Summary
The recently discovered mitochondrial calcium uniporter (MCU) promotes Ca2+ accumulation into the mitochondrial matrix [1, 2]. We identified in silico miR-25 as a cancer-related MCU-targeting microRNA family and demonstrate that its overexpression in HeLa cells drastically reduces MCU levels and mitochondrial Ca2+ uptake, while leaving other mitochondrial parameters and cytosolic Ca2+ signals unaffected. In human colon cancers and cancer-derived cells, miR-25 is overexpressed and MCU accordingly silenced. miR-25-dependent reduction of mitochondrial Ca2+ uptake correlates with resistance to apoptotic challenges and can be reversed by anti-miR-25 overexpression. Overall, the data demonstrate that microRNA targeting of mitochondrial Ca2+ signaling favors cancer cell survival, thus providing mechanistic insight into the role of mitochondria in tumorigenesis and identifying a novel therapeutic target in neoplasia.
Highlights
► miR-25 regulates intracellular calcium homeostasis ► Mitochondrial calcium uniporter (MCU) is a target of miR-25 ► MCU plays a critical role in apoptosis and tumorigenesis ► MCU is downregulated in different cancer cell lines and in human colonic adenocarcinoma
Results and Discussion
miR-25 Downregulates MCU and Protects from Ca2+-Dependent Apoptosis
In the last two decades, mitochondrial Ca2+ homeostasis has been shown to participate in the control of the intrinsic pathway of apoptosis and to be influenced by oncogenes [3–6], thus suggesting that it is a signaling checkpoint in tumorigenesis. However, direct evidence and mechanistic insight were still lacking. The recent identification of the mitochondrial Ca2+ channel (mitochondrial calcium uniporter, MCU) [1, 2] and of the associated regulator MICU1 (also known as CBARA1) [7] now allow molecular investigation of the process, including the regulation of their expression by microRNAs (miRNAs). miRNAs are a class of small (19–25 nt), noncoding regulatory RNAs that regulate gene expression, causing target mRNA degradation or suppressing mRNA translation [8]. In human cancers, specific miRNAs are up- or downregulated, with consequent alteration in the expression of target proteins [9, 10].
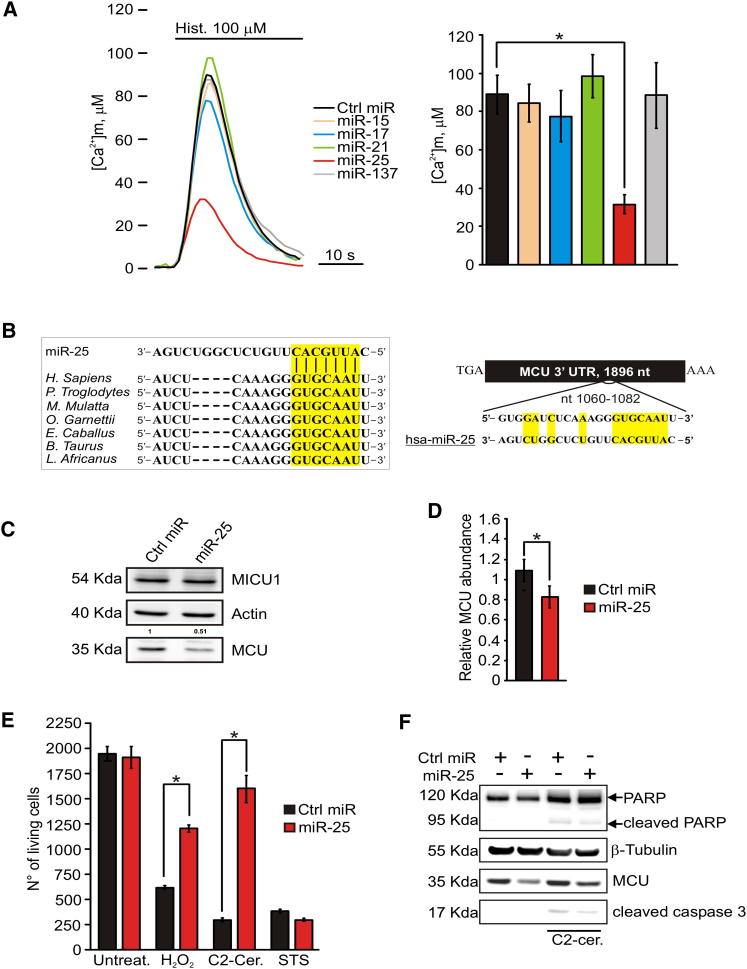
By filtering the output of four target prediction algorithms (TargetScan [11], MicroT [12], MicroCosm [13], and miRanda [14]; see Table S1 available online), we identified five cancer-related miRNA families (miR-15, miR-17, miR-21, miR-25, and miR-137) that could be predicted to target MCU and/or MICU1. We thus tested their effect on mitochondrial Ca2+ homeostasis by expressing them in HeLa cells and measuring mitochondrial [Ca2+] with a targeted aequorin-based Ca2+ probe (mtAEQ) [15]. The data (Figure 1A) showed that only miR-25 caused a marked reduction in the [Ca2+]m rise evoked by cell stimulation with 100 μM histamine, an agonist coupled to the generation of inositol 1,4,5-trisphosphate (InsP3) and the release of Ca2+ from the endoplasmic reticulum (ER). Accordingly, overexpression of an anti-miR-25 increases the mitochondrial Ca2+ uptake to agonist stimulation (Figure S1A), with a slight decrease in cytosolic [Ca2+] ([Ca2+]c), probably due to increased Ca2+ clearance by mitochondria (Figure S1B).
