Abstract
The autophagy factor ATG12~ATG5 conjugate exhibits E3 ligase-like activity by which the lipidation of members of the LC3 family is facilitated. The crystal structure of the human ATG12~ATG5 conjugate bound to the amino-terminal region of ATG16L1, the factor that recruits the conjugate to autophagosomal membranes, reveals an integrated architecture in which ATG12 docks onto ATG5 through conserved residues. ATG12 and ATG5 are oriented such that other conserved residues on each molecule, including the conjugation junction, form a continuous patch. Mutagenesis data support the importance of both the ATG12–ATG5 interface and the continuous patch for E3 activity. The ATG12~ATG5 conjugate interacts with the E2 enzyme ATG3 with high-affinity through another surface location that is exclusive to ATG12, suggesting a different role of the continuous patch in E3 activity. These findings provide a foundation for understanding the mechanism of LC3 lipidation.
INTRODUCTION
Autophagy is a catabolic process that contributes to nutrition homeostasis and damage control in eukaryotic cells1–4. During autophagy, cytoplasmic materials, such as proteins, protein aggregates, damaged organelles, and invasive bacteria, are encapsulated in double-membrane-bound vesicles called autophagosomes and transported to lysosomes for degradation2,5–9. Unusual for a vesicular system, autophagosomes form de novo in the cytoplasm, but current descriptions of this process are limited to the morphological level. At the molecular level, many protein factors are known to participate in autophagosome formation9. One category of these proteins includes the ubiquitin-like proteins (Ublps) of the LC3 family consisting of 7 members in mammals (Atg8 in yeast) and the conserved ATG12 (Atg12 in yeast)10,11. Upon induction of autophagy, LC3 becomes conjugated to phosphatidylethanolamine at autophagosome-forming sites, where this conjugate plays crucial roles in the control of membrane dynamics and in substrate recruitment12–15. In contrast, ATG12 is constitutively conjugated to ATG516, a structural protein consisting of two ubiquitin-like-fold domains (UFDs) and an α-helical bundle region (HBR) on which Lys130, the lysine used for conjugation, is located17. The conjugation between ATG12 and ATG5 is essential for LC3 lipidation and therefore for autophagosome formation13,16.
LC3 and ATG12 are conjugated to their respective targets by enzymatic cascades that are analogous to ubiquitination and involve a common E1 activation enzyme, ATG7, and E2 conjugation enzymes, ATG3 and ATG10, for LC3 and ATG12, respectively10,18. Although these cascades lack canonical E3 ligases such as RING domain-containing proteins, a recent biochemical study showed that yeast Atg12~Atg5 (where ~ indicates conjugation) facilitates Atg8 transfer from Atg3 to phosphatidylethanolamine in vitro19. This report, together with the requirement of ATG12~ATG5 for LC3 lipidation in cells, has led to the recognition of ATG12~ATG5 as the E3 factor for LC3 lipidation. However, much less is understood about the physical role of the covalent linkage between ATG12 and ATG5 and about determinants required for E3 activity.
ATG3 has been reported to interact with ATG1220–22. ATG5 binds to ATG16L1 (Atg16 in yeast), the factor that recruits ATG12~ATG5 to sites of autophagosome formation13,14,22–25. Because these interactions can occur independently of the conjugation between ATG5 and ATG12, the conjugation provides a simple means of recruiting ATG3 to autophagosomal membranes. This idea is supported by a recent report showing that forced localization of an artificial ATG16 construct at the plasma membrane resulted in LC3 lipidation at the plasma membrane22. However, given that ATG12 is conjugated to the specific lysine of ATG5, ATG3 recruitment is unlikely to be the sole purpose of conjugation, unless the specific site is required only for formation of but not for the function of ATG12~ATG5. Canonical Ublps modulate the function of target proteins often by conjugation to specific lysine sites. In several cases, structural evidences has suggested that Ublp conjugation also causes conformational changes of the target proteins26,27, or that the conjugate provides a new surface for the recruitment of other proteins to specific sites of the target protein28. In the case of ATG12~ATG5, it remains to be established whether similar mechanisms operate in addition to ATG3 recruitment.
Here we set out to identify the role of the conjugation between ATG12 and ATG5 in LC3 lipidation. We demonstrate that the native conjugation moeity is crucial for E3 activity using artifical covalent linkages. The crystal structure of human ATG12~ATG5 in complex with the N-terminal region of ATG16L1 (referred to as ATG16N) shows that the conjugate forms an integrated architecture through covalent and non-covalent contacts. Structural and mutational analyses suggest that both ATG12 and ATG5 are directly involved in E3 activity through residues that are assembled into a continuous patch upon conjugation. ATG12 also possesses another surface patch that is responsible for ATG3 binding. These findings establish the structural role of the covalent linkage in building an architecture required for E3 activity and provide insights into how the E2–E3 interaction occurs.
RESULTS
Native conjugation moiety is critical for E3 activity
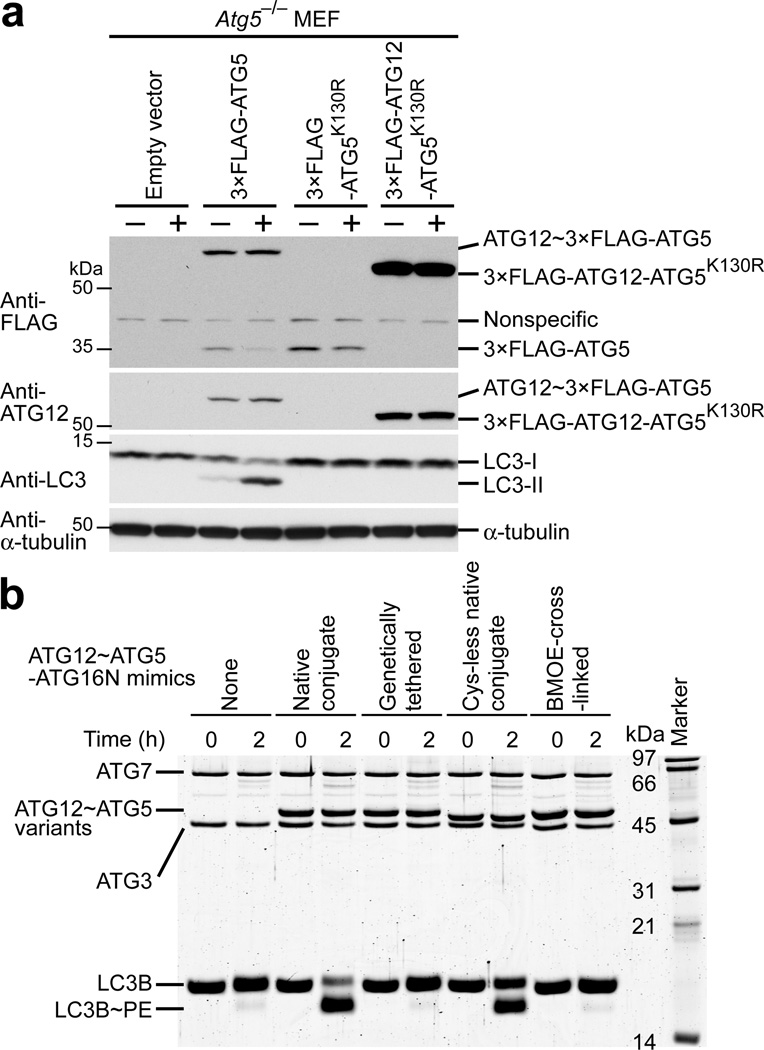
To examine the importance of the specific conjugation site of ATG12~ATG5 for E3 activity, we tested the activity of a conjugate mimic generated by genetic tethering of ATG12 and ATG5. It has previously been reported that in Atg5 knockout (Atg5−/−) mouse embryonic fibroblasts (MEFs) LC3~phosphatidylethanolamine (also referred to as LC3-II in cellular contexts) is not detected13. This defect could be restored by expression of wild-type ATG5 but not by the conjugation-incompetent mutant ATG5K130R, indicating that ATG12~ATG5 is necessary for LC3-II formation13. We obtained the same results with the stable expression of 3×FLAG-ATG5 using a retrovirus system (Fig. 1a). We then expressed 3×FLAG-ATG12-ATG5K130R fusion construct, in which the C terminus of ATG12 is fused to the N terminus of ATG5K130R through a 2×GlyGlySer linker (K130R mutation prevents conjugation to endogenous ATG12), and assayed LC3-II formation. Despite expression of the expected fusion protein in cells, LC3-II was not detected (Fig. 1a), even when autophagy was induced by starvation in the presence of chloroquine, a compound that prevents lysosomal degradation of LC3-II and therefore allows more sensitive detection of LC3-II29. We also tested the same fusion protein in an in vitro LC3 lipidation assay and confirmed that it was not active (Fig. 1b). These data indicate that the attachment of ATG12 to the N terminus of ATG5 does not generate a functional E3.
