Abstract
Protective immunity against Salmonella infection is known to require CD4 Th1 cells and B cells, but the role of MHC class-I-restricted CD8 T cells is less clear. Previous studies have suggested that CD8 T cells participate in secondary, but not primary, bacterial clearance. However, these studies have used experimental models that are difficult to interpret and do not clearly isolate the role of MHC class-I-restricted CD8 T cells from other cell populations. Here, we examined the role of class-I-restricted T cells in protection against Salmonella infection using mice lacking all classical MHC class-Ia molecules, perforin, or granzyme B. Immunized KbDb-, perforin-, granzyme B-, or perforin/granzyme B-deficient mice were able to resolve secondary infection with virulent Salmonella, demonstrating that class-I-restricted CTLs are not required for acquired immunity. However, during primary infection with attenuated bacteria, bacterial clearance was delayed in each of these mouse strains when compared to Wild-type mice. Taken together, these data demonstrate that CD8 T cells are not required for acquired immunity to Salmonella, but can play a protective role in resolving primary infection with attenuated bacteria.
Keywords: Salmonella, Infection, CD8 T cells, Acquired immunity
1. Introduction
Salmonella enterica serovar Typhi is a facultative intracellular bacterial pathogen that causes human typhoid in parts of the developing world [1]. Other Salmonella serovars cause gastroenteritis or a more serious bacteremia that is often associated with immune deficiency [2–4]. Although there are two licensed typhoid vaccines, neither is highly effective or widely used in endemic areas [5,6]. Greater understanding of host immunity to Salmonella infection is required to assist the generation of new vaccines against typhoid and non-typhoidal Salmonellosis.
Although serovar Typhi does not infect mice, immunity to serovar Typhimurium infection can be studied using susceptible or resistant inbred mouse strains [7,8]. Mice expressing a functional Nramp gene are able to resist Salmonella infection, however, the study of cellular immunity in these strains is complicated by the fact that antibody is sufficient for protection [9,10]. In contrast, highly susceptible mouse strains expressing a non-functional Nramp allele succumb rapidly to primary infection with virulent Salmonella [9]. However, prior immunization with attenuated bacteria confers protective immunity that is highly dependent on both CD4 Th1 cells and antibody [11–14]. Thus, the role of T cells in protective immunity is usually studied in the context of secondary infection of vaccinated susceptible mice [15]. A complementary approach is to examine the ability of immune-deficient mice to resolve primary infection with highly attenuated Salmonella strains. Resolution of primary infection with attenuated bacteria has been shown to require a functional adaptive immune system [16]. In particular, MHC class-II-restricted CD4 Th1 cells play an essential protective role [17–19], while B cells are dispensable [20,21]. Taken together, the available evidence suggests that MHC class-II-restricted CD4 Th1 cells participate in both primary and secondary Salmonella clearance, while B cells are essential for protection against secondary infection.
Salmonella-specific CD8 T cells are also readily induced after infection of mice or humans, suggesting that this population of T cells may play a protective role [22]. Human volunteers vaccinated with live attenuated Salmonella have detectable CD8 responses in peripheral blood and produce IFN-γ upon restimulation [23–25]. Salmonella infection in mice also induces activation of CD8 T cells [26,27], although the kinetics of this response appear to be delayed when compared to Salmonella-specific CD4 responses [28,29]. However, it is not yet clear whether class-I-restricted CD8 T cells participate in protective immunity during Salmonella infection.
Several studies have used CD8 antibody depletion to demonstrate a modest protective role for CD8 T cells during secondary Salmonella infection of vaccinated susceptible mice [12–14,30]. However, interpretation of these studies is complicated by the fact that CD8 is expressed by other cell types, including γδ cells, MHC class-Ib restricted CD8 T cells, innate CD8 T cells and some dendritic cell subsets [31–34]. Therefore, it is not clear from these studies whether class-I-restricted CD8 T cells actually participate in immunity to Salmonella infection. Other studies have infected β2-microglobulin (β2μ)-deficient mice with attenuated Salmonella [17,35], and since β2μ forms part of cell surface MHC-class-I, assumed that any deficiency in bacterial clearance is due to a lack of CD8 T cells. Indeed, β2μ-deficient mice resolve primary infection with Salmonella [17], but display moderately enhanced susceptibility to secondary infection [35], confirming the experiments using antibody depletion. However, these experiments are also difficult to interpret since β2μ-deficient mice lack expression of non-classical MHC molecules and CD1 [36], and are also known to express free MHC class-Ia molecules in the absence of β2μ [37]. Thus, the role of class-I-restricted CD8 T cells in primary and secondary immunity to Salmonella remains unclear.
Activated CD8 T cells can differentiate into cytotoxic T lymphocytes (CTL), which lyse pathogen-infected target cells via granule exocytosis or programmed cell death via Fas-FasL interactions [38,39]. During granule-mediated lysis, a cytotoxic CD8 T cell contacts an infected target cell, induces degranulation, and releases perforin, a pore-forming molecule, and the protease enzymes granzyme A and B, into the infected cell. However, despite many studies examining the role of CD8 T cells, the role of perforin and granzyme-mediated lysis during Salmonella infection has not previously been examined.