Figure 1.
miR-25 Reduces [Ca2+]m and Protects from Apoptosis by Downregulation of MCU mRNA and Protein Levels
(A) Mitochondrial and Ca2+ homeostasis in HeLa cells after expression of different miRNAs. Where indicated, mitochondrially targeted aequorin (mtAEQmut)-transfected cells were treated with 100 μM histamine (Hist.). Mitochondrial Ca2+ concentration ([Ca2+]m) peaks: negative control (Ctrl miR): 88.92 ± 10.05 μM; miR-15: 84.47 ± 9.96 μM; miR-17: 77.49 ± 13.23 μM; miR-21: 98.32 ± 11.09 μM; miR-25: 31.64 ± 5.06 μM; miR-137: 88.52 ± 17.12 μM. miR-25 induces an ∼65% reduction of Ca2+ response. n = 18 independent experiments.
(B) The miR-25 seed sequence and its target in seven species; its target site resides at nt 1060–1082 of the MCU 3′ UTR. The middle seven nucleotides of miR-25 and its target region have been highlighted.
(C) Immunoblot for MCU and MICU1 after miR-25 expression in HeLa cells. Quantification of MCU protein is reported.
(D) MCU mRNA expression was assessed by quantitative real-time PCR in HeLa cells transfected with miR-25 or Ctrl miR. GAPDH expression was used to normalize MCU expression results for each sample. miR-25-enforced expression caused a 30% decrease in MCU mRNA levels, as compared to control transfected cells. n = 3 independent experiments.
(E) Microscopy counts of cell viability after treatment with hydrogen peroxide (H2O2; 500 μM for 2 hr) and C2-ceramide (C2-cer.; 40 μM for 2 hr) revealed that miR-25-expressing HeLa cells were protected from apoptosis, as compared to control (Ctrl miR). The number of living cells after staurosporine (STS; 10 μM for 1 hr) treatment appears unaffected by miR-25 expression. n = 3 independent experiments.
(F) Immunoblot shows reduced levels of cleaved PARP and cleaved caspase-3 in miR-25-expressing HeLa cells after treatment with C2-ceramide (C2-cer.; 40 μM for 2 hr).
See also Figure S1. In this and following figures, experiments are representative of more than three trials, and conditions are given in Experimental Procedures. ∗p < 0.05; error bars correspond to mean ± SEM.
The effects were predicted to depend on MCU downregulation. Indeed, the bioinformatics analysis of the 1,896 nt 3′ UTR of MCU revealed a 100% match target seed sequence for miR-25 at nt 1075–1081, highly conserved across seven species (Figure 1B), and insertion of the 759 nt 3′ UTR of MCU (but not of the 569 nt 3′ UTR of MICU1) downstream of the luciferase gene in a reporter plasmid led to significant miR-25-dependent decrease of reporter activity (Figures S1C and S1D). We thus tested MCU expression by immunoblotting and detected a marked reduction in the protein level upon miR-25 overexpression (Figure 1C) and an increase in anti-miR-25-expressing cells (Figure S1E). As expected, MCU mRNA abundance was significantly decreased by miR-25 (Figure 1D), whereas anti-miR-25 increased it (Figure S1F). MCU downregulation was also evident using an immunofluorescence technique: Figure S1G shows that miR-25 expression drastically decreased MCU antibody reactivity.
The effect of miR-25 is shared by the other members of the miRNA family: miR-92a and miR-363 target MCU mRNA and reduce MCU protein levels and, accordingly, inhibit mitochondrial Ca2+ uptake, without affecting [Ca2+]c and [Ca2+]er (data not shown).
