was then determined from 200 individual fits within the experimental error. c) ATPase activity of p60S131E relative to wild-type at 1mM ATP and 5 μM microtubules with propogated error. Data for basal and 250nM are the same as in part a and b, respectively. All data is from at least 3 independent experiments with two separate preparations of each protein.
The N-terminal domain containing S131 is predicted to be important for interacting with microtubules. Since microtubules have been shown to stimulate the ATPase activity of p60 we tested whether the phospho-regulatory site could modulate this activity. The rates of microtubule stimulated ATP hydrolysis, corrected for basal activity, were compared between the two proteins in the presence of increasing microtubule concentrations up to 5 μM. Using p60s at nM, at which the severing activity of the p60S131E mutant is most different, wild-type p60 displayed the expected strong hyperbolic stimulation in response to increasing microtubule concentrations (VmaxMT of 0.16±0.02 s−1 and KmMT of 0.91±0.26 μM). P60S131E, however, showed very minimal stimulation (Vmax MT of 0.06±0.02 s−1 and KmMT of 2.0±1.45 μM) with a potentially increased KmMT though the distribution of possible values in this fitted parameter is large preventing a strong conclusion (Fig. 3b).
Since the severing activity of p60S131E was restored to near wild type level at higher concentrations we sought to determine whether the microtubule-stimulated ATPase activity of was also restored at higher p60 levels. We compared the ATPase activities of 500 nM and 1 μM p60 and p60S131E in the presence of 5 μM microtubules. At this concentration of microtubules, the proteins showed the greatest difference in ATPase activity, imposing the most stringent condition for recovering activity (Fig. 3b). Relative to wild-type activity, the low microtubule-stimulation of p60S131E increased with increasing p60 concentration. The relative activity of p60S131E was only 32.5± 4% of p60 wild type at 250 nM. Interestingly, at 1 μM we observed a complete restoration of microtubule stimulation with p60S131E being 122 ± 1% of wild type p60. An intermediate level of stimulation at 500 nM p60 S131E was found (67 ± 4% of wild type). Microtubule-stimulated ATPase activity of p60S131E therefore mirrors its increasing severing activity between 250 nM and 1 μM. Overall, these results indicate that phospho-regulation at Serine 131 specifically affects the ATPase cycle to control severing activity, not by rearranging the fundamental enzymatic machinery within a monomer, but rather by affecting a mechanism that can be overcome by increasing protein concentration.
Discussion
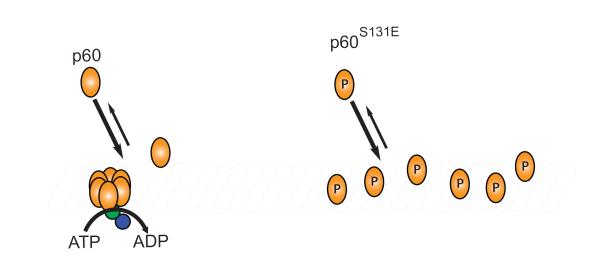
Given that basal ATP hydrolysis and microtubule binding are unaffected by the S131E mutation, we reasoned that the lack of microtubule-stimulated ATP hydrolysis must be due to either perturbed oligomerization or a disruption of an allosteric mechanism that normally stimulates ATPase activity upon microtubule binding. It is currently unknown whether ATPase stimulation occurs directly upon microtubule binding or indirectly as a consequence of microtubule stimulated oligomerization. Our result that both the severing activity and microtubule stimulated ATPase activity of p60S131E are restored to wild-type levels at high concentrations suggests that the modulating effect is associated with oligomerization, because sufficiently high concentrations of p60 would be expected to drive hexamerization and restore wild-type severing activity as observed. However, we have been unable to identify hexameric assemblies in solution using the Xenopus laevis p60 proteins (data not shown) and therefore suggest that transient oligomerization likely occurs only on the microtubule. This is similar to sea urchin p60 that can oligomerize in solution but its oligomerization is stimulated in the presence of microtubules9.
One attractive hypothesis that has been proposed is that p60 monomer binding to microtubules drives hexamerization passively, simply through induced proximity of monomers on the microtubule lattice9. By this model, a population of p60 monomers that previously existed in the three-dimensional volume of the cytosol is collapsed to effectively one-dimension on the microtubule lattice. In support of this model D. melanogaster p60, once bound to a microtubule rapidly diffuses along the lattice providing a mechanism to search the one-dimensional length and promote oligomerization23. Importantly, locking p60 into an ATP bound state prevented diffusion on the lattice23 and seems to indicate that ATP hydrolysis could be required to establish the diffusive state and again be required in a separate reaction for severing. The poor microtubule-stimulated ATPase activity we observed at low concentrations of p60S131E could reflect an important modulation in this process, resulting in less oligomerization and consequently, less stimulation of ATP hydrolysis (Fig. 4). In this model, regulation could occur either through decreasing the affinity of microtubule bound monomers for each other or slowing the diffusional search of p60 monomers on the microtubule. It seems clear, however, that the effect must occur after binding to the lattice, given the nearly identical microtubule-binding affinities we observed for wild type p60 and the S131E mutant (Fig. 2 and Fig. 4). In either case, increasing the local density of monomers could be sufficient to drive hexamerization. Consistent with this idea, our observation that p60S131E severing activity is restored to near wild-type levels at high concentrations could indicate that oligomerization can be restored if monomer density on the microtubule lattice is driven high enough.