In this current study we directly examined the role of class-I-restricted CD8 T cells and granule exocytosis during primary resolution of attenuated bacteria and in acquired protection against secondary virulent Salmonella infection. Surprisingly, we found no deficiency in secondary protection against Salmonella using mice lacking all classical MHC-class-Ia molecules, perforin, or granzyme B. In contrast, we detected a protective role for class-Ia-restricted CD8 T cells and granule exocytosis during the resolution of primary infection with attenuated Salmonella, but only at the late stages of bacterial clearance. Together, our data suggest that the primary role of Salmonella-specific class-I-restricted CD8 T cells is to restrict bacterial growth during primary bacterial clearance.
2. Materials and Methods
2.1. Mouse and bacterial strains
C57BL/6 were purchased from the National Cancer Institute (Frederick, MD) while perforin-deficient (C57BL/6-Prf1tm1Sdz) [40] and granzyme B-deficient (B6.129S2-Gzmbtm1Ley) mice were purchased from the Jackson Laboratory (Bar Harbor, ME). Perforin/granzyme B-deficient mice were generated in our laboratory by intercrossing strains. KbDb-deficient mice were obtained from Dr. Jameson, University of Minnesota. All mice were used at 6–12 weeks of age and were cared for in accordance with University of California Davis and University of Minnesota Research Animal Resource guidelines. Attenuated serovar Typhimurium strain BRD509 (aroA−D−), derived from virulent Salmonella SL1344 [41], was kindly provided by Dr. D. Xu, University of Glasgow, U.K.
2.2. Salmonella infection and bacterial counts
Salmonella (BRD509 and SL1344) were grown overnight in LB broth without shaking and diluted in PBS after estimation of bacterial concentration using a spectrophotometer. Mice were infected intravenously in the lateral tail vein with 5×105 BRD509, and monitored daily for signs of infection. In all experiments the actual bacterial dose administered was confirmed by plating serial dilutions of the original culture onto MacConkey agar plates. Primary infection with BRD509 was resolved around 35–42 days post infection and at this point no detectable bacteria are found in the spleen and liver. On day 60 post-infection with BRD509, mice that had resolved primary infection were infected intravenously with 1×103 SL1344 and monitored daily for signs of disease. Infected mice were determined to be moribund if they were unresponsive to gentle prodding. To determine bacterial colonization in vivo, spleens and livers from infected mice were homogenized in PBS and serial dilutions plated onto MacConkey agar plates. After overnight incubation at 37°C, bacterial numbers were counted and bacterial burdens calculated for each individual organ.
2.3. Statistical analysis
Statistical differences between groups of normally distributed data were examined using Prism (GraphPad Software, La Jolla, CA). For bacterial burdens, Log10 CFUs are normally distributed and were also compared using Prism. Data in each group were compared using an unpaired t test and were considered significantly different with a p value of <0.05.
3. Theory
The role of MHC class-I-restricted CD8 T cells during Salmonella infection is unclear. In this study, we sought to examine the role of class-Ia-restricted CD8 T cells and granule exocytosis during the primary and secondary Salmonella infection using various gene-deficient mice.
4. Results
4.1. MHC class-Ia-restricted CD8 T cells are not required to resist secondary Salmonella infection
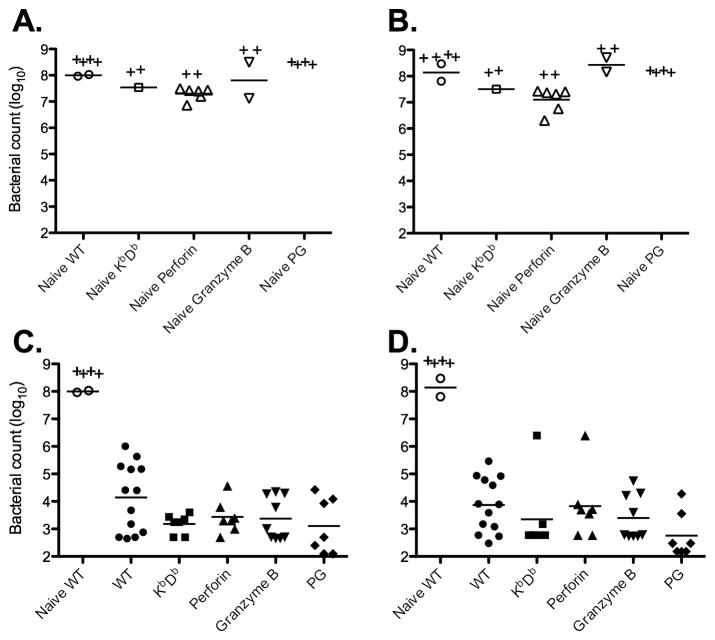
KbDb-deficient C57BL/6 mice lack expression of MHC class-Ia molecules, and have vastly reduced numbers of peripheral CD8 T cells [42]. However, expression of β2μ and class-Ib molecules is normal and these mice have functional NK cells and non-classically restricted CD8 T cells [42], thus allowing direct testing of the protective role of MHC class-Ia-restricted CD8 T cells during Salmonella infection. Previous studies have suggested a protective role for CD8 T cells only during secondary infection [12–14,30,35]. In order to confirm these observations, Wild-type or KbDb-deficient C57BL/6 mice were immunized with an attenuated strain of Salmonella and on day 60 subsequently challenged with a virulent strain of Salmonella, SL1344. Naïve KbDb-deficient mice had similar bacteria burdens to naïve Wild-type mice and some succumbed to death before day 6 (Fig 1A and 1B), demonstrating that they carry they are Nramp susceptible. In contrast, both Wild-type and KbDb-deficient mice survived secondary challenge and had lower bacterial loads than naïve mice (Fig. 1C and 1D), indicating the development of acquired immunity. Surprisingly, KbDb-deficient mice did not display any deficiency in bacterial clearance from the spleen or liver after secondary infection (Fig. 1C and 1D). Thus, class-Ia-restricted CD8 T cells do not appear to contribute to secondary bacterial clearance.