We investigated whether miR-25-dependent reduction in mitochondrial Ca2+ uptake correlates with increased resistance to apoptotic challenges. Microscopy counts of cell viability after treatment with H2O2, C2-ceramide, or staurosporine (STS) revealed that miR-25-expressing HeLa cells were strongly protected from death caused by C2-ceramide and H2O2 (Figure 1E), apoptotic challenges for which mitochondrial Ca2+ loading acts as a sensitizing factor [16–18], whereas the sensitivity to STS was unaffected. Accordingly, PARP and caspase-3 cleavage upon C2-ceramide treatment were markedly reduced in miR-overexpressing cells (Figure 1F). These results were also confirmed by cellular positivity to the apoptotic marker annexin V (Figure S1H).
miR-25 Induces Reduction of Mitochondrial Ca2+ Uptake Exclusively through MCU
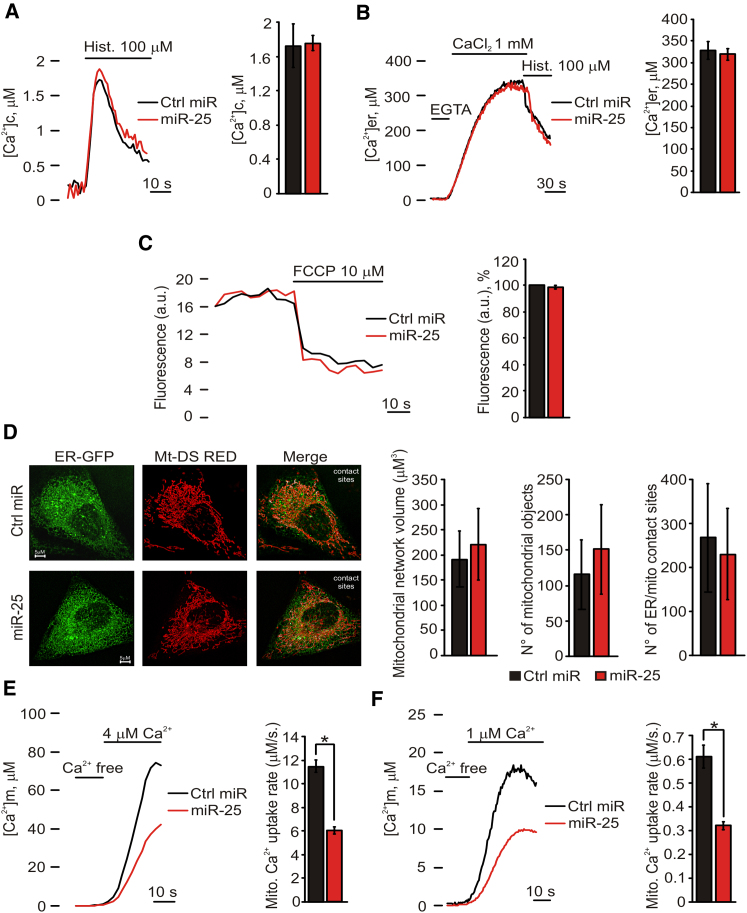
We then proceeded to rule out that the effect on [Ca2+]m was secondary to alterations of global Ca2+ signaling patterns or to morphological or functional dysregulation of mitochondria. On the former aspect, we investigated the cytosolic [Ca2+] changes and the state of filling and release kinetics of the ER. The results showed that miR-25, when expressed in HeLa cells, caused no difference in the amplitude of the [Ca2+]c rise evoked by histamine (Figure 2A), nor in the steady state [Ca2+]er or in the release caused by the agonist (Figure 2B). Thus, the effect of miR-25 on Ca2+ homeostasis is exclusively mitochondrial.
Figure 2.
miR-25 Inhibits Mitochondrial Ca2+ Uptake without Causing Morphological Rearrangement or Changes in the Electrochemical Gradient
(A) Cytosolic Ca2+ concentration peaks: Ctrl miR: 1.72 ± 0.25 μM; miR-25: 1.726 ± 0.08 μM. n = 12 independent experiments.
(B) Reticular Ca2+ concentration levels: Ctrl miR: 328.2 ± 19.68 μM; miR-25: 319.4 ± 13.18 μM. n = 12 independent experiments.
(C) TMRM fluorescence measurements: miR-25-expressing HeLa cells show no difference in TMRM loading (−2.25 ± 1.18% compared to control cells). a.u., arbitrary units. n = 32 independent experiments.
