SUMMARY
During endochondral bone development, the first osteoblasts differentiate in the perichondrium surrounding avascular cartilaginous rudiments; the source of trabecular osteoblasts inside the later bone is, however, unknown. Here, we generated tamoxifen-inducible transgenic mice bred to Rosa26R-LacZ reporter mice to follow the fates of stage-selective subsets of osteoblast lineage cells. Pulse-chase studies showed that osterix-expressing osteoblast precursors, labeled in the perichondrium prior to vascular invasion of the cartilage, give rise to trabecular osteoblasts, osteocytes, and stromal cells inside the developing bone. Throughout the translocation, some precursors were found to intimately associate with invading blood vessels, in pericyte-like fashion. A similar coinvasion occurs during endochondral healing of bone fractures. In contrast, perichondrial mature osteoblasts did not exhibit perivascular localization and remained in the outer cortex of developing bones. These findings reveal the specific involvement of immature osteoblast precursors in the coupled vascular and osteogenic transformation essential to endochondral bone development and repair.
INTRODUCTION
Most mammalian bones develop through endochondral ossification. This process depends on the coordinate proliferation, differentiation, and interaction of various cell types but ultimately relies on osteoblast lineage cells for the actual synthesis and mineralization of the bone matrix. The origin of osteoblasts and osteogenic stromal cells in vivo is, however, incompletely understood.
At the start of endochondral ossification, mesenchymal condensations that prefigure the future long bones first develop into cartilage. When cells in the central area differentiate into hypertrophic chondrocytes, osteoblasts appear in the surrounding perichondrium and generate the “bone collar,” the provisional bone cortex. At this time, the initial vascular invasion of the cartilaginous template occurs, triggering the formation of the primary ossification center inside the bone (see Figure 1A). Endothelial cells and osteoclasts first accumulate in the perichondrial region and are attracted to invade and erode the cartilage (Karsenty and Wagner, 2002; Kronenberg, 2007). Concomitantly, osteoblasts and marrow cells populate the newly excavated, highly vascularized area.
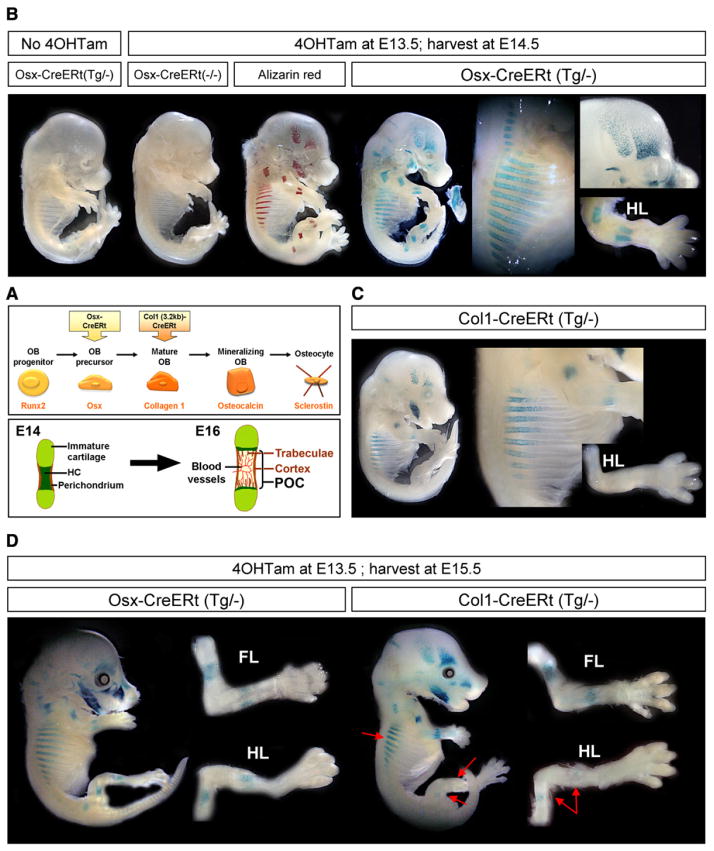
Figure 1. Transgenic Mice with Inducible CreERt Expression in Osteoblast Lineage Cells.
(A) Simplified representation (top) of the line of progression of osteoblast (OB) differentiation and the typical gene product by which each stage is characterized (orange, below). The Osx-CreERt and Col1(3.2 kb)-CreERt transgenes employed in this study start to be expressed in osteoblast precursors and mature oste-oblasts, respectively. Schematic outline (bottom) of the initiation of bone formation during development, occurring between E14 and E16 in the stylopod and zeugopod bones of mice. The initial invasion of the cartilaginous bone model is associated with its transformation into the primary ossification center (POC) that contains bone trabeculae and is surrounded by cortical bone. HC, hypertrophic cartilage.
(B) Whole-mount X-gal staining of E14.5 Osx-CreERt embryos carrying a Rosa26R reporter transgene. (left to right) Embryos unexposed to 4OHTam or not carrying the Osx-CreERt transgene, Alizarin red mineralization staining and corresponding pattern of LacZ activity in Osx-CreERt(Tg/−) embryos exposed to 4OHTam at E13.5. The mandible became disconnected from the embryo proper. Magnifications (right) highlight X-gal staining in the bony regions of the ribs, calvaria and hind limb (HL).
(C) X-gal staining of Col1-CreERt(Tg/−) embryos harvested similarly at E14.5 after 4OHTam exposure at E13.5. Right, magnified ribs and HL.