Figure 1. CD8 T cells are not required for immunity to secondary Salmonella infection.
C57BL6 (B6) Wild-type, KbDb-, perforin-, granzyme B- and perforin/granzyme B-deficient (PG) mice were intravenously infected with 5X105 BRD509. On day 60 post-infection, all the mice that resolved the primary infection with BRD509 as well as naïve strains were rechallenged intravenously with 1000 virulent Salmonella SL1344. Infected mice were monitored daily for signs for disease and sacrificed after the development of a moribund state. Bacterial burden was tested at day 6 after SL1344 infection. Spleens and livers from infected mice were homogenized in PBS and serial dilutions plated on MacConkey agar plates to determine bacterial burden. Scatter plots represent the bacterial burden of individual mice in spleens (left) or livers (right) from naïve strains (top) or mice that resolved BRD509 infection (bottom). “+” Indicates a dead mouse in this group before day 6. Data with KbDb-, perforin-, and granzyme B-deficient mice are pooled from two different experiments. Bacterial burden was analyzed by one-way ANOVA followed by Bonferroni’s Multiple Comparison Test (***: p<0.001).
4.2. Perforin or granzyme B are not required to resist secondary Salmonella infection
Next, we examined whether perforin or granzyme B expression was required for secondary protective immunity to Salmonella infection. Wild-type, perforin-, granzyme B-, or perforin/granzyme B-deficient mice were immunized with attenuated Salmonella and on day 60 subsequently challenged with a virulent bacterial strain. All naïve perforin-, granzyme B-, or perforin/granzyme B-deficient mice had similar bacteria burdens to Wild-type mice, typical of an Nramp-susceptible background, and some mice succumbed to death before day 6 (Figure 1A and 1B). As with KbDb-deficient mice, perforin-, granzyme B-, or perforin/granzyme B-deficient mice all had similar bacterial loads to Wild-type controls after secondary infection (Fig. 1C and 1D). Thus, expression of perforin or granzyme B is not required for secondary protection against Salmonella infection.