(D) Fluorescence images of mtDsRed- and erGFP-labeled mitochondria and ER, respectively, in control- and miR-25-expressing HeLa cells. Mitochondrial volume and number were deduced by calculating object size (Ctrl miR: 191.49 ± 54.64 μm3; miR-25: 221.16 ± 74.4 μm3) and number (Ctrl miR: 115.56 ± 49 μm3; miR-25: 151.67 ± 63.2 μm3). ER/mitochondria colocalization was estimated by the average volume of overlapping areas (Ctrl miR: 267.89 ± 123.93 μm3; miR-25: 230.7 ± 103.26 μm3). n = 10 independent experiments.
(E and F) [Ca2+]m in permeabilized cells stimulated with 4 μM (mitochondrial Ca2+ uptake rate: Ctrl miR: 11.44 ± 0.49 μM/s; miR-25: 6.01 ± 0.31 μM/s; E) or 1 μM (mitochondrial Ca2+ uptake rate: Ctrl miR: 0.61 ± 0.04 μM/s; miR-25: 0.32 ± 0.01 μM/s; F) EGTA-buffered fixed [Ca2+]. n = 14 independent experiments.
∗p < 0.05; error bars correspond to mean ± SEM. See also Figure S2.
We then investigated the mitochondrial membrane potential (ΔΨm), the driving force for Ca2+ accumulation, and the morphology of mitochondria, i.e., both the contacts with the ER (which were shown to be a critical determinant of rapid Ca2+ transfer between the two organelle [19–21]) and the formation of largely interconnected tubules, which favors Ca2+ diffusion within mitochondria. On the former aspect, measurements with the ΔΨm-sensitive fluorescent dye tetramethylrhodamine methyl ester (TMRM) revealed no difference between miR-overexpressing and control HeLa cells (Figure 2C). As to morphology, mitochondrial labeling with the fluorescent probe mtDsRed showed that miR-25 overexpression causes no significant difference in mitochondrial volume or number (Figure 2D). Similarly, cotransfection with mtDsRed and an ER-targeted GFP showed no difference in the number of contact sites (Figure 2D, contact sites in white).
Overall, the data reveal that the [Ca2+]m reduction caused by miR-25 should be ascribed to reduction of mitochondrial Ca2+ uptake through MCU. To further confirm this notion, we measured mitochondrial Ca2+ accumulation in permeabilized cells. For this purpose, HeLa cells were perfused with a solution mimicking the intracellular milieu (IB), supplemented with 2 mM EGTA, and permeabilized with digitonin for 1 min. The perfusion buffer was then changed to IB with an EGTA-buffered [Ca2+] of 4 μM (Figure 2E) or 1 μM (Figure 2F), eliciting a gradual rise in [Ca2+]m that reached a plateau value of ∼80 and ∼20, respectively. At both buffered [Ca2+], miR-25 overexpression causes a marked reduction in the rate of Ca2+ accumulation into mitochondria.
Mitochondrial Ca2+ alterations induced by miR-25 could be reverted by MCU re-expression in miR-25-expressing cells (Figure S2A) and, accordingly, this rescued Ca2+ affinity was mirrored in enhanced susceptibility to Ca2+-dependent apoptosis (Figure S2B). Moreover, 22Rv1 prostatic cells, which possess very high levels of miR-25 (see Figure 3), were strongly sensitized to apoptosis after MCU overexpression (Figure S2C). The increased ability of mitochondria to accumulate Ca2+ is a fundamental aspect in MCU-related promotion of cell death: indeed, apoptosis induction observed in MCU-overexpressing HeLa cells was almost abolished in the presence of intracellular Ca2+ buffer BAPTA (Figure S2D).
Figure 3.