Figure 1.
Artificial mimics of ATG12~ATG5 do not promote LC3 lipidation. (a) Western blots of LC3-II formation assays carried out in Atg5−/− MEFs stably transduced with the pMX-puro retrovial vectors expressing the indicated 3×FLAG-tagged constructs under non-starvation (−) and starvation (+) conditions. (b) SDS-PAGE of in vitro LC3 lipidation assays carried out with the indicated ATG12~ATG5 conjugate mimics complexed with ATG16N.
We next generated another conjugate mimic by chemically cross-linking ATG12 and ATG5 molecules at the native conjugation site and examined its E3 activity. To this end, we constructed ATG12 and ATG5 mutants whose residues used for native conjugation were replaced by cysteine (ATG12G140C and ATG5K130C), while native cysteine residues were changed to non-cysteine amino acids, and cross-linked these mutant proteins with bismaleimidoethane (BMOE). As a control, we made a mutant conjugate containing the same substitutions for the native cysteine residues. While this Cys-less mutant exhibited an E3 activity in vitro (Fig. 1b), albeit at a slightly lower level than the wild-type, the BMOE-cross-linked conjugate mimic failed to do so (Fig. 1b), indicating that BMOE cross-linking caused a too large structural deviation from the functional form. Overall, these data support the importance of the native conjugation moiety and rationalize the need for structural investigation on ATG12~ATG5.
Structure determination of the ATG12~ATG5–ATG16N complex
To gain structural insights into the role of the covalent linkage, we crystalized human ATG12~ATG5 in complex with a 33-residue long ATG16N construct. Crystals appeared under two different solution conditions and exhibited space groups, P212121 and C2. We solved the native data sets of these crystals by molecular replacement using a model derived from a multi-wavelength anomalous diffraction data set collected on a selenomethionine-incorporated complex (see Methods). The structures were refined to resolutions of 2.7 Å and 2.9 Å for the P212121 and C2 data sets, respectively (Table 1). The P212121 and C2 crystals contained two and one complex, respectively, in the asymmetric unit. Despite the different crystal packing, the structures are overall very similar, as revealed by the low 1.0 ± 0.1 Å root mean square distance (r.m.s.d.) of all the Cα atoms among the three complex molecules (Supplementary Fig. 1a).
Table 1.
Data collection, phasing and refinement statistics
| ATG12~ATG5– ATG16N: crystal Ia |
ATG12~ATG5– ATG16N: crystal IIa |
ATG12~ATG5–ATG16N(I17M L21M I36M L43M)b | |||
|---|---|---|---|---|---|
| Data collection | |||||
| Space group | P212121 | C2 | C2 | ||
| Cell dimensions | |||||
| a, b, c (Å) | 43.8, 113.2, 210.7 | 135.1, 58.9, 90.8 | 102.8, 45.4, 100.7 | ||
| 90, 90, 90 | 90, 129.1, 90 | 90, 102.6, 90 | |||
| Peak | Inflection | Remote | |||
| Wavelength (Å) | 1.0000 | 1.0092 | 0.9796 | 0.9799 | 0.9724 |
| Resolution (Å)c | 50.0–2.7 (2.80–2.70) | 50.0–2.9 (3.00–2.90) | 50.0–2.9 (3.00–2.90) | 50.0–2.9 (3.00–2.90) | 50.0–2.9 (3.00–2.90) |
| Rmergec | 10.0 (63.6) | 5.6 (54.1) | 6.7 (33.5) | 6.1 (34.5) | 6.0 (31.6) |
| I / Ic | 18.5 (2.7) | 22.6 (1.9) | 24.0 (5.3) | 27.9 (5.2) | 17.8 (3.7) |
| Completeness (%)c | 99.9 (100.0) | 98.6 (92.4) | 99.9 (99.4) | 100.0 (99.8) | 99.7 (99.2) |
| Redundancyc | 6.9 (6.1) | 4.4 (3.7) | 7.2 (6.6) | 7.2 (6.5) | 3.6 (3.4) |
| Refinement | |||||
| Resolution (Å) | 44.1–2.7 | 36.2–2.9 | |||
| No. reflections | 29598 | 12465 | |||
| Rwork / Rfree | 17.6 / 23.6 | 21.5 / 25.4 | |||
| No. atoms | |||||
| Protein | 6469 | 3226 | |||
| Ligand/ion | 2 | 1 | |||
| Water | 76 | 0 | |||
| B-factors | |||||
| Protein | 67.5 | 138.8 | |||
| Ligand/ion | 58.9 | 121.6 | |||
| Water | 50.1 | n/a | |||
| R.m.s. deviations | |||||
| Bond lengths (Å) | 0.003 | 0.006 | |||
| Bond angles (°) | 0.66 | 0.51 | |||
Native.
Selenomethionine.
Values in parentheses are for the highest-resolution shell. Each data set was collected from one crystal.