4.3. Role of MHC class Ia-restricted T cells, perforin and granzyme B during primary Salmonella infection
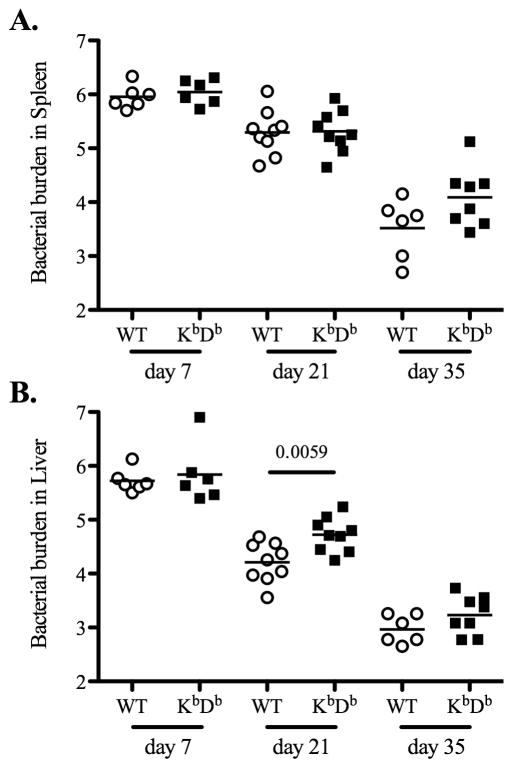
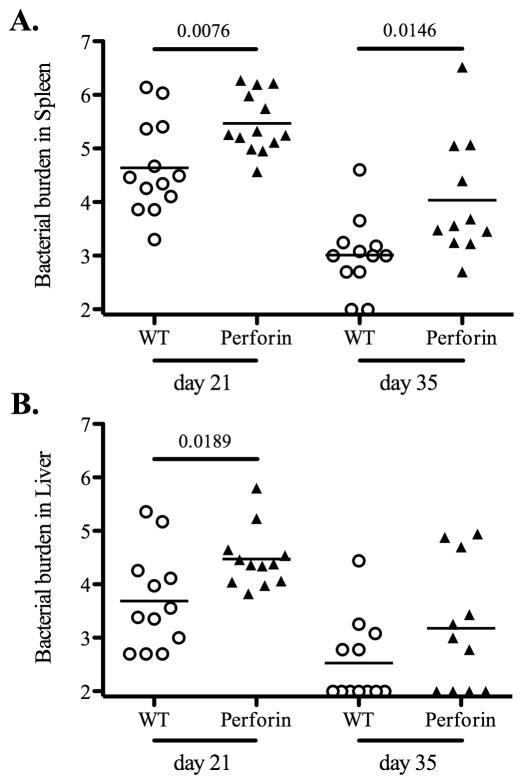
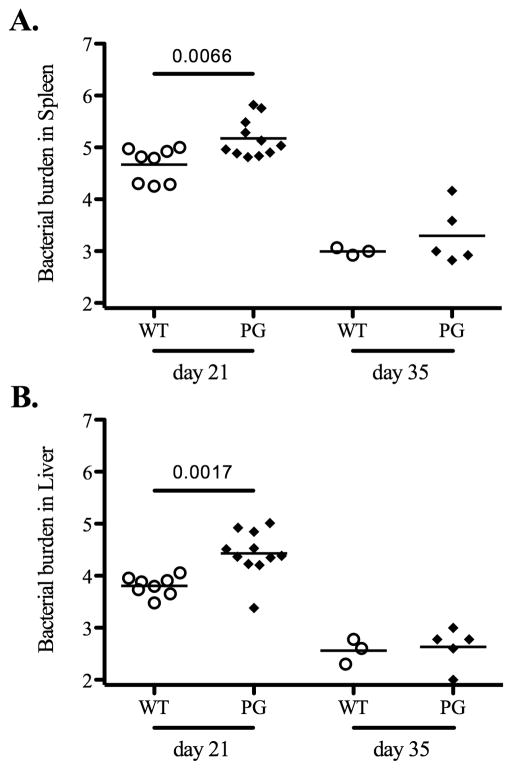
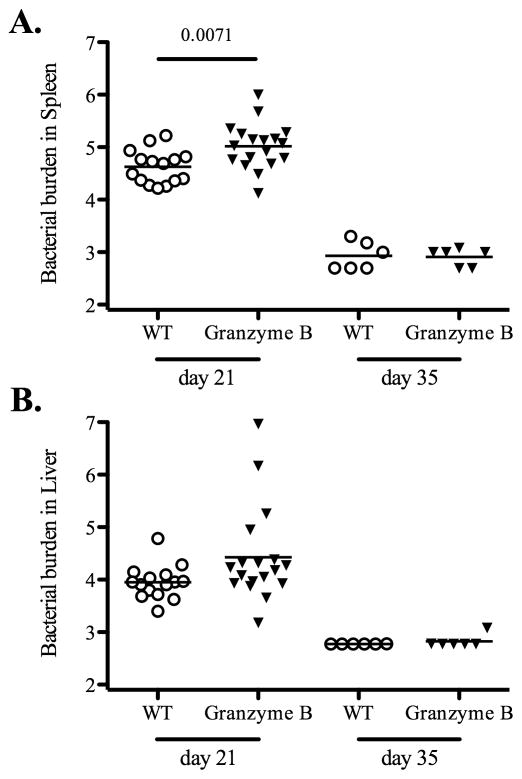
Next, we examined primary clearance of attenuated bacteria in each gene-deficient mouse strain. Wild-type and KbDb-deficient mice were infected with attenuated Salmonella and bacterial loads were quantified in spleens and livers at day 7, 21, and 35 post-infection. At day 7 post-infection, there was no difference in bacterial loads in spleens or livers of Wild-type or KbDb-deficient mice (Fig. 2). However, at day 21 post-infection, bacterial loads were significantly higher in the liver of KbDb-deficient mice, while at day 35 post-infection, KbDb-deficient mice appeared to have a slightly higher bacterial load in both organs, although this was not statistically significant (Fig. 2). Together, these data suggested a modest protective role for MHC class Ia-restricted CD8 T cells during the late stage of clearing attenuated Salmonella. Given this finding, we also carefully examined the late stages of primary bacterial clearance in perforin-, granzyme B-, and perforin/granzyme B-deficient mice. Similarly, each of these strains demonstrated a modest but significant deficiency in primary bacterial clearance from the spleen at 21 days post-infection, and both perforin-, and perforin/granzyme B-deficient mice displayed deficient clearance in the liver at this time point (Fig. 3–5). However, by day 35 post-infection all these mice had reduced bacterial burdens although bacterial loads in perforin-deficient mice remained slightly higher than Wild-type controls (Fig. 3–5). All of these strains had completely cleared bacteria from the spleen and liver by day 60 post-infection (data not shown). Taken together, these data suggest that MHC class-I-restricted CD8 T cells play a minor role in the late stages of primary attenuated bacterial clearance and that perforin and granzyme B expression are also required at this time.