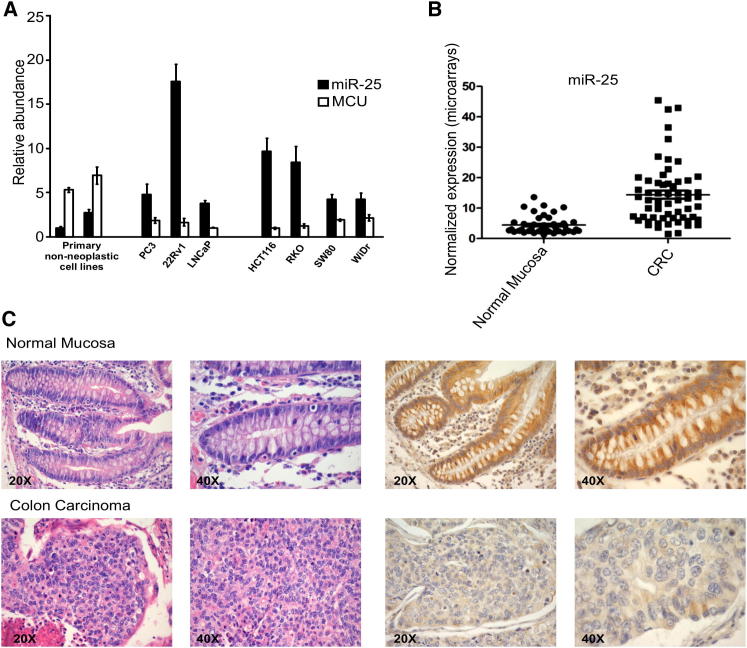
Inhibition of MCU Levels by miR-25 Is a Key Aspect in Human Colon Cancer Progression
(A) miR-25 and MCU mRNA expression levels were analyzed by quantitative real-time PCR in three prostate cancer (PC3, 22Rv1, LnCaP), four colon cancer (HCT116, RKO, SW80, WiDr), and primary nonneoplastic cell lines. RNU6B and 18S expression were used to normalize miR-25 and MCU expression results, respectively, for each sample. Primary nonneoplastic cells present very low abundance of miR-25 and high MCU levels, whereas cancer lines are characterized by inverse correlation between miR-25 levels and MCU mRNA expression. Error bars correspond to mean ± SEM of n = 3 independent experiments.
(B) miRNA expression was assessed in 44 normal mucosa samples and 59 stage 2–3 CRC samples via microarray. The graph shows the average expression level of miR-25 in both groups. miR-25 was significantly overexpressed in cancer samples, as compared to normal mucosa (p < 0.0001).
(C) Upper row: normal colonic mucosa (routinely stained with hematoxylin and eosin, at left) demonstrated strong cytoplasmic granular reactivity with the anti-MCU antibody (immunoperoxidase staining performed on formalin-fixed paraffin-embedded tissue sections, at right). Lower row: poorly differentiated colonic adenocarcinoma with solid pattern of growth (hematoxylin and eosin, at left) showing low level of reactivity with the anti-MCU antibody (immunoperoxidase staining, at right). Two neoplastic cells with cytoplasmic immunoreactivity of moderate intensity can be observed.
See also Figure S3.
Finally, although miR-25 has also been reported to exert antiapoptotic effects via interference with the expression of proapoptotic proteins, such as Bim [22], TRAIL [23], and PTEN [24], these results show how MCU can be considered a fundamental target of miR-25-dependent apoptosis inhibition.
Inhibition of MCU Levels by miR-25 Is a Key Aspect in Human Colon Cancer Progression
We then extended the analysis to cancer cells and tissues. We first evaluated cell lines derived from human carcinomas, in which miR-25 was reported to be highly expressed [24–26]. Both in PC3, LnCaP, and 22Rv1 (derived from prostate cancer) and in HCT116, RKO, SW80, and WiDr (derived from colon cancer) cell lines, we detected an inverse correlation between miR-25 levels and MCU mRNA expression, with high miR-25 levels and low MCU expression levels in cancer lines, compared to primary nonneoplastic cells (Figure 3A). We then directly investigated human poorly differentiated colonic adenocarcinoma samples by immunohistochemistry and microarray. Also in this case, a significant difference in miR-25 expression levels was detected (Figure 3B), which correlates with a downregulation of MCU expression. Indeed, in colonic adenocarcinoma samples with high miR-25 expression levels, MCU was virtually undetectable by immunohistochemistry in cancerous tissues, compared to relatively high protein abundance in the normal mucosa (Figure 3C).
To validate that miR-25 exerts its biological activity through its effect on MCU, we transfected HeLa cells with short hairpin RNA (shRNA) targeting MCU: as for miR-25, shRNA-MCU decreases MCU abundance and increases proliferation (Figure S3A), indicating that MCU targeting is important for the growth-promoting activity of miR-25. We also tested the ability of MCU to inhibit the proliferation. We generated PC3 cells that stably expressed a MCU-FLAG-tagged construct (MCU-FLAG), in which MCU level and activity was increased relative to that in empty vector (pcDNA3) stable clones (Figures S3B and S3C), and found that they formed lower numbers of colonies in soft agar compared to control pcDNA3 stable clones (Figure S3D).