Figure 4. P60 is regulated after monomer enrichment on the microtubule.
Binding to the microtubule is unaffected by modification at serine 131 but microtubule severing and microtubule stimulated ATPase activity is inhibited. The point of regulation by phosphophorylation at serine 131 must come after monomers of p60 are bound to the microtubule. We hypothesize that oligomerization is regulated to control levels of severing activity.
Our results reveal a novel, potentially general mode of AAA regulation. The concentration-dependent inhibition we observed for p60 could enable the cell to down-regulate the activity of bulk, diffuse protein while still allowing for unperturbed function at sites of local enrichment where its activity is absolutely necessary. For example, high local concentrations in the spindle might allow katanin to severe microtubules efficiently at spindle poles where it is enriched but prevent severing of microtubules elsewhere in the spindle. This may be important in length control of the spindle or for driving organization of the microtubule architecture. Regulation of this type might therefore effectively uncouple the activities of otherwise indistinct subcellular AAA populations as a function of local concentration. This is in contrast to the more simple case of a regulatory modification that simply “turns off” the protein by blocking its catalytic activity. Such a modification would, in the most basic case, result in a spatially uniform down-regulation of the entire population independent of local concentrations, and would therefore be impractical if specific local activities are necessary to tune functional outputs.
Our experiments here demonstrate the direct biochemical effect of a physiologically relevant regulatory phosphorylation event on p60 katanin. That a phosphorylation event distant from the AAA domain can inhibit substrate-stimulated ATPase activity may have implications for many hexameric AAA proteins, particularly the closely related severing proteins spastin and fidgetin. Additionally, the switch-like behavior observed over a range of concentrations of p60S131E suggests a way for the cell to target AAA activities exclusively to sites of local enrichment.
Highlights.
The N-terminal domain of p60 katanin is a hotspot for potential mitotic kinase regulation
Phosphomimetic p60 katanin displays lower activity with pure microtubules compared to wild-type
The phosphomimetic mutation reduces microtubule-stimulated ATPase activity
P60 katanin’s affinity for microtubules is unchanged by the phosphomimetic mutation
P60 katanin is regulated at the point of assembly on the microtubule
Acknowledgements
We thank Andy Martin and members of the Heald lab for helpful discussions, and Dr. Liam Holt, Dr. Magdalena Strzelecka, and Dr. Marina Ellefson for critical reading of the manuscript. R.H. is supported by NIH R01GM098766.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dougan DA, Mogk A, Zeth K, Turgay K, Bukau B. AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 2002;529:6–10. doi: 10.1016/s0014-5793(02)03179-4. [DOI] [PubMed] [Google Scholar]
- 2.Hanson PI, Whiteheart SW. AAA+ proteins: have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–29. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 3.White SR, Lauring B. AAA+ ATPases: achieving diversity of function with conserved machinery. Traffic. 2007;8:1657–67. doi: 10.1111/j.1600-0854.2007.00642.x. [DOI] [PubMed] [Google Scholar]
- 4.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 5.Mori-Konya C, Kato N, Maeda R, Yasuda K, Higashimae N, Noguchi M, Koike M, Kimura Y, Ohizumi H, Hori S, Kakizuka A. p97/valosin-containing protein (VCP) is highly modulated by phosphorylation and acetylation. Genes Cells. 2009;14:483–97. doi: 10.1111/j.1365-2443.2009.01286.x. [DOI] [PubMed] [Google Scholar]
- 6.Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–6. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- 7.Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J Cell Biol. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–29. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- 9.Hartman JJ, Vale RD. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 1999;286:782–5. doi: 10.1126/science.286.5440.782. [DOI] [PubMed] [Google Scholar]
- 10.Yang HY, McNally K, McNally FJ. MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C elegans. Dev Biol. 2003;260:245–59. doi: 10.1016/s0012-1606(03)00216-1. [DOI] [PubMed] [Google Scholar]