Figure 2. KbDb-deficient mice have slightly higher bacteria loads at the late stage of primary Salmonella infection.
B6 Wild-type and KbDb-deficient mice were intravenously infected with 5X105 attenuated Salmonella, BRD509. Bacterial burden was examined at day 7, 21, and 35 post-infection. Spleens and livers from infected mice were homogenized in PBS and serial dilutions plated onto MacConkey agar plates to determine bacterial numbers at each time points. Data show mean bacterial burden in spleen (top) and liver (bottom) with individual mice shown as scatter plots. Data are pooled from at least three separate experiments. Numbers above each group indicate statistical significance and show the p value of a comparison between groups.
Figure 3. Perforin-deficient mice have moderately higher bacterial burden at late stage of primary Salmonella infection.
B6 Wild-type and perforin-deficient mice were intravenously infected with 5×105 BRD509. Bacterial burden was examined at day 21 and 35 post-infection. Spleens and livers from infected mice were homogenized in PBS and serial dilutions plated on MacConkey agar plates to determine bacterial burden at each time points. Data show mean bacterial burden in spleen (top) and liver (bottom) with individual mice shown as scatter plots. Data are pooled from at least three separate experiments. Numbers above each group indicate statistical significance and show the p value of a comparison between groups.
Figure 5. Perforin/granzyme B-deficient mice have the ability to resolve the primary Salmonella infection.
B6 Wild-type and perforin/granzyme B-deficient (PG) mice were intravenously infected with 5×105 BRD509. Bacterial burden was tested at day 21 and 35 post-infection. Spleens and livers from infected mice were homogenized in PBS and serial dilutions plated on MacConkey agar plates to determine bacterial burden at each time points. Data show mean bacterial burden in spleen (top) and liver (Bottom) with individual mice shown as scatter plots. Day 21 data are combined from two different experiments and day 35 data is from one experiment. Numbers above each group indicate statistical significance and show the p value of a comparison between groups.
5. Discussion
We have examined the role of CD8 T cells during Salmonella infection using KbDb-, perforin-, granzyme B-, and perforin/granzyme B-deficient mice. Surprisingly, secondary protective immunity against Salmonella infection did not require class Ia-restricted CD8 T cells, perforin, or granzyme B. This finding conflicts somewhat with previous studies that have pointed to a protective role for CD8 cytotoxic lymphocytes in clearing secondary infections [12–14,30]. However, these experiments involved antibody depletion of CD8 T cells, an approach that affects other CD8-expressing populations. Thus, it seems likely that these previous observations are due to a protective role for MHC class-Ib-restricted CD8 T cells, or another non-classically restricted CD8 population during secondary Salmonella infections. Indeed, Qa-1-restricted CD8 T cells specific for the heat-shock protein GroEL have previously been reported [35,43], and an H2-M3-restricted CD8 T cell line has been generated [44]. Studies in Listeria infection also point to a protective role for class-Ib-restricted CD8 T cells in the absence of KbDb [45]. Further studies are therefore required to examine the role of MHC class Ib-restricted CD8 T cells in secondary Salmonella infection.
Our studies also have detected a modest but significant role for MHC class-I-restricted CD8 T cells in the resolution of primary attenuated Salmonella infection, whereas previous studies examining β2μ-deficient mice failed to detect a role for MHC class-I-restricted T cells in primary clearance [17,35]. However, the expression of MHC class-I molecules is maintained in the absence of β2μ [37], meaning that studies using β2μ-deficient mice underestimate any protective role for class-I-restricted CD8 T cells during primary infection. Interestingly, the protective effect of class-I-restricted T cells was only evident at the late stages of primary infection. Indeed, it has previously been reported that Salmonella-specific CD8 T cell activation is delayed during primary infection [28,46]. Thus, it seems likely that the late contribution of CD8 T cells to pathogen clearance is simply due to their delayed activation after Salmonella infection. It also possible that this protective role is only found in primary immunity to slow-growing attenuated bacteria and indeed none of these mouse strains show any enhanced susceptibility to virulent bacteria, although these studies would need to be repeated on an Nramp-resistant background to examine this more rigorously.
We detected a protective role for perforin and granzyme B in the late stages of primary Salmonella infection. Although it is formally possible that this represents the activity of other cytolytic cell populations, the close correspondence with MHC-class-I-deficient mice suggest that MHC class-I restricted CD8 T cells mediate their protective effect via cytolytic granule release. It is not clear why these Salmonella-specific CD8 T cells would not contribute significantly during the secondary response. It is possible that the primary role of CD8 T cell lysis of Salmonella-infected cells is not to directly limit bacterial replication, but to release Salmonella antigens for presentation in the MHC class-II pathway, and thus amplify the CD4 response. This contribution would be more likely to be important during a primary infection where CD4 T cell activation is more dependent on antigen availability due to the inhibitory effects of replicating bacteria on class-II antigen presentation or CD4 T cell survival [47–50]. It is clear from our data that secondary protection against Salmonella can progress normally in the absence of both perforin and granzyme B. Indeed, previous studies from our laboratory and others show that this secondary response is heavily dependent on CD4 Th1 cells and antibody [14,19–21].