(D) E15.5 Osx- and Col1-CreERt(Tg/−) embryos injected with 4OHTam at E13.5, and magnified HL and forelimb (FL). Red arrows, reduced staining in the ribs and HL of Col1-CreERt embryos, compared with Osx-CreERt embryos.
See also Figure S1 on the generation and characterization of the osteoblastic CreERt mice.
Osteoblasts differentiate from mesenchymal progenitors that express Runx2, a transcription factor required for full osteoblastic differentiation (Nakashima et al., 2002; Komori et al., 1997; Otto et al., 1997). Osterix (Osx), genetically downstream of Runx2, is another early transcription factor required for osteoblastogenesis (Nakashima et al., 2002). As they further differentiate, osteoblasts produce matrix proteins, including the main bone constituent, type I collagen (Col1). Mature osteoblasts that become embedded in the bone matrix differentiate into osteocytes (Marie, 2008; Karsenty and Wagner, 2002; see Figure 1A).
Fusion of Cre recombinase to a modified estrogen receptor ligand-binding domain (ERt) allows the temporal induction of its activity by administration of tamoxifen or 4-hydroxytamoxifen (4OHTam) (Metzger et al., 1995). The clearance of tamoxifen from the cell mediates the return of the CreERt complex to the inactive state, thus creating a window of time for gene modulation (Feil et al., 1996). A conditional reporter gene, such as the Rosa26R β-galactosidase (LacZ) transgene (Soriano, 1999), can indicate the recombination by the blue staining of the cell with the LacZ substrate 4-chloro-3-indolyl-β-D-galactopyrano-side (X-gal). Thus, this system allows genetic marking of CreERt-expressing cells at the time of tamoxifen injection, and the subsequent tracing of these cells and their progeny (see Maes et al., 2007 and references therein).
Here, we used the CreERt system to develop mouse models for osteoblast lineage tracing in vivo. By employing the Osx-and Collagen I(α1) (Col1) gene promoters, we could label and follow stage-selective osteogenic cells during bone development. Our data reveal that specific pools of osteoblast lineage cells display different fates and capacities to initiate new sites of bone formation.
RESULTS
CreERt Mice Driven by Stage-Specific, Osteoblast Lineage Promoters
Osx-CreERt and Col1-CreERt mice were generated (see Supplemental Experimental Procedures available online) and crossed to Rosa26R LacZ reporter mice (Soriano, 1999) (Figure S1A). X-gal staining of the progeny retrieved from pregnant mice injected with 4OHTam at specific times identified cells that had undergone CreERt-mediated recombination and the descendants thereof (Figure S1B). To assess the fates of osteoblast lineage cells originating in the perichondrium, we genetically labeled such cells prior to the development of the primary ossification center, a process initiating at or beyond E14.5 (Figure 1A). We first confirmed that the CreERt-activity was ligand-dependent and specific throughout embryogenesis. In the absence of 4OHTam, no leaky recombination occurred in either of the mouse lines, as evidenced by the lack of X-gal staining at any age between E14.5 and E17.5, and no nonspecific LacZ-activity was seen in the skeleton of embryos lacking the Osx- or Col1-CreERt transgene (Figure 1B; data not shown).
When pregnant mice were pulsed with 4OHTam at E13.5, both Osx-CreERt(Tg/−) and Col1-CreERt(Tg/−) embryos showed intense X-gal staining after sacrifice at E14.5 (Figures 1B and 1C). Macroscopic and histological analysis confirmed the skeleton-specificity of the induced LacZ expression (Figures S1C–S1F). X-gal staining in E14.5 Osx-CreERt(Tg/−) embryos that received 4OHTam at E13.5 was manifest in the mandible, the lateral calvarial bones, the proximal part of the ribs and the proximal limb bones (Figure 1B), whereas time-matched Col1-CreERt(Tg/−) embryos showed roughly this pattern but a reduced distribution throughout the skeleton (Figure 1C, note, for instance, the absence of staining in the hind limbs). These observations verified the tightly tamoxifen- and Cre-dependent LacZ activation specifically in the skeleton of Osx-and Col1-CreERt(Tg/−) embryos and also suggested subtle differences between the mouse lines.
To further compare the labeling patterns in Osx- and Col1-CreERt mice, we next analyzed embryos at E15.5, after pulsing the mother at E13.5 (Figure 1D). The X-gal staining patterns were analogous to those observed at E14.5. Hence, Col1-CreERt(Tg/−) embryos again had a reduced extent of LacZ activity compared with Osx-CreERt(Tg/−) mice (Figure 1D). Thus, as expected, Osx-CreERt was expressed earlier in development than Col1-CreERt.