- 11.Dymek EE, Lefebvre PA, Smith EF. PF15p is the chlamydomonas homologue of the Katanin p80 subunit and is required for assembly of flagellar central microtubules. Eukaryot Cell. 2004;3:870–9. doi: 10.1128/EC.3.4.870-879.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dymek EE, Smith EF. PF19 encodes the catalytic subunit of katanin, p60, and is required for assembly of the flagellar central apparatus in Chlamydomonas. J Cell Sci. doi: 10.1242/jcs.096941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lohret TA, McNally FJ, Quarmby LM. A role for katanin-mediated axonemal severing during Chlamydomonas deflagellation. Mol Biol Cell. 1998;9:1195–207. doi: 10.1091/mbc.9.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rasi MQ, Parker JD, Feldman JL, Marshall WF, Quarmby LM. Katanin knockdown supports a role for microtubule severing in release of basal bodies before mitosis in Chlamydomonas. Mol Biol Cell. 2009;20:379–88. doi: 10.1091/mbc.E07-10-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma N, Bryant J, Wloga D, Donaldson R, Davis RC, Jerka-Dziadosz M, Gaertig J. Katanin regulates dynamics of microtubules and biogenesis of motile cilia. J Cell Biol. 2007;178:1065–79. doi: 10.1083/jcb.200704021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panteris E, Adamakis ID, Voulgari G, Papadopoulou G. A role for katanin in plant cell division: microtubule organization in dividing root cells of fra2 and lue1Arabidopsis thaliana mutants. Cytoskeleton (Hoboken) 68:401–13. doi: 10.1002/cm.20522. [DOI] [PubMed] [Google Scholar]
- 17.Stoppin-Mellet V, Gaillard J, Vantard M. Plant katanin, a microtubule severing protein. Cell Biol Int. 2003;27:279. doi: 10.1016/s1065-6995(02)00324-4. [DOI] [PubMed] [Google Scholar]
- 18.Uyttewaal M, Burian A, Alim K, Landrein B, Borowska-Wykret D, Dedieu A, Peaucelle A, Ludynia M, Traas J, Boudaoud A, Kwiatkowska D, Hamant O. Mechanical stress acts via katanin to amplify differences in growth rate between adjacent cells in Arabidopsis. Cell. 149:439–51. doi: 10.1016/j.cell.2012.02.048. [DOI] [PubMed] [Google Scholar]
- 19.Ahmad FJ, Yu W, McNally FJ, Baas PW. An essential role for katanin in severing microtubules in the neuron. J Cell Biol. 1999;145:305–15. doi: 10.1083/jcb.145.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loughlin R, Wilbur JD, McNally FJ, Nedelec FJ, Heald R. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell. 2011;147:1397–407. doi: 10.1016/j.cell.2011.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eckert T, Link S, Le DT, Sobczak JP, Gieseke A, Richter K, Woehlke G. Subunit Interactions and Cooperativity in the Microtubule-severing AAA ATPase Spastin. J Biol Chem. 2012 doi: 10.1074/jbc.M111.291898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maisel T, Joseph S, Mielke T, Burger J, Schwarzinger S, Meyer O. The CoxD Protein, a Novel AAA+ ATPase Involved in Metal Cluster Assembly: Hydrolysis of Nucleotide-Triphosphates and Oligomerization. PLoS One. 2012;7:e47424. doi: 10.1371/journal.pone.0047424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz-Valencia JD, Morelli MM, Bailey M, Zhang D, Sharp DJ, Ross JL. Drosophila katanin-60 depolymerizes and severs at microtubule defects. Biophys J. 2011;100:2440–9. doi: 10.1016/j.bpj.2011.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwaya N, Kuwahara Y, Fujiwara Y, Goda N, Tenno T, Akiyama K, Mase S, Tochio H, Ikegami T, Shirakawa M, Hiroaki H. A common substrate recognition mode conserved between katanin p60 and VPS4 governs microtubule severing and membrane skeleton reorganization. J Biol Chem. 2010;285:16822–9. doi: 10.1074/jbc.M110.108365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNally KP, Bazirgan OA, McNally FJ. Two domains of p80 katanin regulate microtubule severing and spindle pole targeting by p60 katanin. J Cell Sci. 2000;113(Pt 9):1623–33. doi: 10.1242/jcs.113.9.1623. [DOI] [PubMed] [Google Scholar]
- 26.Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics. 2008;7:1598–608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feller SE, Blaich CF. Error Estimates for Fitted Parameters: Application to HCl/DCl Vibrational–Rotational Spectroscopy. J Chem Ed. 2001;78:409–12. [Google Scholar]
- 28.Huang TG, Hackney DD. Drosophila kinesin minimal motor domain expressed in Escherichia coli. Purification and kinetic characterization. J Biol Chem. 1994;269:16493–501. [PubMed] [Google Scholar]