Overall, our data support a re-evaluation of protective CD8 T cell responses in Salmonella infection. Previous studies have suggested that class-I-restricted CD8 T cells are functional during secondary, but not primary infection [12–14,30,35]. In contrast, our data indicate that class-Ia-restricted CD8 T cells can enhance bacterial clearance during primary infection with attenuated bacteria but are not required for the development of secondary protection.
Figure 4. Granzyme B-deficient mice are competent to resolve the primary bacteria infection.
B6 Wild-type and granzyme B-deficient mice were intravenously infected with 5X105 BRD509 Salmonella. Bacteria colonization in vivo was tested at day 21 and 35 post-infection. Spleens and livers from infected mice were homogenized in PBS and serial dilutions plated on MacConkey agar plates to specify bacterial burden at each time points. Data show mean bacterial burden in spleen (top) and liver (bottom) with individual mice shown as scatter plots. Data are combined from at least three different experiments. Numbers above each group indicate statistical significance and show the p value of a comparison between groups.
Highlight.
MHC class-I-restricted CD8 T cells were not involved in the acquired immunity to Salmonella infection
The death pathway via perforin or/and granzyme B was not required for the acquired immunity after Salmonella infection
CD8 T cells played a role during primary Salmonella infection
Acknowledgments
The authors would like to acknowledge helpful discussions with members of the McSorley laboratory and the laboratory of Dr. S. Way in completion of these experiments. This work was supported by grants from the National Institutes of Health AI56172 and AI055743.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Crump JA, Mintz ED. Global trends in typhoid and paratyphoid Fever. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2010;50:241–246. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordon MA. Salmonella infections in immunocompromised adults. J Infect. 2008;56:413–422. doi: 10.1016/j.jinf.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Reddy EA, Shaw AV, Crump JA. Community-acquired bloodstream infections in Africa: a systematic review and meta-analysis. Lancet Infect Dis. 2010;10:417–432. doi: 10.1016/S1473-3099(10)70072-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Typhoid immunization. Recommendations of the Immunization Practices Advisory Committee (ACIP) MMWR Recomm Rep. 1990;39:1–5. [PubMed] [Google Scholar]
- 6.Fraser A, Paul M, Goldberg E, Acosta CJ, Leibovici L. Typhoid fever vaccines: systematic review and meta-analysis of randomised controlled trials. Vaccine. 2007;25:7848–7857. doi: 10.1016/j.vaccine.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 7.Santos RL, Zhang S, Tsolis RM, Kingsley RA, Adams LG, Baumler AJ. Animal models of salmonella infections: enteritis versus typhoid fever. Micrtobes Infect. 2001;3:1335–1344. doi: 10.1016/s1286-4579(01)01495-2. [DOI] [PubMed] [Google Scholar]
- 8.Griffin AJ, McSorley SJ. Development of protective immunity to Salmonella, a mucosal pathogen with a systemic agenda. Mucosal Immunol. 2011;4:371–382. doi: 10.1038/mi.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisenstein TK, Angerman CR. Immunity to experimental Salmonella infection: studies on the protective capacity and immunogenicity of lipopolysaccharide, acetone-killed cells, and ribosome-rich extracts of Salmonella typhimurium in C3H/HeJ and CD-1 mice. J Immunol. 1978;121:1010–1014. [PubMed] [Google Scholar]
- 10.Johanns TM, Law CY, Kalekar LA, O’Donnell H, Ertelt JM, Rowe JH, et al. Early eradication of persistent Salmonella infection primes antibody-mediated protective immunity to recurrent infection. Microbes and infection / Institut Pasteur. 2011;13:322–330. doi: 10.1016/j.micinf.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoiseth SK, Stocker BAD. Aromatic-dependent salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 12.Nauciel C. Role of CD4+ T cells and T-independent mechanisms in aquired resistance to Salmonella typhimurium infection. J Immunol. 1990;145:1265–1269. [PubMed] [Google Scholar]
- 13.Mastroeni P, Villarreal-Ramos B, Hormaeche CE. Role of T cells, TNF alpha and IFN gamma in recall of immunity to oral challenge with virulent salmonellae in mice vaccinated with live attenuated aro- Salmonella vaccines. Microb Pathog. 1992;13:477–491. doi: 10.1016/0882-4010(92)90014-f. [DOI] [PubMed] [Google Scholar]