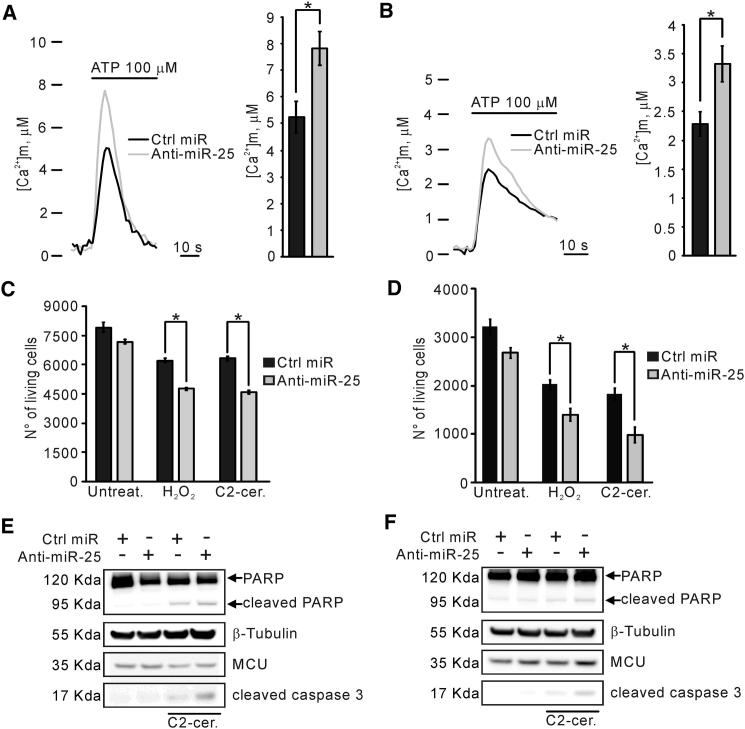
We then investigated whether miR-25-dependent inhibition of mitochondrial Ca2+ uptake, and the ensuing resistance to apoptosis, could be specifically reversed in cancer cells. For this purpose, we overexpressed anti-miR-25 in the PC3 and HCT116 cells lines investigated in Figure 3. In both cell types, anti-miR-25 expression caused an ∼40% increase in the [Ca2+]m rise evoked by 100 μM ATP (Figures 4A and 4B). Accordingly, sensitivity to C2-ceramide and H2O2 were enhanced, as revealed by the lower viability (Figures 4C and 4D) and increased PARP and caspase-3 cleavage (Figures 4E and 4F) detected in anti-miR-25-expressing cells. These data were also confirmed measuring cellular positivity to annexin V (Figures S4A and S4B).
Figure 4.
Regulation of miR-25 Levels Strongly Sensitizes Cells to Ca2+-Dependent Apoptotic Stimuli
(A) [Ca2+]m peaks in PC3 cells: Ctrl miR: 5.25 ± 0.59 μM; anti-miR-25: 7.81 ± 0.64 μM. n = 16 independent experiments.
(B) [Ca2+]m peaks in HCT116 cells: Ctrl miR: 2.28 ± 0.21 μM; anti-miR-25: 3.32 ± 0.31 μM. n = 16 independent experiments.
(C and D) Microscopy counts of cell viability in PC3 (C) and HCT116 (D) cells. Treatments with H2O2 (500 μM for 2 hr) and C2-ceramide (C2-cer.; 40 μM for 2 hr) reveal a more efficient apoptosis induction after anti-miR-25 transfection. n = 3 independent experiments.
(E and F) Immunoblot shows increased levels of cleaved PARP and cleaved caspase-3 in anti-miR-25-expressing PC3 (E) and HCT116 (F) cells after treatment with C2-ceramide (C2-cer.; 40 μM for 2 hr).
∗p < 0.05; error bars correspond to mean ± SEM. See also Figure S4.
Overall, the data identify a microRNA (miR-25), highly expressed in cancer cells, that by targeting the newly discovered calcium channel of mitochondria reduces the sensitivity of cancer cells to apoptotic agents. This not only represents conclusive evidence of the key role of organelle Ca2+ accumulation in the mitochondria-dependent apoptotic routes but also highlights a novel, unexpected target in cancer therapy. Now, the exciting task of unveiling the structural and functional properties of this long-awaited component of the calcium signaling machinery of the cell finds an immediate translational application in a disease area of paramount importance.
Experimental Procedures
Cell Culture and Transient Transfection
HeLa, Hek293, HCT116, and RKO cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum (FCS), L-glutamine, and penicillin/streptomycin in 75 cm2 Falcon flasks. PC3, 22Rv1, and LnCaP cells were cultured in RPMI 1640, supplemented with 10% FCS, 2 mM L-glutamine, and penicillin/streptomycin, in 75 cm2 Falcon flasks.