Architecture of the ATG12~ATG5–ATG16N complex
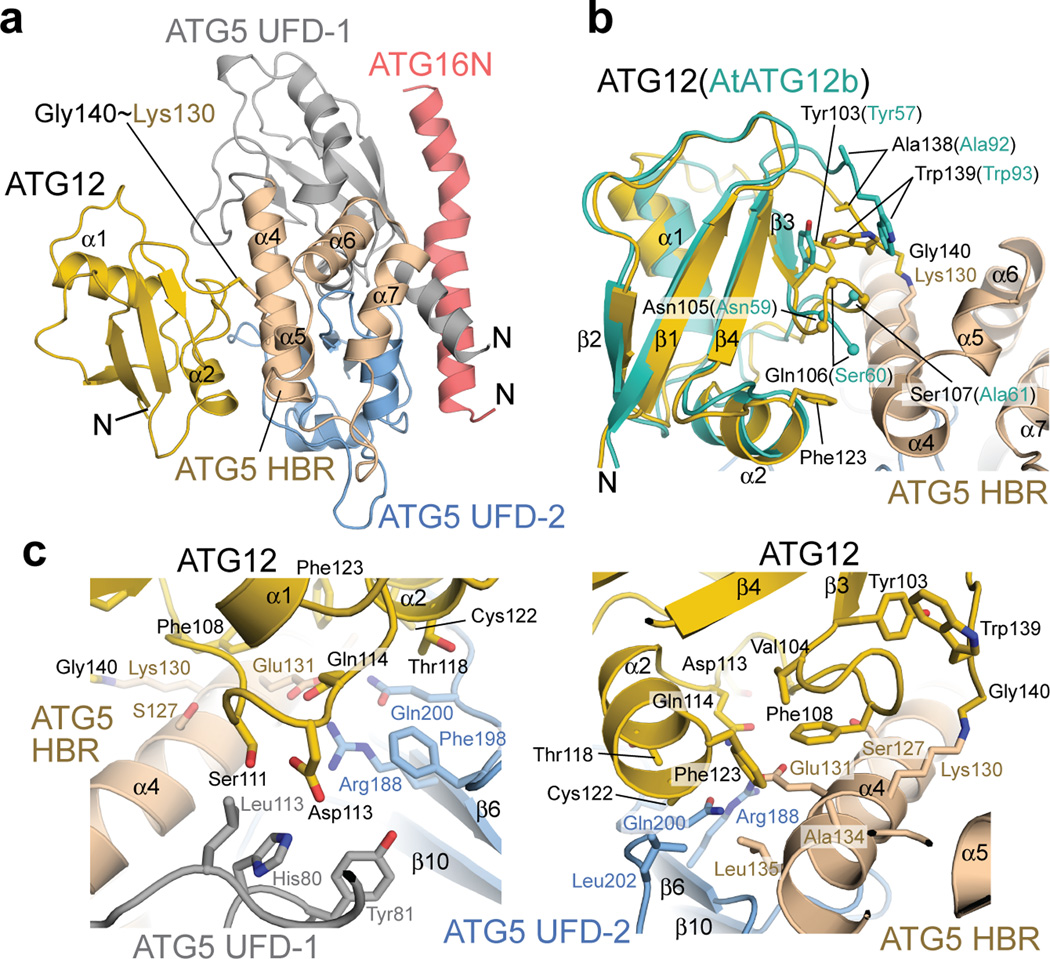
In the crystals, ATG12~ATG5–ATG16N formed a compact structure (Fig. 2a). As previously observed in the structure of Saccharomyces cerevisiae Atg5–Atg16N17,30, ATG16N is bound to a surface of ATG5 consisting of two UFDs (referred to as UFD-1 and -2, which corresponds to UblA and UblB, respectively, in the previous report17) in an α–helical conformation with a slight kink in the middle. ATG12 is located on ATG5 at the side opposite of ATG16N and there are no contacts between ATG12 and ATG16N. The electron density for the C-terminal residue Gly140 of ATG12 and that for Lys130 of ATG5 are clearly connected, validating our structure as the conjugated form (Supplementary Fig. 1b). There are no major conformational changes in ATG12 and ATG5 upon conjugation (Fig. 2b and Supplementary Fig. 1c). We note that although Arabidopsis thaliana (At) ATG12b in the previous crystal structure formed a domain-swapped dimer30, ATG12 in our structure is monomeric. At a local level, there is a small change in ATG12 upon conjugation. In unconjugated AtATG12b, the penultimate tryptophan residue in the C-terminal tail makes contacts with the other monomer in the domain-swapped dimer but not with its own UFD30. In our structure, the C-terminal tail of ATG12 folds back onto its own UFD through rearranged side chain–side chain interactions that include contacts between the penultimate tryptophan Trp139 and Tyr103 of its own UFD (Fig. 2b). Such conformation of the C-terminal tail of ATG12 has not been observed among Ublp structures; the C terminus of canonical Ublps lacks such an aromatic residue and is considered to be flexible31.
Figure 2.
The crystal structure of human ATG12~ATG5–ATG16N. (a) Cartoon representation of the ATG12~ATG5–ATG16N structure. ATG12 and ATG16N are colored yellow and red, respectively. ATG5 is shown in three colors to represent three subdomains: gray, wheat, and blue for UFD-1, HBR, and UFD-2, respectively. The N terminus of each protein is indicated with the letter N. This view is defined as the front view throughout the rest of the figures. (b) Superimposition of the “monomeric form” of AtATG12b (PDB ID: 1WZ3, shown in turquoise) onto ATG12 (yellow) of the ATG12~ATG5–ATG16N structure. The residues around the C-terminal tail of ATG12 are shown with stick models for their side chains. The residues in the first turn of the “turn-loop-turn-α2” segment of ATG12 and the corresponding ones of AtATG12b are indicated as spheres at the Cα positions. Ser60 and Ala61 of AtATG12b are not connected because they belong to different chains in the domain-swapped AtATG12b dimer. (c) Enlarged approximately top (left) and bottom (right) views of the interface.
The non-covalent contacts between ATG12 and ATG5 bury a total of 1300 Å2 of solvent-accessible surface area (Fig. 2c). The aliphatic atoms of the side chain of Lys130 are buried within this interface, indicating that Lys130 not only provides its ε-amino group for isopeptide bonding to ATG12 but also has a structural role as part of the interface. A patch on ATG12 comprising the turn-loop-turn-α2 (Asn105–Phe123) segment is associated with the interaction surface on ATG5 that is formed by residues from several regions in its primary structure, including UFD-1 (Gly14–Lys105), the loop (S106–S117) connecting UFD-1 and HBR, α4 (Lys118–His137) of HBR, and the loop (Phe198–Leu202) of UFD-2. Of note, this turn-loop-turn-α2 segment is the region that was exchanged between the two molecules in the AtATG12b dimer (Fig. 2b and Supplementary Fig. 1d), indicating that ATG12 may be stabilized upon conjugation. This idea is supported by our observation in which unconjugated ATG12 tended to form oligomers slowly during purification and storage in solution (Supplementary Fig. 2). Phe108 in the turn-loop-turn-α2 segment of ATG12, whose corresponding residue in yeast Atg12 has previously been suggested to be important for autophagy32, is located in the center of the patch of ATG12 and makes contacts with Glu131 in α4 of ATG5. There is an intermolecular hydrogen bond between the side chains of this Glu131 and Gln114 in α4 of ATG12 as well as a salt bridge between the side chains of Asp113 of ATG12 and His80 on the UFD-1 of ATG5, which are located at a distal site from the conjugation site (Fig. 2c). Evaluation of the properties of this interface using PDBePISA33 revealed that the interface is hydrophobic, favorable for association, and unlikely to be a specific interaction site, as indicated by a ΔiG (change in the total solvation energy upon complexation) of −6.8 kcal mol−1, a ΔiG P-value (P-value of the observed solvation free energy gain) of 0.25, and a complex formation significance score of 0, respectively. Consistent with this result, we have not been able to demonstrate an interaction between unconjugated ATG12 and ATG5 molecules. However, the interface between ATG12 and ATG5 is unlikely to be a crystallographic artifact because essentially the same structure has been observed in two different space groups; the only appreciable difference between the two crystal structures is at the loop between β1 and β2 in UFD-1 of ATG5, which is involved in crystal packing in the C2 but not in the P212121 crystal and is far away from the ATG12–ATG5 interface (Supplementary Fig. 1a). Based on these analyses and considerations, we propose that the architecture of ATG12~ATG5 is based on the stabilization of weak and less-specific but favorable hydrophobic interfaces by the covalent linkage and by the C-terminal tail folding of ATG12.