- 14.Mastroeni P, Villarreal-Ramos B, Hormaeche CE. Adoptive transfer of immunity to oral challenge with virulent salmonella in innately susceptible BALB/c mice requires both immune serum and T cells. Infect Immun. 1993;61:3981–3984. doi: 10.1128/iai.61.9.3981-3984.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moon JJ, McSorley SJ. Tracking the dynamics of salmonella specific T cell responses. Curr Top Microbiol Immunol. 2009;334:179–198. doi: 10.1007/978-3-540-93864-4_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinha K, Mastroeni P, Harrison J, de Hormaeche RD, Hormaeche CE. Salmonella typhimurium aroA, htrA, and AroD htrA mutants cause progressive infections in athymic (nu/nu) BALB/c mice. Infect Immun. 1997;65:1566–1569. doi: 10.1128/iai.65.4.1566-1569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hess J, Ladel C, Miko D, Kaufmann SH. Salmonella typhimurium aroA- infection in gene-targeted immunodeficient mice: major role of CD4+ TCR-alpha beta cells and IFN-gamma in bacterial clearance independent of intracellular location. J Immunol. 1996;156:3321–3326. [PubMed] [Google Scholar]
- 18.VanCott JL, Chatfield SN, Roberts M, Hone DM, Hohmann EL, Pascual DW, et al. Regulation of host immune responses by modification of Salmonella virulence genes. Nat Med. 1998;4:1247–1252. doi: 10.1038/3227. [DOI] [PubMed] [Google Scholar]
- 19.Ravindran R, Foley J, Stoklasek T, Glimcher LH, McSorley SJ. Expression of T-bet by CD4 T cells is essential for resistance to Salmonella infection. Journal of immunology. 2005;175:4603–4610. doi: 10.4049/jimmunol.175.7.4603. [DOI] [PubMed] [Google Scholar]
- 20.McSorley SJ, Jenkins MK. Antibody is required for protection against virulent but not attenuated Salmonella enterica serovar typhimurium. Infect Immun. 2000;68:3344–3348. doi: 10.1128/iai.68.6.3344-3348.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mastroeni P, Simmons C, Fowler R, Hormaeche CE, Dougan G. Igh-6(−/−) (B-cell-deficient) mice fail to mount solid aquired resistance to oral challenge with virulent Salmonella enterica serovar typhimurium and show impaired Th1 T-cell responses to Salmonella antigens. Infect Immun. 2000;68:46–53. doi: 10.1128/iai.68.1.46-53.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ravindran R, McSorley SJ. Tracking the dynamics of T-cell activation in response to Salmonella infection. Immunology. 2005;114:450–458. doi: 10.1111/j.1365-2567.2005.02140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lundin BS, Johansson C, Svennerholm AM. Oral immunization with a Salmonella enterica serovar typhi vaccine induces specific circulating mucosa-homing CD4(+) and CD8(+) T cells in humans. Infection and immunity. 2002;70:5622–5627. doi: 10.1128/IAI.70.10.5622-5627.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salerno-Goncalves R, Pasetti MF, Sztein MB. Characterization of CD8(+) effector T cell responses in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J Immunol. 2002;169:2196–2203. doi: 10.4049/jimmunol.169.4.2196. [DOI] [PubMed] [Google Scholar]
- 25.Salerno-Goncalves R, Wyant TL, Pasetti MF, Fernandez-Vina M, Tacket CO, Levine MM, et al. Concomitant induction of CD4+ and CD8+ T cell responses in volunteers immunized with Salmonella enterica serovar typhi strain CVD 908-htrA. J Immunol. 2003;170:2734–2741. doi: 10.4049/jimmunol.170.5.2734. [DOI] [PubMed] [Google Scholar]
- 26.Mittrucker H, Kohler A, Kaufmann SH. Characterization of the murine T-lymphocyte response to salmonella enterica serovar typhimurium infection. Infect Immun. 2002;70:199–203. doi: 10.1128/IAI.70.1.199-203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srinivasan A, Foley J, McSorley SJ. Massive number of antigen-specific CD4 T cells during vaccination with live attenuated Salmonella causes interclonal competition. Journal of immunology. 2004;172:6884–6893. doi: 10.4049/jimmunol.172.11.6884. [DOI] [PubMed] [Google Scholar]
- 28.Luu RA, Gurnani K, Dudani R, Kammara R, van Faassen H, Sirard JC, et al. Delayed expansion and contraction of CD8+ T cell response during infection with virulent Salmonella typhimurium. Journal of immunology. 2006;177:1516–1525. doi: 10.4049/jimmunol.177.3.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McSorley SJ, Asch S, Costalonga M, Rieinhardt RL, Jenkins MK. Tracking Salmonella-specific CD4 T cells in vivo reveals a local mucosal response to a disseminated infection. Immunity. 2002;16:365–377. doi: 10.1016/s1074-7613(02)00289-3. [DOI] [PubMed] [Google Scholar]
- 30.Nauciel C, Espinasse-Maes F. Role of gamma interferon and tumor necrosis factor alpha in resistance to Salmonella typhimurium infection. Infection and immunity. 1992;60:450–454. doi: 10.1128/iai.60.2.450-454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishikawa H, Naito T, Iwanaga T, Takahashi-Iwanaga H, Suematsu M, Hibi T, et al. Curriculum vitae of intestinal intraepithelial T cells: their developmental and behavioral characteristics. Immunological reviews. 2007;215:154–165. doi: 10.1111/j.1600-065X.2006.00473.x. [DOI] [PubMed] [Google Scholar]