For aequorin experiments, cells were seeded onto 13 mm glass coverslips and allowed to grow to 75% confluence; for microscopy counts of cell viability, mitochondrial/reticular morphology analysis, and mitochondrial membrane potential measurements, cells were seeded on 24 mm glass coverslip in the same growth conditions.
Lipofectamine 2000 was used as transfectant according to the manufacturer’s recommendations. For aequorin measurements, we used mtAEQmut for HeLa cells and mtAEQ for PC3 and HCT116 cells. All measurements were performed 24 hr after transfection. All miR and anti-miR molecules (hsa-miRNA precursor and hsa-miRNA inhibitor) were purchased from Ambion. shRNA targeting MCU (TRCN0000133861) was purchased from Sigma-Aldrich.
Vectors and Luciferase Assay
Portions of 3′ UTR of human MCU and MICU1 genes, containing miR-25 putative target regions, were amplified through PCR; primers are indicated in Supplemental Experimental Procedures.
Real-Time RT-PCR to Evaluate miRNA and mRNA Expression
Total RNA was extracted from cells with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions (see Supplemental Experimental Procedures).
Aequorin Measurements
Probes employed were chimeric aequorins targeted to the endoplasmic reticulum (erAEQmut), cytosol (cytAEQ), and mitochondria (mtAEQmut). “AEQ” refers to wild-type aequorin, and “AEQmut” refers to a low-affinity D119A mutant of aequorin, as described previously (see Supplemental Experimental Procedures and [15]).
Immunoblotting
Total cell lysates were prepared in RIPA buffer, and standard immunoblotting procedures were used (Supplemental Experimental Procedures).
Apoptosis Assay
After 24 hr transfection with the indicated miR, cells were treated with apoptotic stimuli (H2O2, C2-ceramide, or staurosporine), washed three times in PBS, and then fixed with 4% formaldehyde for 10 min at room temperature (RT). Cells were rinsed with PBS, and 0.1 μg/ml DAPI was added for 10 min at RT. After washing with PBS, the cells were detected with fluorescence microscopy, and cells with condensed and/or fragmented chromatin indicative of apoptosis were not counted as living cells. 250 fields per well were counted using a Scanˆr high-content-throughput system (Olympus).
Immunohistochemistry
Sections (4 μm thick) were cut from formalin-fixed paraffin-embedded blocks. One section for each block was routinely stained with hematoxylin and eosin for histological examination (Supplemental Experimental Procedures).
Microarray and Data Analysis
RNA labeling and hybridization on microRNA microarray chips (ArrayExpress accession number A-MEXP-258) were performed as described previously [25]. Raw data were normalized and analyzed in GeneSpring GX software version 7.3 (Silicon Genetics or Agilent Technologies). Values below 0.01 were set to 0.01. Each measurement was divided by the 50th percentile of all measurements in that sample. GeneSpring software generated a unique value for each miRNA, performing the average of four probes. Graphs and statistical analyses were performed using GraphPad Prism 5 software.
Acknowledgments
We thank E. Magri for technical assistance. This research was supported by the Italian Association for Cancer Research (AIRC); Telethon (GGP09128 and GGP11139B); local funds from the University of Ferrara; the Italian Ministry of Education, University and Research (COFIN, FIRB, and Futuro in Ricerca); the Italian Cystic Fibrosis Research Foundation and Italian Ministry of Health to P.P.; the Italian Ministry of Health to A.R.; grants from the Italian Ministry of Health and Ministry of Education, University and Research, the European Union (ERC mitoCalcium, 294777 and FP7 “MyoAGE,” 223576), the National Institutes of Health (#1P01AG025532-01A1), the Cariparo Foundation (Padua), AIRC, and Telethon-Italy (GPP1005A, GGP11082) to R.R.; and grants from AIRC, the Italian Ministry of Education, University and Research, FIRB program 2011 (RBAP11BYNP), and Regione Emilia Romagna to M.N. S. Marchi was supported by a FIRC fellowship.