Lineage Tracing Reveals Different Fates for Perichondrial Osteoblast Subsets In Vivo
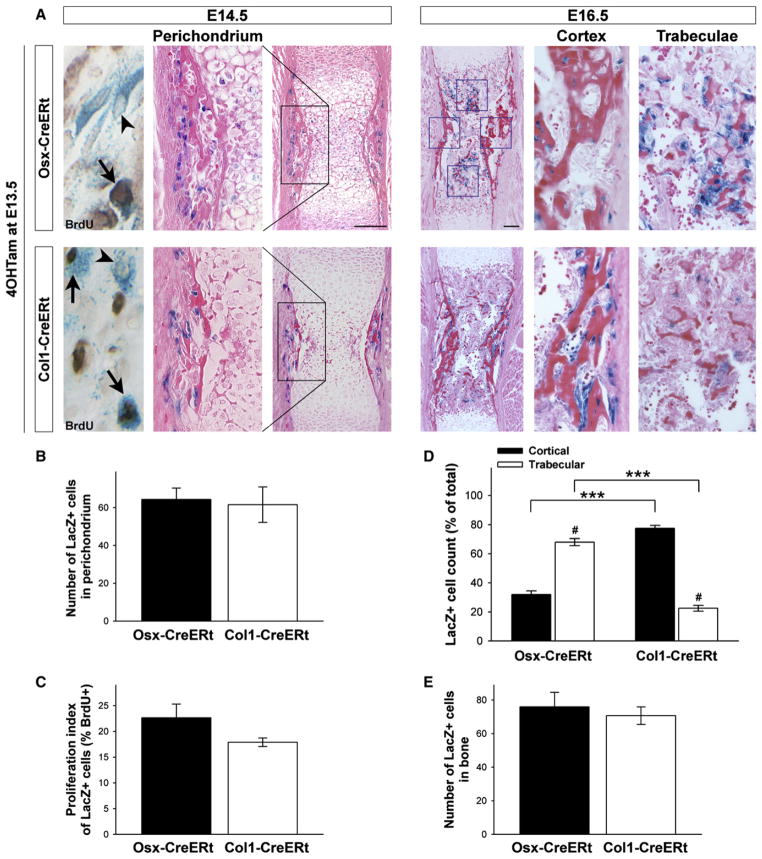
In line with the forelimbs running developmentally ahead of the hind limbs, the humerus, radius and ulna stained blue in Osx-as well as Col1-CreERt(Tg/−) embryos exposed to 4OHTam at E13.5. We, therefore, focused our tracing experiments on the humerus. Histology confirmed its cartilaginous state at E14.5 (Figure 2A). Both Osx- and Col1-CreERt(Tg/−) embryos displayed mosaic LacZ labeling of cells in the perichondrium (Figure 2A, left). The number of labeled cells, designated Osx/LacZ+ and Col1/LacZ+ cells, respectively, was not significantly different (Figure 2B) (p = 0.84). Both sets of labeled cells contained BrdU-positive cells (Figure 2A, far left) and their proliferation indices were not significantly different (Figure 2C) (p = 0.16).
Figure 2. Perichondrial Osteoblast Lineage Cells Marked by Osx- or Col1-CreERt Expression Display Differential Fates in Developing Bones.
(A) Osx-CreERt (top) and Col1-CreERt (bottom) humerus sections of mice pulsed with 4OHTam at E13.5 and sacrificed at E14.5 (left) or at E16.5 (right) and stained for X-gal and eosin or BrdU (far left; arrowheads indicate single LacZ+ cells, arrows point at LacZ+/BrdU+ double-positive cells). Scale bars, 100 μm. (B and C) Quantification of the perichondrial labeling (B) and the proliferation index of the labeled cells (C) in Osx- and Col1-CreERt(Tg/−) mice at E14.5. The number of LacZ+ cells and the percentage that additionally stained positive for BrdU were determined in a defined region, encompassing the central portion of the perichondrium corresponding to the black boxed area in (A). Bars, mean ± SEM; n = 3.
(D and E) Distribution (D) and total number (E) of traced Osx/LacZ+ and Col1/LacZ+ cells at E16.5. Cells were counted in fixed cortical and trabecular bone areas (blue squares in A). ***p < 0.001 for comparison between genotypes; #p < 0.001 between locations. Bars, mean ± SEM; n = 5–9.
See also Figure S2 on the lineage tracing kinetics of the system.
In order to trace the fates of the labeled cells throughout the next steps of endochondral bone development, we needed to define the time window of labeling and verify that no further de novo labeling of cells occurred beyond this period. We, therefore, assessed the kinetics of LacZ induction in the embryos after administration of 4OHTam (Figure S2). X-gal staining became detectable as early as 8 hr after 4OHTam injection and its pattern of skeletal distribution remained similar thereafter. By using the differential developmental timing of the various skeletal elements as an internal clock, we could ascertain that the induced activity of both Osx-CreERt (Figure S2) and Col1-CreERt (data not shown) was fast and transient in vivo, with an activation window of less than 24 hr following 4OHTam administration.
Interestingly, as we traced cells labeled at E13.5 for 3 days, the staining patterns in the two transgenic mouse lines became markedly different (Figure 2A, right). Osx/LacZ+ cells sparsely localized along the outer cortical bone, while they abundantly occupied the trabecular bone formed inside the shaft (Figure 2A, upper right). In sharp contrast, Col1/LacZ+ cells localized almost exclusively on the cortical surfaces and were hardly detected inside the bone cavity (Figure 2A, lower right). These differing patterns were confirmed by quantification in designated trabecular and cortical areas (blue squares in Figure 2A): 70% of the counted Osx/LacZ+ cells were located in the trabecular regions, in contrast to only 20% of the Col1/LacZ+ cells (Figure 2D). The total cell counts were similar between the two mouse lines (Figure 2E), suggesting that subtle differences in the proliferative potential of the respective cell populations were unlikely to account for the observed positional diversity. When 4OHTam was given after the formation of the primary ossification center (at or beyond E15.5), the Col1-CreERt transgene was capable of labeling trabecular osteoblasts inside the bone (not shown). These data indicate that perichondrial osteoblast lineage cells marked by differentiation stage-selective gene activity display differential destinies in developing bones. Cells of the Osx/LacZ+ pool appear predominantly to move inside the bone to the trabecular regions, whereas Col1/LacZ+ cells predominantly remain on the cortex.