Perturbation of the ATG12–ATG5 interface affects E3 activity
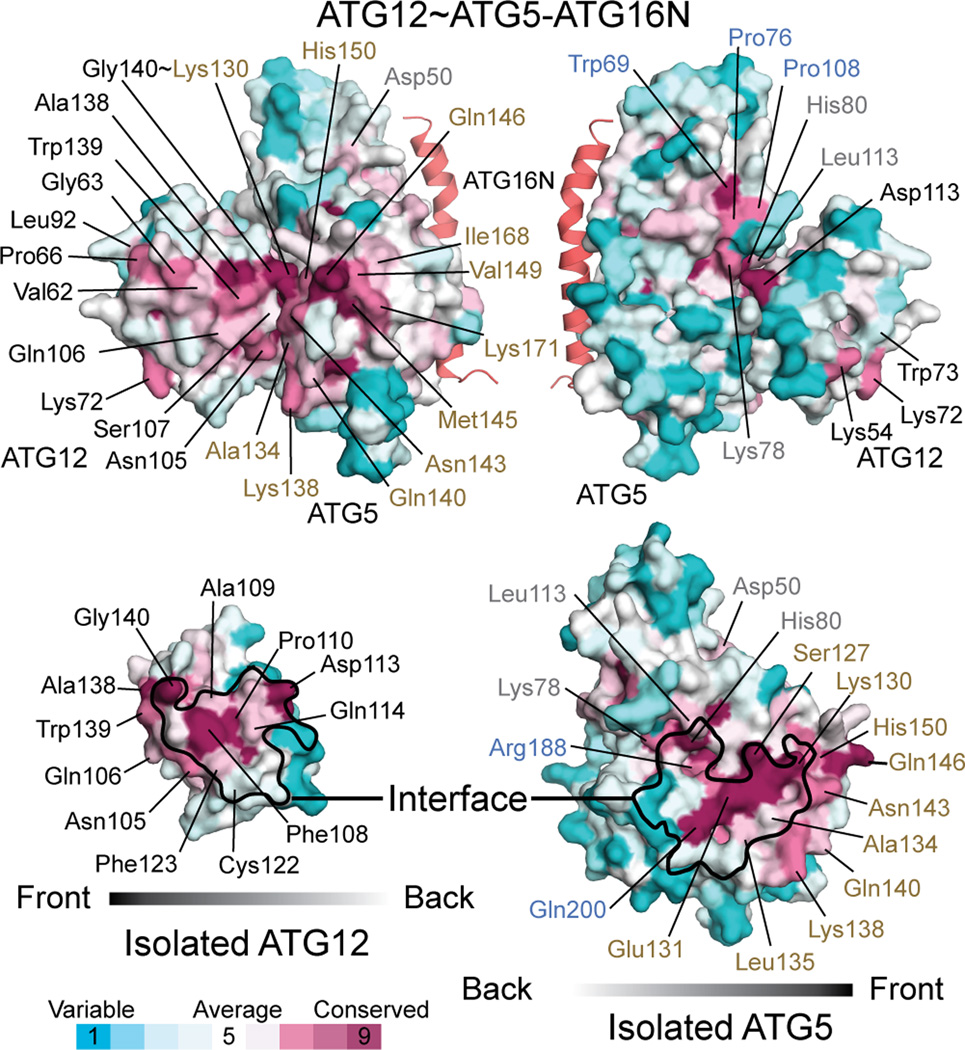
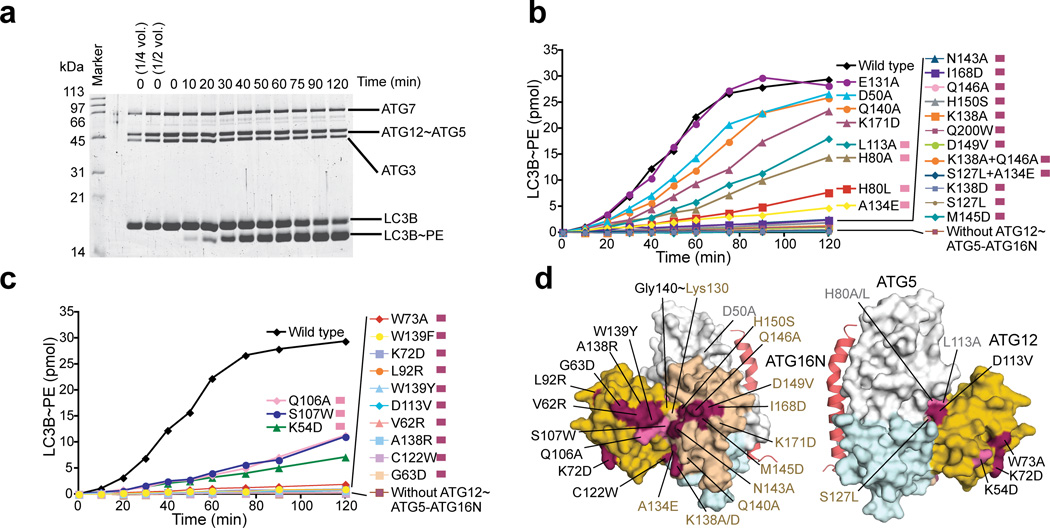
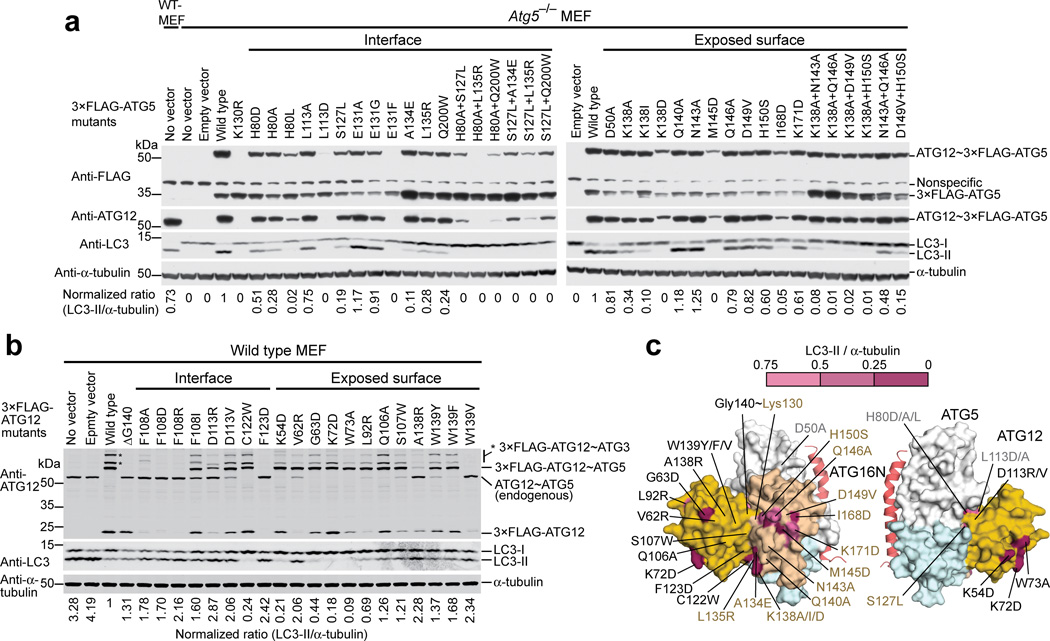
Having revealed the architecture of ATG12~ATG5, we sought to establish its functional significance. As shown in Figure 3, conservation mapping on the surface of each molecule revealed that the ATG12–ATG5 interface contains highly conserved residues, including Phe108 and Asp113 of ATG12 and His80, Leu113, Ser127, Glu131, Arg188, and Gln200 of ATG5; this high level of conservation at the interface suggests that there is evolutionary pressure on the ATG12~ATG5 architecture. To confirm the importance of the interface, we mutated these conserved residues and some additional less-conserved peripheral residues and tested E3 activity of the mutants in vitro (Fig. 4a). Amino acids that are bulkier than or have opposite chemical properties of the native ones were included in the choices for mutagenesis because we predicted that such amino acids would interfere with the packing at the interface and thus sterically disrupt the architecture more than alanine would. The results agree well with this prediction (Fig. 4b,c and Supplementary Fig. 3); the mutations D113V and C122W in ATG12 and H80L, S127L, and A134E in ATG5 severely impaired E3 activity, whereas the effects of the mutations to alanine such as H80A and L113A in ATG5 were moderate and E131A in ATG5 had no effect. In addition to those mutations, we attempted to test F108A and F108D in ATG12 and E131F in ATG5 but failed to do so because these mutations impaired the production of the conjugate in E.coli.
Figure 3.
Identification of potential functional patches on ATG12~ATG5 by conservation mapping. Surface representations of ATG12~ATG5 (left top and right top for the front and approximately backside view, respectively) as well as ATG12 (left bottom) and ATG5 (right bottom) that are isolated from the ATG12~ATG5 structure are shown with colors according to the conservation scores obtained from the ConSurf server44. Fifty and 45 sequences of ATG12 and ATG5, respectively, were used in the analysis (Supplementary Fig. 5). On the surfaces of the isolated ATG12 and ATG5, the interfaces in ATG12~ATG5 are indicated by black borderlines. Highly conserved residues as well as less-conserved residues that were tested in mutagenesis experiments are indicated.
Figure 4.
Conserved residues of ATG12~ATG5 are important for E3 activity. (a) SDS-PAGE of an in vitro kinetic LC3 lipidation assay carried out with the wild-type ATG12~ATG5–ATG16N. (b and c) The quantified data of the in vitro kinetic LC3 lipidation assays carried out with ATG12~ATG5–ATG16N containing mutations in ATG5 (b) or ATG12 (c). The raw data for (b) and (c) are shown in Supplementary Figure 3. (d) Mapping of the results of the in vitro mutagenesis data onto the structure. The residues that have been affected by mutation are shown in the same color as indicated on the right side of the description for each mutation in (b) and (c).