- 32.Lara-Tejero M, Pamer EG. T cell responses to Listeria monocytogenes. Current opinion in microbiology. 2004;7:45–50. doi: 10.1016/j.mib.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Lee YJ, Jameson SC, Hogquist KA. Alternative memory in the CD8 T cell lineage. Trends Immunol. 2011;32:50–56. doi: 10.1016/j.it.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shortman K, Liu YJ. Mouse and human dendritic cell subtypes. Nat Rev Immunol. 2002;2:151–161. doi: 10.1038/nri746. [DOI] [PubMed] [Google Scholar]
- 35.Lo WF, Ong H, Metcalf ES, Soloski MJ. T cell responses to Gram-negative intracellular bacterial pathogens: a role for CD8+ T cells in immunity to Salmonella infection and the involvement of MHC class Ib molecules. Journal of immunology. 1999;162:5398–5406. [PubMed] [Google Scholar]
- 36.Brigl M, Brenner MB. CD1: antigen presentation and T cell function. Annual review of immunology. 2004;22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608. [DOI] [PubMed] [Google Scholar]
- 37.Bix M, Raulet D. Functionally conformed free class I heavy chains exist on the surface of beta 2 microglobulin negative cells. The Journal of experimental medicine. 1992;176:829–834. doi: 10.1084/jem.176.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harty JT, Tvinnereim AR, White DW. CD8+ T cell effector mechanisms in resistance to infection. Annual review of immunology. 2000;18:275–308. doi: 10.1146/annurev.immunol.18.1.275. [DOI] [PubMed] [Google Scholar]
- 39.Hoves S, Trapani JA, Voskoboinik I. The battlefield of perforin/granzyme cell death pathways. J Leukoc Biol. 2010;87:237–243. doi: 10.1189/jlb.0909608. [DOI] [PubMed] [Google Scholar]
- 40.Kagi D, Ledermann B, Burki K, Seiler P, Odermatt B, Olsen KJ, et al. Cytotoxicity mediated by T cells and natural killer cells is greatly impaired in perforin-deficient mice. Nature. 1994;369:31–37. doi: 10.1038/369031a0. [DOI] [PubMed] [Google Scholar]
- 41.Strugnell R, Dougan G, Chatfield S, Charles I, Fairweather N, Tite J, et al. Characterization of a Salmonella typhimurium aro vaccine strain expressing the P. 69 antigen of Bordetella pertussis. Infection and immunity. 1992;60:3994–4002. doi: 10.1128/iai.60.10.3994-4002.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vugmeyster Y, Glas R, Perarnau B, Lemonnier FA, Eisen H, Ploegh H. Major histocompatibility complex (MHC) class I KbDb −/− deficient mice possess functional CD8+ T cells and natural killer cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:12492–12497. doi: 10.1073/pnas.95.21.12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lo WF, Woods AS, DeCloux A, Cotter RJ, Metcalf ES, Soloski MJ. Molecular mimicry mediated by MHC class Ib molecules after infection with gram-negative pathogens. Nature medicine. 2000;6:215–218. doi: 10.1038/72329. [DOI] [PubMed] [Google Scholar]
- 44.Ugrinovic S, Brooks CG, Robson J, Blacklaws BA, Hormaeche CE, Robinson JH. H2-M3 major histocompatibility complex class Ib-restricted CD8 T cells induced by Salmonella enterica serovar Typhimurium infection recognize proteins released by Salmonella serovar Typhimurium. Infection and immunity. 2005;73:8002–8008. doi: 10.1128/IAI.73.12.8002-8008.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seaman MS, Perarnau B, Lindahl KF, Lemonnier FA, Forman J. Response to Listeria monocytogenes in mice lacking MHC class Ia molecules. Journal of immunology. 1999;162:5429–5436. [PubMed] [Google Scholar]
- 46.Jones-Carson J, McCollister BD, Clambey ET, Vazquez-Torres A. Systemic CD8 T-cell memory response to a Salmonella pathogenicity island 2 effector is restricted to Salmonella enterica encountered in the gastrointestinal mucosa. Infection and immunity. 2007;75:2708–2716. doi: 10.1128/IAI.01905-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Griffin AJ, McSorley SJ. Generation of Salmonella-specific Th1 cells requires sustained antigen stimulation. Vaccine. 2011;29:2697–2704. doi: 10.1016/j.vaccine.2011.01.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bueno SM, Gonzalez PA, Schwebach JR, Kalergis AM. T cell immunity evasion by virulent Salmonella enterica. Immunology letters. 2007;111:14–20. doi: 10.1016/j.imlet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 49.Srinivasan A, Nanton M, Griffin A, McSorley SJ. Culling of activated CD4 T cells during typhoid is driven by Salmonella virulence genes. Journal of immunology. 2009;182:7838–7845. doi: 10.4049/jimmunol.0900382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ertelt JM, Johanns TM, Mysz MA, Nanton MR, Rowe JH, Aguilera MN, et al. Selective culling of high avidity antigen-specific CD4(+) T cells after virulent Salmonella infection. Immunology. 2011;134:487–497. doi: 10.1111/j.1365-2567.2011.03510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]