Colabeling of Osx-CreERt-Expressing Chondrocytes Does Not Contribute Much to the Trabecular Osteoblast Pool
Col1-CreERt labeling was exclusive to the perichondrium, whereas Osx-CreERt(Tg/−) embryos also showed mosaic labeling in the pre- and early hypertrophic chondrocyte zones (Figure 3A), in line with the endogenous expression of Osx in prehypertrophic chondrocytes (Hilton et al., 2005) (see also Figure S4A). No self-renewing population was targeted, as Osx/LacZ+ chondrocytes completely disappeared by 3 days postinjection (E16.5) (Figure 3A). To assess whether these cells had died by apoptosis or had transdifferentiated into osteogenic cells, we employed a cartilage-directed Col2-CreERt mouse line (Nakamura et al., 2006) (Figure 3B).
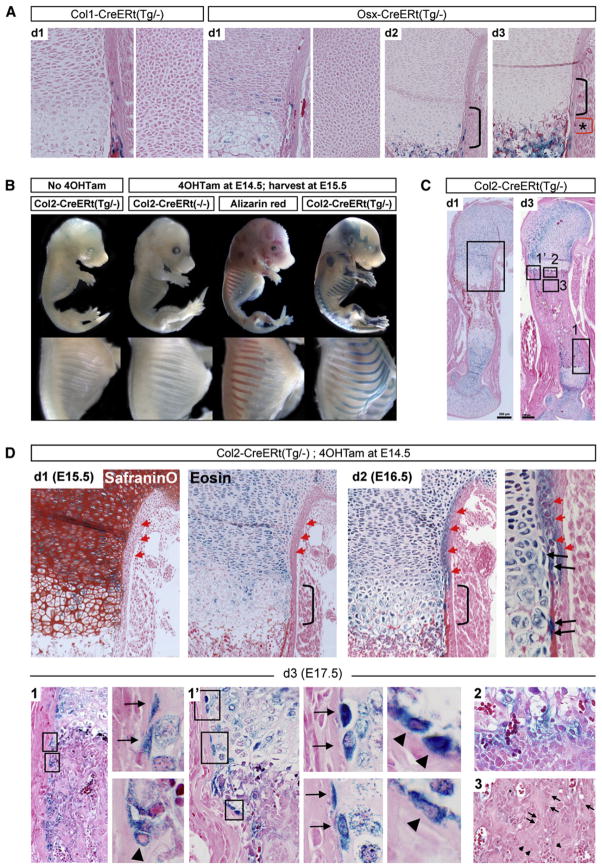
Figure 3. Colabeling of Osx-CreERt-Expressing Chondrocytes Does Not Contribute Detectably to the Trabecular Osteoblast Pool.
(A) Growth cartilage of the proximal humerus on 3 successive days (d1–d3) after giving 4OHTam at E13.5. Osx/LacZ+ cells are detected in the (pre-)hypertrophic zones (left) at d1, but not in the round chondrocyte zone (right). Few Osx/LacZ+ hypertrophic chondrocytes remain at d2 (bracket). By d3, the cartilage is devoid of blue cells, while Osx/LacZ+ cells are abundant in the metaphysis beyond the chondro-osseous junction (asterisk-marked red bracket).
(B) X-gal staining of E15.5 Col2-CreERt embryos either or not exposed to 4OHTam at E14.5. Note cartilage-confined X-gal staining, complementing Alizarin red staining.
(C) Col2-CreERt humeri stained by X-gal at d1 and d3 after 4OHTam at E14.5. Note cartilage-specific labeling at d1 and a more complex staining pattern by d3, analyzed in detail in the indicated areas in (D). Scale bar, 200 μm.
(D) SafraninO and eosin staining (upper left) on consecutive d1 sections showing abundant Col2/LacZ+ cells in the immature cartilage, infiltration in the hypertrophic layer (bracket), and absence from the perichondrium (red arrowheads). Increased hypertrophic and perichondrial labeling at d2 (upper right), including stretched cells near the cartilage-perichondrium interface, arranged perpendicular to the bulk of the more central chondrocytes (black arrows). (Lower panels) (d3) Area 1 (rotated) and 1′ (as indicated in C, with magnified views on the right) showing Col2/LacZ+ cells at the cartilage-perichondrium interface (arrows) and on the endosteal bone surface (arrowheads). Area 2, diffuse blue coloration at the chondro-osseous interface, likely explained by residual LacZ activity released from eroded hypertrophic chondrocytes. Area 3, central metaphyseal regions occupied by trabecular osteoblasts (arrows) and osteocytes (arrowheads) that are generally not blue. Representative sections of n = 3–7 forelimbs analyzed at 15–50 μm intervals are shown. Col2-CreERt(−/−) sections were devoid of X-gal staining (n = 2 per time point).
See also Figure S3.
This Col2-Cre(ERt) line, like many others (Hilton et al., 2007; Sakai et al., 2001) marks both chondrocytes and perichondrial cells and their osteoblastic descendants when active early in bone development, probably by targeting a population of chondro-perichondrial progenitors (Figure S3). By injecting 4OHTam later, at E14.5, this limitation could be largely circumvented in the humerus. We stepwise traced the labeled cells over a 3 day period. Our data (Figures 3C and 3D) suggest that specific cells at the lateral cartilage-perichondrium interface contribute to the endosteal osteoblast pool (osteoblasts on the inner surface of the cortex) (Figure 3D, bottom, 1 and 1′), but terminally matured chondrocytes do not detectably contribute to the osteoblast/osteocyte pool in the central metaphyseal regions beneath the growth cartilage (away from the endosteal surface) (Figure 3D, bottom, 2 and 3). Thus, we conclude that the osteoblasts of the primary ossification center are primarily derived from translocating perichondrial cells, as represented by the Osx/LacZ+ cells.