We extended the mutagenesis study to the cellular context to confirm our in vitro results. To test ATG5 mutations in cells, 3×FLAG-ATG5 mutants were expressed in Atg5−/− MEFs. As shown in Figure 5a, mutations at the interface, such as H80A, H80L, S127L, A134E, L135R, and Q200W lowered LC3-II formation, and some combinations of these single mutations had more severe effects. The reduced amounts of some of ATG12~3×FLAG-ATG5 mutants, such as H80L and of a few double mutants, are unlikely to affect the interpretation, because it has previously been demonstrated that amounts of ATG12~ATG5 as low as the detection limit of Western blotting are sufficient for LC3-II formation34. The mutations E131A and E131G showed little effect in MEFs. Overall, these data are consistent with the in vitro results. The Mutations L113D, E131F, and H80A+L135R abolished the formation of ATG12~3×FLAG-ATG5. We presume that these mutations destabilize the interface in a way that is incompatible with the formation of ATG12~ATG5 or the protein folds.
Figure 5.
Residues of ATG12~ATG5 important for E3 activity in vitro are also important in MEFs. (a and b) The Western blots of LC3-II formation assays in Atg5−/− MEFs stably expressing 3×FLAG-ATG5 mutants (a) or in wild-type MEFs stably expressing 3×FLAG-ATG12 mutants (b). The ratio of LC3-II over α-tubulin in each lane was normalized against that in the wild-type lane and is shown at the bottom of each lane. The bands indicated by asterisks in (b) are 3×FLAG-ATG12~ATG3 as reported previously35. (c) Mapping of the cell-based data onto the structure. The residues that were affected by mutation are colored according to the ratio of LC3-II over α-tubulin reported in (a) and (b).
To test ATG12 mutations in cells, we used wild-type MEFs. When we stably expressed 3×FLAG-ATG12 in wild-type MEFs, we detected 3×FLAG-ATG12~ATG5 and additional 3×FLAG-ATG12-containing bands as well as LC3-II (Fig. 5b). This observation is consistent with a previous report in which stable expressions of ATG12 in mammalian cells using a retrovirus system similarly generated multiple ATG12-containing species but did not affect autophagy35. However, we found that the expression of 3×FLAG-ATG12 resulted in approximately 75% reduction in the level of LC3-II compared to the controls (Fig. 5b). This reduced amount of LC3-II was likely due to the presence of unconjugated 3×FLAG-ATG12 in the cells because the expression of a conjugation-incompetent form, 3×FLAG-ATG12ΔG140 also showed a reduction at a similar level as wild-type. This interpretation is consistent with another previous report in which transient overexpression of ATG12 in mammalian cells abolished LC3-II formation22. While performing this experiment, we noticed that the expression of ATG12 caused a downregulation of the endogenous ATG12~ATG5 to a level undetectable in Western blots (Fig. 5b), an effect not discussed previously. These observations suggested that despite the smaller dynamic range described above, the effect of a mutation in ATG12 on LC3-II formation could be examined in wild-type MEFs, as long as the expressed 3×FLAG-ATG12 mutant conjugates with the endogenous ATG5 efficiently enough to knock down the native ATG12~ATG5 as observed with wild-type ATG12 expression. Mutation of most of residues at the interface, such as Phe108, Asp113, and Phe123 in ATG12, failed to knock down the endogenous ATG12~ATG5 (Fig. 5b), presumably due to inefficient conjugation to ATG5; as a result, we could not obtain information on the effects of these mutations on LC3-II production. However, the C122W mutant efficiently conjugated to ATG5 and abolished the endogenous ATG12~ATG5. LC3-II formation was severely inhibited by C122W, consistent with the in vitro data described above. The amounts of the unconjugated mutants F108A, F108D, F108R, and F123D were not at detectable levels (Fig. 5b), indicating that these mutations destabilized the protein. In contrast, the F108I mutant was detectable and conjugated to ATG5. Thus, the hydrophobic nature of Phe108 seems to be necessary for protein stability and conjugation to ATG5. But the conjugation was not efficient enough to knock down the endogenous ATG12~ATG5. These observations contrast to a previous report in which the mutation of Phe154 in Atg12, the residue corresponding to Phe108 in ATG12, to alanine or aspartic acid did not affect Atg5 conjugation but impaired Atg8 lipidation in yeast32. Although such previous data support our structure, the loss of the function by the Phe154 mutations could also have been a consequence of the aggregation of the mutant conjugates as shown in the same report32. Indeed, we found that recombinant ATG12F108D protein aggregates to a much greater degree than the wild-type (Supplementary Fig. 2). Based on these findings, together with our structure, in which the Phe108 phenyl ring is sandwiched between the hydrophobic core of ATG12 and the interaction surface of ATG5 (Fig. 2c), we propose that Phe108 is important for the stability of both ATG12 and ATG12~ATG5.
Identification of functional patches on ATG12~ATG5
We next sought to identify potential functional sites on the surface of ATG12~ATG5. Conservation mapping revealed a continuous patch across ATG12 and ATG5 (Fig. 3). This patch contains the conjugation site and the C-terminal residues of ATG12, including Ala138, Trp139, and Gly140. Other residues such as Val62, Gly63, Leu92, Gln106, and Ser107, which are located on various segments of ATG12, and residues such as Lys138, Asn143, Met145, Gln146, His150, and Ile168 in the HBR of ATG5 also participate in this patch. In vitro, ATG5 mutations including N143A, Q146A, M145D, K138A, K138D, H150S, D149V, and I168D and the K138A+Q146A double mutation impaired E3 activity severely, whereas Q140A and K171D affected E3 activity only slightly (Fig. 4b,d, and Supplementary Fig. 3). In MEFs, the mutations K138I, K138D, M145D, and I168D severely impaired LC3-II formation (Fig. 5a), whereas other single mutants affected LC3-II formation less. Double mutants based on these less effective mutations severely affected the activity. The in vitro data for ATG12 show that LC3 lipidation was appreciably reduced by the mutations S107W or Q106A and was abolished by V62R, G63D, A138R, W139F, or W139Y (Fig. 4c,d, and Supplementary Fig. 3). In MEFs, only the G63D and Q106A mutants efficiently conjugated with ATG5 and knocked down the endogenous ATG12~ATG5 (Fig. 5b). Consistent with the in vitro data, G63D impaired LC3-II formation in MEFs, whereas Q106A did not. Taken together, these data support the importance of the residues in this patch in the E3 activity of this complex (Fig. 5c).
In addition to the continuous patch, two highly conserved residues of ATG12, Lys54 and Lys72, are located at the side opposite of the ATG12–ATG5 interface (Fig. 3). Strikingly, the mutation of these residues, as well as of Trp73, located adjacent to these two residues on the surface, abolished E3 activity both in vitro and in MEFs (Figs. 4c,d and 5b,c); therefore, these residues make up another functional patch.