Supplemental Information
References
- 1.Baughman J.M., Perocchi F., Girgis H.S., Plovanich M., Belcher-Timme C.A., Sancak Y., Bao X.R., Strittmatter L., Goldberger O., Bogorad R.L. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Stefani D., Raffaello A., Teardo E., Szabò I., Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clapham D.E. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- 4.Pinton P., Giorgi C., Siviero R., Zecchini E., Rizzuto R. Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene. 2008;27:6407–6418. doi: 10.1038/onc.2008.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giorgi C., Baldassari F., Bononi A., Bonora M., De Marchi E., Marchi S., Missiroli S., Patergnani S., Rimessi A., Suski J.M. Mitochondrial Ca(2+) and apoptosis. Cell Calcium. 2012;52:36–43. doi: 10.1016/j.ceca.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roderick H.L., Cook S.J. Ca2+ signalling checkpoints in cancer: remodelling Ca2+ for cancer cell proliferation and survival. Nat. Rev. Cancer. 2008;8:361–375. doi: 10.1038/nrc2374. [DOI] [PubMed] [Google Scholar]
- 7.Perocchi F., Gohil V.M., Girgis H.S., Bao X.R., McCombs J.E., Palmer A.E., Mootha V.K. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature. 2010;467:291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 9.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lovat F., Valeri N., Croce C.M. MicroRNAs in the pathogenesis of cancer. Semin. Oncol. 2011;38:724–733. doi: 10.1053/j.seminoncol.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Maragkakis M., Reczko M., Simossis V.A., Alexiou P., Papadopoulos G.L., Dalamagas T., Giannopoulos G., Goumas G., Koukis E., Kourtis K. DIANA-microT web server: elucidating microRNA functions through target prediction. Nucleic Acids Res. 2009;37(Web Server issue):W273–W276. doi: 10.1093/nar/gkp292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong H., Paquette M., Williams A., Zoeller R.T., Wade M., Yauk C. Thyroid hormone may regulate mRNA abundance in liver by acting on microRNAs. PLoS ONE. 2010;5:e12136. doi: 10.1371/journal.pone.0012136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinton P., Rimessi A., Romagnoli A., Prandini A., Rizzuto R. Biosensors for the detection of calcium and pH. Methods Cell Biol. 2007;80:297–325. doi: 10.1016/S0091-679X(06)80015-4. [DOI] [PubMed] [Google Scholar]
- 16.Scorrano L., Oakes S.A., Opferman J.T., Cheng E.H., Sorcinelli M.D., Pozzan T., Korsmeyer S.J. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 2003;300:135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 17.Pinton P., Ferrari D., Rapizzi E., Di Virgilio F., Pozzan T., Rizzuto R. The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: significance for the molecular mechanism of Bcl-2 action. EMBO J. 2001;20:2690–2701. doi: 10.1093/emboj/20.11.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garrido C., Galluzzi L., Brunet M., Puig P.E., Didelot C., Kroemer G. Mechanisms of cytochrome c release from mitochondria. Cell Death Differ. 2006;13:1423–1433. doi: 10.1038/sj.cdd.4401950. [DOI] [PubMed] [Google Scholar]
- 19.Giorgi C., De Stefani D., Bononi A., Rizzuto R., Pinton P. Structural and functional link between the mitochondrial network and the endoplasmic reticulum. Int. J. Biochem. Cell Biol. 2009;41:1817–1827. doi: 10.1016/j.biocel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm S. The ER-mitochondria interface: the social network of cell death. Biochim. Biophys. Acta. 2012;1823:327–334. doi: 10.1016/j.bbamcr.2011.11.018. [DOI] [PubMed] [Google Scholar]
- 21.Simmen T., Lynes E.M., Gesson K., Thomas G. Oxidative protein folding in the endoplasmic reticulum: tight links to the mitochondria-associated membrane (MAM) Biochim. Biophys. Acta. 2010;1798:1465–1473. doi: 10.1016/j.bbamem.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H., Zuo Z., Lu X., Wang L., Wang H., Zhu Z. MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol. Rep. 2012;27:594–598. doi: 10.3892/or.2011.1530. [DOI] [PubMed] [Google Scholar]
- 23.Razumilava N., Bronk S.F., Smoot R.L., Fingas C.D., Werneburg N.W., Roberts L.R., Mott J.L. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology. 2012;55:465–475. doi: 10.1002/hep.24698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poliseno L., Salmena L., Riccardi L., Fornari A., Song M.S., Hobbs R.M., Sportoletti P., Varmeh S., Egia A., Fedele G. Identification of the miR-106b∼25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci. Signal. 2010;3:ra29. doi: 10.1126/scisignal.2000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lanza G., Ferracin M., Gafà R., Veronese A., Spizzo R., Pichiorri F., Liu C.G., Calin G.A., Croce C.M., Negrini M. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol. Cancer. 2007;6:54. doi: 10.1186/1476-4598-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishida N., Nagahara M., Sato T., Mimori K., Sudo T., Tanaka F., Shibata K., Ishii H., Sugihara K., Doki Y., Mori M. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin. Cancer Res. 2012;18:3054–3070. doi: 10.1158/1078-0432.CCR-11-1078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.