Differentiation Stage Selectivity of Osx/LacZ+ Cells and Col1/LacZ+ Cells
Since perichondrial Osx/LacZ+ cells move into the bone, whereas Col1/LacZ+ cells hardly do, these cell pools may differ substantially. We closely examined the cells and their direct environment shortly after labeling at E13.5 by transmission electron microscopy (EM), using immediately adjacent sections to identify the LacZ+ cells (blue X-gal staining) within the tissue outline (toluidine blue) (Figures 4A1–4A3) (see Experimental Procedures). Labeled cells analyzed by EM were identified based on their blue coloration in the consecutive thin section throughout the study. The validity of this cell-specific alignment of the EM and LacZ information was confirmed by the detection of X-gal reaction product as dark cytoplasmic deposits on EM in several of the analyzed samples (Figure 4A4).
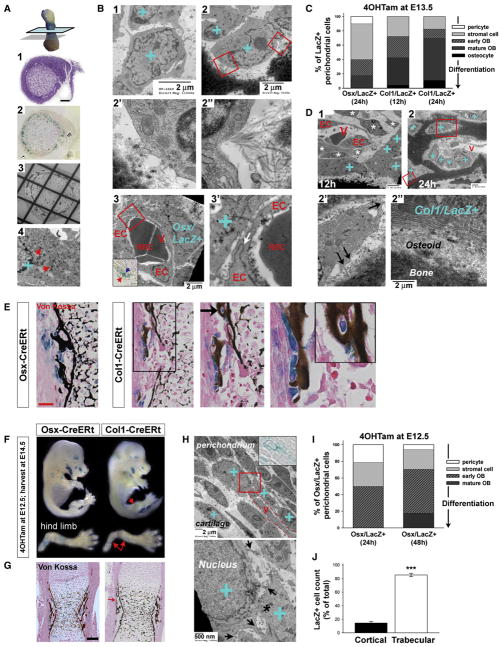
Figure 4. Perichondrial Osx/LacZ+ and Col1/LacZ+ Cells Represent Increasing Stages of Osteogenic Differentiation.
(A) EM methodology. Consecutive middiaphyseal, transversal thin sections analyzed by (A1) toluidine blue, (A2) X-gal, and (A3) EM were aligned. Scale bar, 50 μm. (A4), dark accumulations of X-gal deposits (arrows) as often detected among the granular ER confirmed that blue stained cells identified on A2 could be specifically designated as X-gal+ (blue “+”) on the EM images of the consecutive section.
(B) EM images of perichondrial Osx/LacZ+ cells (blue “+”) 1 day after 4OHTam-induced labeling at E13.5, appearing as loosely focally connected immature stromal cells (B1), early osteoblasts (B2 and boxes magnified in B2′ and B2′), or pericytic cells surrounding the endothelial cells (EC) of perichondrial blood vessels (V) (B3). The inset demonstrates the blue Osx/LacZ+ cell at the light microscopic level as used throughout the study as identification criterion. The high power (B3′) reveals a tight junction (arrow) between the ECs of the immature capillary lacking a basement membrane and containing RBCs. Scale bars, 2 μm.
(C) Scoring analysis by EM of Osx/LacZ+ and Col1/LacZ+ cells at 12 or 24 hr after labeling at E13.5 (40–47 cells analyzed per group).
(D) Representative Col1/LacZ+ cells at 12 (D1) and 24 hr (D2). V, perichondrial capillary vessels. (D1) Typically, stromal and perivascular cells (asterisks) are not labeled whereas the Col1/LacZ+ cells present as mature, cuboidal osteoblasts. (D2 and large box magnified in D2′) A Col1/LacZ+ osteocyte with cell dendrites (arrows). (D2 and small box in D2″) Mature osteoblasts. Scale bars, 2 μm.
(E) Histology at E14.5 showing the relationship between E13.5-labeled LacZ+ perichondrial cells and the bone collar (Von Kossa, black). Note Osx/LacZ+ cells that are unrelated to the mineralized bone, whereas Col1/LacZ+ cells are mostly located on the bone surface (magnification) or embedded in the bone (arrow and inset). Scale bar, 50 μm.
(F) E14.5 embryos exposed to 4OHTam at E12.5. Note X-gal staining in Osx- but not in Col1-CreERt hind limbs (arrows).
(G) Histology of the corresponding humeri stained by Von Kossa. Col1/LacZ+ cells are only rarely found (red arrow). Scale bar, 100 μm.
(H) Representative EM images of Osx/LacZ+ cells labeled at E12.5 and analyzed at E13.5, showing their appearance as immature stromal cells in the loose connective tissue, often in close juxtaposition with capillaries (V, dotted line). Inset, aligned X-gal view. Bottom, magnified view of boxed area, showing cytoplasmatic processes (arrows) and gap junctions (asterisk).
(I) Morphological appearance of Osx/LacZ+ cells scored by EM analysis at 24 or 48 hr after labeling at E12.5, revealing the enrichment of immature cells (14–17 cells analyzed per group).
(J) E12.5-labeled Osx/LacZ+ cell counts in the cortical and trabecular bone regions of E16.5 humeri. Bars, mean ± SEM; n = 2; ***p < 0.001. See also Figure S4.