ATG12 is responsible for high-affinity binding to ATG3
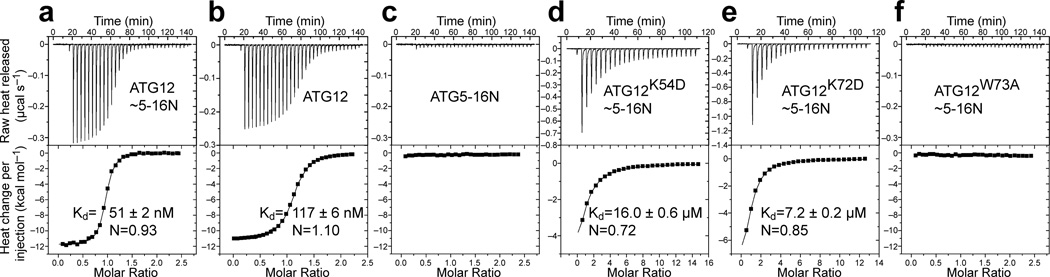
Understanding how ATG12~ATG5 interacts with ATG3 is essential for a mechanistic description of the E3 activity of the conjugate. Previous data suggest that both ATG12~ATG5 and the unconjugated ATG12 can bind to ATG319–22. However, how these interactions are related to the E3 activity of ATG12~ATG5 and how ATG5 is involved in this interaction are unclear. To probe the role of ATG5 in the interaction between ATG12~ATG5 and ATG3, we examined the binding abilities of conjugated and unconjugated proteins using isothermal titration calorimetry (ITC). As shown in Figure 6a,b, ATG12~ATG5–ATG16N binds tightly to ATG3 with a Kd of 51 nM, whereas unconjugated ATG12 binds to ATG3 with a slightly higher Kd of 117 nM. However, under our experimental conditions, the unconjugated ATG5–ATG16N did not show detectable binding to ATG3 (Fig. 6c). These results establish that the direct contribution of ATG5 to ATG3 binding is very small, if such a contribution exists at all. To identify the high-affinity interaction site, we assayed the binding of ATG3 to ATG12~ATG5–ATG16N complexes containing mutations in ATG12. The results show that mutations in the surface patch exclusively on ATG12 affected the binding markedly; the K54D and K72D mutations resulted in greater than 100-fold increases in Kd values and the W73A mutant showed no detectable binding (Figs. 6d–f). In contrast, ATG12 mutations in the interface to ATG5 (D113V, C122W, and A134E) or in the continuous patch (V62R, G63D, S107W, A138R, and W139Y) resulted in only minor (less than 2.5-fold) differences in Kd values (Supplementary Fig. 4). We also examined some of the ATG5 mutations that had severely impaired the E3 activity both in vitro and in MEFs (H80L, K138D, M145D, S127L+A134E, and K138A+Q146A) and confirmed that these ATG5 mutations do not affect the binding (Supplementary Fig. 4).
Figure 6.
Point mutations in the binding patch in ATG12 reduce the affinity for ATG3. (a–f) ITC data obtained by injecting ATG3 into a solution of (a) ATG12~ATG5–ATG16N, (b) ATG12, (c) ATG5–ATG16N, or (d–f) ATG12~ATG5–ATG16N containing point mutation K54D (d), K72D (e), or W73A (f) in ATG12 are shown. The binding constants (Kd) and stoichiometry (N) were determined by curve fitting.
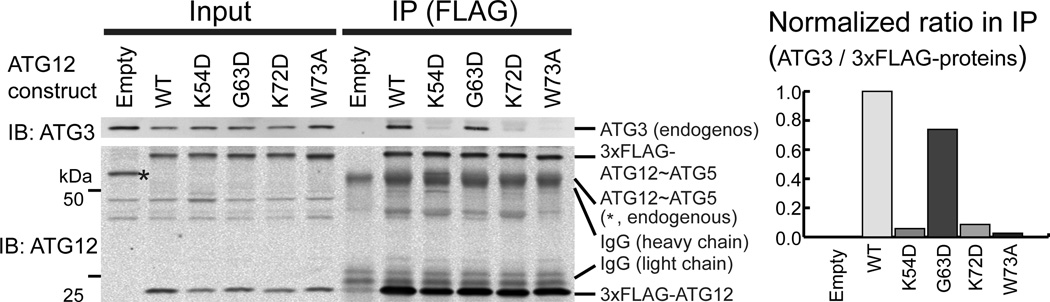
The effects of the mutations in ATG12 on ATG3 interaction were further verified in cells using immunoprecipitation. Lysates of the MEFs expressing ATG12 constructs described above were subjected to anti-FLAG immunoprecipitation. For this assay, we choose the mutants whose mutational sites are located on the conjugate surface and that impaired LC3-II formation in cells. As shown in Figure 7, both 3×FLAG-ATG12~ATG5 and 3×FLAG-ATG12 were detected in the immunoprecipitates. Endogenous ATG3 was co-precipitated with wild-type ATG12 but not with the control, demonstrating the interaction between these proteins. Remarkably, the immunoprecipitates containing the mutant K54D, K72D or W73A, showed markedly less amounts of ATG3, while a similar amount of ATG3 as wild-type was observed for G63D. The quantification of these immunocomplexes indicates that ATG3 interaction was severely weakened by the former three mutations but not much by G63D. These data agree well with the in vitro binding data described above. Thus, we conclude that the ATG12-exclusive patch including Lys54, Lys72 and Trp73 is the high-affinity binding site to ATG3 and the patch across ATG12 and ATG5 plays a different role in the E3 function.
Figure 7.
Point mutations in the binding patch on ATG12 impair ATG3 interaction in MEFs. The Western blot of an anti-FLAG immunoprecipitation assay carried out with lysates of MEFs expressing ATG12 constructs is shown. The ratios of the amount of ATG3 to the sum of 3×FLAG-ATG12-ATG5 and 3×FLAG-ATG12 in the immunoprecipitates were normalized against the wild-type and are shown as a bar graph.
DISCUSSION
Our structural and mutational analyses on ATG12~ATG5–ATG16N revealed that the conjugation generates a patch across ATG12 and ATG5 required for E3 activity. This finding explains why the ATG12-ATG5 fusion construct was not functional; the ATG12 in the fusion that was attached to the N terminus of ATG5 was too far away from its original position in ATG12~ATG5 to form such functional patch even with a flexible 2×GlyGlySer linker inserted between ATG12 and ATG5. The BMOE-cross-linked conjugate mimic was incompetent perhaps because it could not form the native ATG12~ATG5 architecture due to the increased number of the chemical bonds in the covalent linkage (8 bonds more than the native isopeptide bond). Alternatively, the extra atoms of BMOE may have caused steric hindrance for a binding partner during the E3 function, as the native conjugation site is located in the continuous patch. Our mutational analyses also suggested that the interface between ATG12 and ATG5 is important for E3 activity. While mutations in the interface that exhibited negative effects are likely to have destabilized the ATG12~ATG5 architecture and thus disrupted the continuous patch, we do not know the precise mechanisms underlying the effects by each mutation. Investigations on these mutants as well as the BMOE-conjugate would provide further insights into the mechanism of the E3 activity of ATG12~ATG5.
We have identified a surface location of ATG12 at the side opposite from the interface to ATG5 as the hot spot for ATG3 interaction and demonstrated that the residues in the hot spot are important for E3 activity. These data, however, challenge a previous report in which Phe108 of ATG12 was shown to be necessary for ATG3 interaction22. This conclusion was drawn from the result of an immunoprecipitation experiment using HEK293A cells over-expressing Phe108 mutants of ATG12. As described above, in our structure Phe108 is located at the interface to ATG5. In principle, it is possible that the mutations of Phe108 allosterically affect ATG3 binding, which occurs at the side opposite of Phe108. However, given the aggregation-prone nature of the Phe108 mutants, we suggest that the previous observation might have resulted from aggregation or destabilization of the expressed mutant proteins in cells. Ultimately, these contradictory data should be clarified by careful characterizations of the Phe108 mutants.
The next question regarding the E3 function of ATG12~ATG5 is exactly how this conjugate interacts with ATG3 or LC3-loaded ATG3 and facilitates the transfer of LC3 to phosphatidylethanolamine. Unlike E3 ubiquitin-ligases, ATG12~ATG5 does not appear to contribute specificity in substrate recognition; ATG3 alone can conjugate LC3, respectively, to phosphatidylethanolamine in vitro, albeit at much slower rates than in the presence of ATG12~ATG519,36–38. Thus, ATG12~ATG5 may function as an E2-stimulating machine analogously to RING domains in ubiquitin ligases. Although ATG12 alone binds tightly to ATG3, ATG12 alone is not sufficient for E3 activity, suggesting that the high-affinity binding site to ATG3 does not stimulate ATG3. Rather, we suggest the possibility that the patch across ATG12 and ATG5 serves this purpose. Our data showing that the affinity of ATG12~ATG5 for ATG3 is only slightly higher than that of ATG12 alone (Fig. 6) indicate that the continuous patch does not contribute much to the affinity for ATG3. However, such low affinity of the patch may be consistent with the typically observed low affinities (Kd~µM to mM) between RING domains and E2 enzymes for ubiquitination39. Alternatively, the continuous patch may function in stabilizing a conformation of LC3-charged ATG3 in an optimal orientation to enhance conjugation, as previously proposed for SUMO and ubiquitin systems40–43. Answers to these questions will be provided by future structural and biochemical studies on larger complexes.