At E14.5, perichondrial Osx/LacZ+ cells constituted a heterogeneous population. The majority (24/40; 60%) were immature stromal cells displaying large nuclei, very sparse endoplasmic reticulum (ER), and cytoplasmic extrusions, embedded in a loose connective tissue matrix, with some adherence to cells of similar morphology (Figure 4B1). A second class was defined as “early osteoblasts” (9/40); such cells had features resembling stromal cells but were found near collagen-containing matrix, and displayed no or limited active bone formation (Figure 4B2–4B2″). Only a minority of the Osx/LacZ+ cells had an appearance consistent with that of mature osteoblasts (see below). Interestingly, almost 20% of the cells with stromal appearance (10% of all Osx/LacZ+ cells examined) were found alongside perichondrial blood vessels. These cells attained a pronounced pericytic localization with cellular processes enfolding the endothelial lining of red blood cell (RBC)-filled capillaries (Figures 4B3–4B3′).
A parallel analysis of Col1/LacZ+ cells performed 12 or 24 hr after labeling revealed that none of these cells were perivascular, pericyte-like (Figure 4C) (n = 46–47 cells examined). Instead, they were primarily mature, matrix-secreting, and mineralizing osteoblasts and osteocytes, each with abundant arrays of granular ER and/or partially or completely surrounded by osteoid matrix and mineralized bone (Figure 4D). Routine light microscopy confirmed these findings, as Osx/LacZ+ cells were found both on and unrelated to the mineralized bone collar, whereas Col1/LacZ+ cells were primarily associated with the bone surface (Figure 4E, Von Kossa staining with black deposits denoting mineralization).
Selective Labeling and Tracing of the Earliest Osx-CreERt-Expressing Cells
The analysis described above confirmed our expectation that, at E13.5, Col1-CreERt expression was associated with maturing osteoblasts, whereas Osx-CreERt expression was associated with a wider range of osteoblast lineage cells, including immature cells. Particularly these early cells may thus be responsible for the osteogenic colonization of the primary ossification center. To test this hypothesis, we next labeled an Osx/LacZ+ population enriched in immature cells. In agreement with the differential timing of activation of the endogenous Osx- and Col1-gene promoters (see Figures S4A–S4C), Osx-CreERt labeling was achieved at an earlier time than Col1-CreERt labeling. Injecting 4OHTam at E12.5 was the earliest time point that led to marked X-gal staining of the proximal bones of the limbs in Osx-, but not in Col1-CreERt (Tg/−) embryos (Figure 4F; data not shown). Histology at E14.5 showed perichondrial labeling in Osx-CreERt limb sections, while Col1/LacZ+ cells could barely be detected, even in the humerus (Figure 4G). The Osx/LacZ+ cells appeared prior to bone collar formation, as evidenced by the lack of mineralization at E13.5 (Figure S4D, left). These data suggest that giving 4OHTam at E12.5 selectively labeled Osx/LacZ+ cells that had not yet matured to become collagen type I-producing osteoblasts.
EM analysis confirmed that the Osx/LacZ+ pool labeled at E12.5 consisted exclusively of cells classified morphologically as immature stromal cells and early osteoblasts 24 hr after labeling, only progressing to contribute to the pool of mature osteoblasts involved in the bone collar after 48 hr (Figures 4H and 4I). The early labeled Osx/LacZ+ pool was further enriched in pericytic cells positioned on blood vessels (20% of the cells screened) (Figure 4I).
We then traced these immature Osx/LacZ+ cells over the next days (Figure S4D). As the primary ossification center developed, blue cells progressively localized inside the bone cavity and increasingly associated with the primary trabeculae and the endosteal surface of the bone collar (Figure S4D). Using a quantification strategy like that used in Figure 2D, the Osx/LacZ+ cells labeled earlier shifted even more to the primary spongiosa by E16.5 (>80%), with less than 20% of the cells residing in the corticalareas (including the endosteum)(Figure 4J).Thus, a subset of the earliest Osx-CreERt-expressing cells represent precursors of the osteoblast pool populating the primary ossification center.
Osx-Expressing Precursors Give Rise to the Full Differentiation Spectrum of Osteoblast Lineage Cells inside the Growing Bone but Also Include Pericyte-like Cells
Histological and EM analysis at E16.5 showed that the Osx/LacZ+ cells presented as peritrabecular stromal cells, cuboidal osteoblasts covering the trabecular bone surfaces, endosteal osteoblasts, and osteocytes entrapped within the bone (Figures 5A–5C). Many, albeit not all, of the Osx/LacZ+ cells now expressed endogenous Col1 mRNA, as detected by in situ hybridization (ISH) (Figure 5B). Of note, all Col1/LacZ+ cells expressed Col1 mRNA (not shown). Thus, these tracing studies revealed that the earliest Osx-CreERt-expressing cells, or at least a subset of them, gave rise to the full differentiation spectrum of osteoblast lineage cells inside the growing bone, and therefore represent true precursors of trabecular osteoblasts.
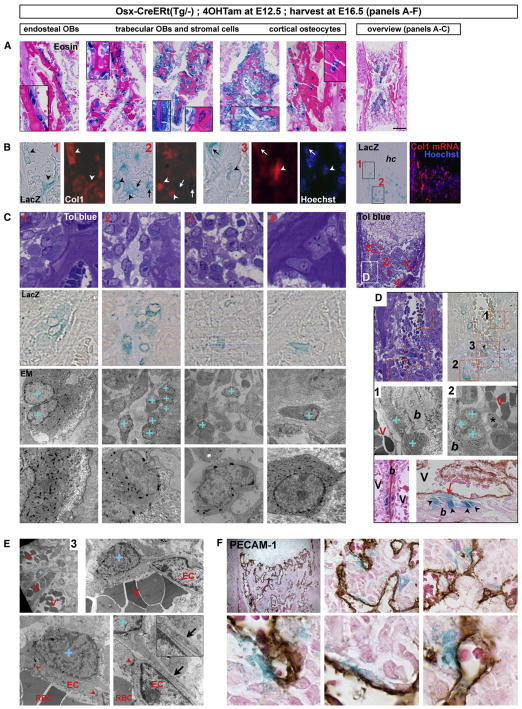
Figure 5. Perichondrial Osx/LacZ+ Cells Include Osteoblast Precursors that Give Rise to Stromal Cells, Osteoblasts, and Osteocytes inside the Developing Bone, and Pericytic Cells.
(A–C) Perichondrial Osx-CreERt-expressing osteoblast precursors labeled at E12.5 give rise to endosteal osteoblasts (OBs), trabecular OBs and peritrabecular stromal cells, and cortical osteocytes by E16.5. Overview sections are shown on the right. (A) Histology with magnifications in insets. Scale bar, 200 μm. (B) Col1 ISH (red signal, overlaid onto contrast or Hoechst images). Arrowheads, Osx/LacZ+ cells expressing Col1 mRNA; arrows, Col1-negative Osx/LacZ+ cells; hc, hypertrophic cartilage. Areas (1) and (2) as from boxes in the overview image; area (3) shows a cortex detail. (C) EM analysis by aligned toluidine blue, LacZ and EM views of the red boxed areas (1–4, as localized in the overview section on the right) with indication of Osx/LacZ+ cells, identified by blue staining, on the parallel EM image by a blue “+”. Bottom, magnified views; note cytoplasmic X-gal deposits in these LacZ+ cells.
(D) Frequent proximity of cuboidal Osx/LacZ+ osteoblasts on mineralized bone (“+” and arrowheads) to RBC-containing blood vessels (V). Top, outline of the region (white box in (C)) and areas analyzed by EM in (D) (1–2) and (E) (3). Bottom, histology by (left) eosin or (right) additional PECAM-1 staining (brown). Note vessels running directly adjacent to Osx/LacZ+ osteoblasts (left panels), or separated only by spindle-shaped perivascular cells (asterisk), some being Osx/LacZ+ (red arrow) (right panels).
(E and F) Pericytic localization of Osx/LacZ+ cells revealed by EM (E) and PECAM-1 IHC (F). (E) Overview and increasing magnifications of area 3 (see D), entry site of vessels into the diaphysis. Arrowheads, tight junctions between ECs; inset and arrow, overlapping cellular extensions of the subendothelial coverage. (F) Top, overview and magnifications of a section stained by X-gal, PECAM-1 (brown), and eosin. Bottom, detailed views of perivascular-located Osx/LacZ+ cells. See also Figure S5, providing molecular characterization of Osx-expressing cells using the Osx-Cre:GFP mouse model.
Analyzing the environment of the Osx/LacZ+ cells at E16.5, we noted that they often resided in close proximity to blood vessels inside the marrow cavity (Figure 5D). Some cells that had become mature cuboidal osteoblasts displayed a prominent polarity with bone matrix on the apical side, lateral neighboring osteoblasts, and blood vessels flanking the basal surface. Sometimes endothelial cells were immediately adjacent to the osteoblasts, while in other cases stretched perivascular cells were seen in between (Figure 5D). Some of these perivascular cells were, in fact, Osx/LacZ+ cells juxtaposed to the endothelium like pericytes (Figures 5D–5F). Such cells displayed a morphology characteristic of undifferentiated cells, with a very prominent nucleus, a small cell body with long and thin processes and sparse granular ER, Golgi apparatus, and organelles. Like pericytes, their cell processes closely enfolded the endothelial lining of the vessel (Figure 5E). Immunohistochemistry (IHC) for the endothelial cell-specific marker platelet/endothelial cell adhesion molecule 1 (PECAM-1) indicated that Osx/LacZ+ cells were abundantly found in these perivascular locations (Figure 5F). Thus, in addition to giving rise to bone-forming osteoblasts in the primary spongiosa, a fraction of the embryonic osteoblast precursors characterized by Osx expression retains an immature identity and a pericytic localization during bone development.
To address the molecular characteristics of Osx-expressing cells, we took advantage of the Osx-Cre:GFP mouse model (Rodda and McMahon, 2006) to allow GFP-based cell sorting (designated as Osx/GFP+ cells) and visual colocalization studies (red fluorescent IHC and ISH) (Figure S5). The same BAC was used to generate both the Osx-Cre:GFP and the Osx-CreERt mice, and the Cre:GFP expression correlates with endogenous Osx mRNA expression and with Osx-CreERt-labeling in terms of pattern and timing in developing bones (see Figures S4 and S5). Like traced Osx/LacZ+ cells, Osx/GFP+ populations encompassed cells of a wide osteoblast differentiation spectrum including non-collagen type I- and osteopontin-expressing immature cells; their genetic profile indicated strong enrichment in Osx- and Runx2-expression (Figures S5A–S5D). Osx/GFP+ cells too could regularly be found in immediate perivascular locations, even in adult bones (Figure S5E). Osx/GFP+ perivascular cells did, however, not fully cover the vascular bed but were rather found scattered on some of the blood vessels. Since qRT-PCR revealed the presence of transcripts for several pericyte markers (RGS5, SM22, and desmin; data not shown) in Osx/GFP+ sorted cell samples, we assessed the potential colocalization of Osx/GFP+ or Osx/LacZ+ cells and pericyte markers in bone sections (Figures S5F–S5I). While α-smooth muscle actin (αSMA), SM22, desmin, and Nerve/Glial antigen 2 (NG2) were indeed detected in a few Osx-expressing cells, the functional significance of their presence is, however, unclear: none of the markers’ expression patterns tracked that of the vascular pattern in bone (PECAM-1 staining on subsequent sections, not shown). Hence, the vasculature inside embryonic and adult bones is unusual in that it largely lacks classical pericyte coverage.