ONLINE METHODS
Preparation of the ATG12~ATG5–ATG16N complex
All proteins used in this work are from human. The ATG16L111–43 (ATG16N) coding DNA sequence was cloned into the NdeI-XhoI site of a modified pET-Duet-1 vector (Novagen); the resulting vector expresses the 6×His-maltose binding protein (MBP)-tobacco etch virus (TEV) protease cleavage sequence (TEVs)-ATG16N fusion. E. coli BL21(λDE3) cells transformed with this vector were grown in Luria broth media and protein expression was induced by adding 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at an optical density (OD600) of 0.8, after which the cells were grown at 37°C for 3 hours and then harvested. The 6×His-MBP-TEVs-ATG16N fusion was purified by nickel affinity chromatography (Qiagen) followed by Source 15S cation-exchange chromatography (GE Healthcare). The purified protein was stored at −80°C until use. The full-length ATG7 and ATG10 coding DNA sequences were cloned into the NcoI-SalI and NdeI-XhoI sites, respectively, of a modified pCDF-Duet-1 vector (Novagen); the resulting vector expresses glutathione S-transferase-fused ATG7 and ATG10. The ATG1252–140 and full-length ATG5 coding DNA sequences were cloned into the BamHI-SalI and NdeI-XhoI sites, respectively, of a modified pACYC Duet-1 vector (Novagen); the resulting vector expresses the MBP-6×His-TEVs-ATG1252–140 fusion and ATG5. BL21(λDE3) cells co-transformed with these two plasmids were grown in Terrific broth media and protein expression was induced with 0.2 mM IPTG at an OD600 of 2.4. Then, the cells were grown at 18°C and harvested after 18 hours. The MBP-6×His-TEVs-ATG12~ATG5 fusion was purified by nickel affinity and Superdex 200 (SD200) size-exclusion chromatography (GE Healthcare), treated with TEV protease followed by Source 15S cation-exchange chromatography. Then, a 1.2-fold molar excess of 6×His-MBP-TEVs-ATG16N was mixed with ATG12~ATG5 and treated with TEV protease. Lastly, the cleaved protein was purified on Source 15S and SD200 columns. Mutations in ATG12 or ATG5 were incorporated into the coding sequences using a two-step PCR method, and mutant complexes were generated similarly as the wild-type. None of the purified mutant complexes showed aggregation in the final gel-filtration step. The cysteine-less ATG12~ATG5 included the mutations C122N and C134S in ATG12 and the mutations C19Q, C115L, C128S, and C223L in ATG5. Additional methods for the preparations of other proteins used in this work are described in Supplementary Note.
Crystallization of the ATG12~ATG5–ATG16N complex
Crystals of the native human ATG12~ATG5–ATG16N complex were grown at 20°C using the sitting-drop vapor diffusion method from drops containing 1 µl of 10 mg ml−1 protein in 10 mM Hepes, pH 7.0, 300 mM NaCl and 1 mM DTT and 1 µl of reservoir solution: 0.1 M Hepes, pH 7.5, 0.2 M lithium citrate, and 20% (w/v) PEG 3350 for crystal I and 0.1 M BICINE, pH 9.0, 0.2 M sodium phosphate dibasic, and 18% (w/v) PEG 3350 for crystal II. Under these conditions, the crystals typically grew to 80~120 µm × 50 µm × 10~20 µm in 5 days. The crystals were cryo-protected with 0.1 M Hepes, pH 7.5, 0.2 M lithium citrate, 21.5% (w/v) PEG 3350, 0.1 M NaCl, and 25% (v/v) ethylene glycol for crystal I and 0.1 M MES, pH 6.5, 0.2 M sodium phosphate dibasic, 19.5% (w/v) PEG 3350, and 20% (v/v) ethylene glycol for crystal II and flash-cooled in liquid nitrogen. The crystals I and II contained two and one complexes in the asymmetric unit, respectively. Crystals of the native ATG12~ATG5 in complex with the selenomethionine-labeled ATG16NI17M L21M I36M L43M mutant were grown at 4°C from drops containing 1 µl of 9 mg ml−1 protein in 10 mM Hepes, pH 7.0, 300 mM NaCl and 5 mM DTT and 1 µl of reservoir solution (0.1 M Hepes, pH 7.0, 0.2 M lithium citrate, and 18% (w/v) PEG 3350) and cryo-protected with 0.1 M Hepes, pH 7.0, 0.2 M lithium citrate, 20% (w/v) PEG 3350, 0.1 M NaCl, and 25% (v/v) ethylene glycol and flash-cooled in liquid nitrogen.
X-ray diffraction data collection and structure determination
The X-ray diffraction data of the native crystals I and II, and the selenomethionine crystal were collected at 100 K at the beamlines APS 22-BM, ALS 5.0.2 and ALS 8.2.1, respectively. All X-ray diffraction data were indexed, integrated and scaled using the HKL-2000 package45. The multi-wavelength anomalous diffraction (MAD) data set was solved and an initial model was built using the AutoSol and AutoBuild modules of the PHENIX package46, respectively. Because the selenomethionine crystal was not isomorphous with respect to the native crystals, the initial model from the MAD data was used for molecular replacement of the native data sets using AutoMR in PHENIX. The structures were further refined by iterative model building in Coot47 and refinement using the PHENIX refinement module. The qualities of the final models were assessed with MolProbity48; 95.3–97.0% of all φ and ψ geometries were in the most favored region, with less than 0.3% outliers in the Ramachandran statistics. All structure figures were generated using PyMOL (Schrödinger).
In vitro LC3 lipidation assay
Synthetic 1,2-dioleoyl-sn-glycero-3-phosphocholine (PC) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (PE), and bovine liver L-α-phosphatidylinositol (blPI) were purchased from Avanti Polar Lipids. Lipids were mixed with a molar ratio of 40% PC, 40% PE, and 20% blPI, and were dried under a nitrogen gas stream and under vacuum for 2 hours. Lipids were hydrated in 20 mM Hepes, pH 7.5, and 100 mM NaCl 1 hour at room temperature, freeze-thawed 3 times using dry-ice and a 42°C water bath, and extruded through a 100 µm polycarbonate filter using a mini-extruder (Avanti Polar Lipids). Liposomes were used within 2 days after extrusion. For the assay, 0.5 µM ATG7, 1 µM ATG3, and 1 µM ATG12~ATG5–ATG16N and 10 µM LC3B1–120 (matured form) were mixed with 1 mM liposomes in 50 mM Hepes, pH 7.5, 100 mM NaCl, 0.3 mM DTT and 1 mM MgCl2. Reactions were initiated by adding 1 mM ATP and were performed at 37°C. The reactions were quenched at various time points by adding SDS-PAGE sample buffer (2×). Proteins were separated by 6 M urea SDS-PAGE and stained with GelCode Blue (Thermo Pierce). The quarter, half, or same volume of the time zero sample as other time points were loaded into three separate lanes to obtain a calibration curve for the quantitation of LC3B in each gel. Gels were scanned on a LI-COR Odyssey infrared imager, and the bands corresponding to LC3B~phosphatidylethanolamine were quantified using the calibration curve obtained for the unconjugated LC3B from each gel using Image Studio 2.0 (LI-COR).
LC3-II formation assay in MEFs
MEFs stably expressing 3×FLAG-ATG5 (full-length), 3×FLAG-ATG12 (full-length), and mutant proteins were prepared using the pMXs-puro vector and Plat-E packaging cells, as described previously49. Atg5+/+ MEFs were used as wild-type in this work. Cells were cultured at 37°C with 5% CO2 in a humidified incubator. For starvation, the cell medium was changed from Dulbecco’s Modified Eagle’s Medium (DMEM, Life Technologies) to Hank’s Balanced Salt Solution (Sigma-Aldrich) containing 20 µM chloroquine. After 90 min, the cells were washed with phosphate-buffered saline on ice and lysed by adding the lysis buffer (20 mM Tris pH 7.5, 150 mM NaCl, 2 mM EDTA, 2% (v/v) TritonX-100) containing Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scienctific). Lysates were centrifuged at 28,500g for 15 min at 4°C. Total protein concentrations of the supernatants were determined using the BCA assay (Pierce). SDS-PAGE sample buffer (2×) was added to the supernatants, and the samples were boiled at 100°C for 5 min. Twenty µg of total protein was separated on 4–20% (w/v) acrylamide gradient gels and subjected to Western blot analyses. The antibodies used were anti-LC3 (MBL International, PM036), anti-ATG12 (Cell Signaling, 4180), anti-FLAG M2 (Sigma-Aldrich, F1804), and anti-α-tubulin (Cell Signaling, 3873). Immunoreactive bands corresponding to LC3-II and α-tubulin were quantified.
Immunoprecipitation assay
MEFs stably expressing ATG12 constructs were cultured in DMEM and cell lysates were prepared as described above. Total 500 µg protein in 300 µl of the lysis buffer was incubated with anti-FLAG M2 antibody-loaded Dynabeads protein G (Life Technologies) for 20 minutes at room temperature. Beads were washed four times with 400 µl of the wash buffer (20 mM Tris pH 7.5, 200 mM NaCl, 2 mM EDTA, 0.1% (v/v) TritonX-100). Input and final beads samples were subjected to SDS-PAGE followed by Western blot analyses using anti-ATG12 and anti-ATG3 (MBL International, M133-3) antibodies. The bands were quantified in Image Studio 2.1.
ITC experiments
All protein samples for ITC experiments were dialyzed against buffer containing 10 mM Hepes, pH 7.0, 200 mM NaCl and 2 mM TCEP overnight. All titrations were carried out at 25°C in a VP-ITC isothermal titration calorimeter (Microcal). Approximately 6.5 µM ATG12, ATG5–ATG16N, or ATG12~ATG5–ATG16N was placed in the cell. Data were fit with the model for one set of binding sites in Origin (OriginLab).
Supplementary Material
ACKNOWLEDGMENTS
We thank Noboru Mizushima (Tokyo Medical and Dental University, Tokyo, Japan) for the kind gift of the Atg5 knockout MEFs; Toshio Kitamura (University of Tokyo, Tokyo, Japan) for the kind gift of the pMXs-puro vector and Plat-E retrovirus packaging cells; Peter E. Wright for his critical reading of the manuscript; Kent Baker, Owen Pornillos, Kentaro Ihara, Mischa Machius, and the beamline staff for assistance and advice on crystallographic data collection; Andrew Ward and Gaya Amarasinghe for technical advice regarding the ITC experiments. This work was supported by National Institutes of Health (NIH) grant GM092740 to T.O. and by funds from Japan Science and Technology Agency through Keio Kanrinmaru Project to G. T. The synchrotron beamlines the Advanced Photon Source (APS) 22-BM operated by the Southeast Regional Collaborative Access Team (SER-CAT) at the Argonne National Laboratory and the Advanced Light Source (ALS) 5.0.2 and 8.2.1 operated by the Berkeley Center for Structural Biology (BCSB) at the Lawrence Berkeley National Laboratory are supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contracts No. W-31-109-Eng-38 and DE-AC02-05Ch11231, respectively. The BCSB is also supported in part by the NIH, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute.
Footnotes
Accession codes.
The structures of human ATG12~ATG5–ATG16N have been deposited to the Protein Data Bank under the accession codes 4GDK and 4GDL.
AUTHOR CONTRIBUTIONS
CO performed protein preparation, crystallization, and cell biology, and assisted biochemistry; ZM assisted structure refinement, biochemical analyses and manuscript preparation; GT managed cell biological assays; TO conceived the project and performed structure determination, in vitro biochemistry, and ITC analyses, and wrote the paper.
REFERENCES
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12:814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rubinsztein DC, Marino G, Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto A, Simonsen A. The elimination of accumulated and aggregated proteins: a role for aggrephagy in neurodegeneration. Neurobiol Dis. 2011;43:17–28. doi: 10.1016/j.nbd.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernales S, McDonald KL, Walter P. Autophagy counterbalances endoplasmic reticulum expansion during the unfolded protein response. PLoS Biol. 2006;4:e423. doi: 10.1371/journal.pbio.0040423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 9.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 10.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 11.Geng J, Klionsky DJ. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. 'Protein modifications: beyond the usual suspects' review series. EMBO Rep. 2008;9:859–864. doi: 10.1038/embor.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 13.Mizushima N, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kraft C, Peter M, Hofmann K. Selective autophagy: ubiquitin-mediated recognition and beyond. Nat Cell Biol. 2010;12:836–841. doi: 10.1038/ncb0910-836. [DOI] [PubMed] [Google Scholar]
- 15.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett. 2010;584:1359–1366. doi: 10.1016/j.febslet.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 17.Matsushita M, et al. Structure of Atg5.Atg16, a complex essential for autophagy. J Biol Chem. 2007;282:6763–6772. doi: 10.1074/jbc.M609876200. [DOI] [PubMed] [Google Scholar]
- 18.Schulman BA, Harper JW. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat Rev Mol Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanada T, et al. The Atg12–Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 20.Uetz P, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 21.Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem. 2002;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- 22.Fujita N, et al. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–3100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizushima N, Noda T, Ohsumi Y. Apg16p is required for the function of the Apg12p–Apg5p conjugate in the yeast autophagy pathway. EMBO J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y, Inagaki F. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem. 2010;285:1508–1515. doi: 10.1074/jbc.M109.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12–Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. J Biol Chem. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 26.Baba D, et al. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 27.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armstrong AA, Mohideen F, Lima CD. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature. 2012;483:59–63. doi: 10.1038/nature10883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki NN, Yoshimoto K, Fujioka Y, Ohsumi Y, Inagaki F. The crystal structure of plant ATG12 and its biological implication in autophagy. Autophagy. 2005;1:119–126. doi: 10.4161/auto.1.2.1859. [DOI] [PubMed] [Google Scholar]
- 31.Winget JM, Mayor T. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol Cell. 2010;38:627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Hanada T, Ohsumi Y. Structure-function relationship of Atg12, a ubiquitin-like modifier essential for autophagy. Autophagy. 2005;1:110–118. doi: 10.4161/auto.1.2.1858. [DOI] [PubMed] [Google Scholar]
- 33.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 34.Hosokawa N, Hara Y, Mizushima N. Generation of cell lines with tetracycline-regulated autophagy and a role for autophagy in controlling cell size. FEBS Lett. 2006;580:2623–2629. doi: 10.1016/j.febslet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 35.Radoshevich L, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ichimura Y, et al. In vivo and in vitro reconstitution of atg8 conjugation essential for autophagy. J Biol Chem. 2004;279:40584–40592. doi: 10.1074/jbc.M405860200. [DOI] [PubMed] [Google Scholar]
- 37.Oh-oka K, Nakatogawa H, Ohsumi Y. Physiological pH and acidic phospholipids contribute to substrate specificity in lipidation of Atg8. J Biol Chem. 2008;283:21847–21852. doi: 10.1074/jbc.M801836200. [DOI] [PubMed] [Google Scholar]
- 38.Nair U, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 40.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO–RanGAP1–Ubc9–Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pruneda JN, et al. Structure of an E3:E2 approximately Ub Complex Reveals an Allosteric Mechanism Shared among RING/U-box Ligases. [8 Aug 2012];Mol Cell. doi: 10.1016/j.molcel.2012.07.001. online publication, http://dx.doi.org/10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–W533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 45.Otwinowski Z, Minor W, Charles W, Carter . Methods in Enzymology. Vol. Volume 276. Academic Press; 1997. Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 46.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitamura T, et al. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp Hematol. 2003;31:1007–1014. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.