Coinvasion of Osteoblast Precursors and Blood Vessels into Developing Long Bones
The finding that Osx/LacZ+ cells anatomically resembling pericytes were observed both at the time of their initial labeling in the perichondrium and after their tracing to the inside of the bone raised the question whether the perivascular localization of Osx/LacZ+ cells was a consistent feature. We therefore examined the spatial relationship between the Osx/LacZ+ cells and the vasculature throughout cartilage invasion.
Osx/LacZ+ cells closely colocalized with blood vessels and osteoclasts as they accumulated in the perichondrium just prior to the invasion of the hypertrophic cartilage (Figure 6A), as well as during initial invasion (Figure 6B). Osx/LacZ+ cells were detected alongside the vessels and at the angiogenic front (Figures 6A–6C,1). Capillaries traversed through openings in the bone collar and entered the hypertrophic cartilage core as perfused vessels (filled with RBCs) (Figure 6C,2) interspersed with Osx/LacZ+ cells (Figure 6C,3). The initial vascular invasion process and formation of a primary ossification center did not depend on the presence osteoclasts (Figure S6).
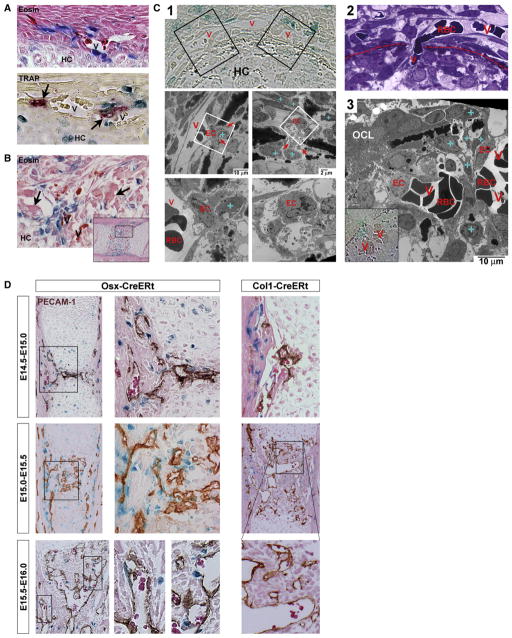
Figure 6. Entry of Osteoblast Precursors into Developing Bones Correlates with Their Colocalization to the Cartilage-Invading Vasculature.
(A and B) Longitudinal sections stained by eosin or TRAP (A, bottom), showing Osx/LacZ+ cells colocalizing with vessels (V, containing RBCs [intense magenta on eosin]) and osteoclasts (arrows) at the time of their accumulation in the middiaphyseal perichondrium just prior to invasion of hypertrophic cartilage (HC) (A) and during the invasion (B).
(C) Analysis of the initial HC invasion process using thin transverse, middiaphyseal sections. Abbreviations: V, vessel; HC, hypertrophic cartilage; EC, endothelial cell; RBC, red blood cell; blue “+”, Osx/LacZ+ cell; OCL, osteoclast. (C1) X-gal view (top) identifying Osx/LacZ+ cells and (below) aligned EM images corresponding to the black boxes on the left and right, respectively. Further magnifications from the respective white boxes are shown below each panel. Note initiation of cartilage invasion with bone-collar-transversing ECs displaying abundant filopodia extensions (red arrows), indicative of sprouting angiogenesis. (C2) Toluidine blue picture of a RBC-filled vessel transversing the bone collar (dotted line). (C3) RBC-filled capillaries budding into the cartilage, interspersed with Osx/LacZ+ cells. (D) PECAM-1 stained Osx-CreERt and Col1-CreERt humeri at the indicated time points. Osx-CreERt embryos were exposed to 4OHTam at E12.5 or E13.5 in (A)–(D), Col1-CreERt mice received 4OHTam at E13.5.
See Figure S6 on the role of osteoclasts in initial invasion.
A pulse-chase analysis employing PECAM-1 staining revealed the persistence of the osteoblast precursor/endothelium juxtaposition throughout the subsequent stages of cartilage excavation, culminating in the establishment of the primary ossification center by E15.5–E16.0 (Figure 6D, left panels). Although Osx/LacZ+ cells were regularly wrapped around the blood vessels, they were never observed inside the lumen. In sharp contrast to the Osx/LacZ+ precursor cells, Col1/LacZ+ mature osteoblasts showed little or no evidence of association with the invading vasculature (Figure 6D, right). These findings indicate that the time of entry of osteoblast precursors into developing bones is intimately related to their invasion by blood vessels, suggesting the possibility of a coupled movement process involving mechanisms that are specific to early cells in the osteoblast lineage and lost upon advanced osteoblastic differentiation.
Coupled Angiogenesis and Osteogenesis in Healing Fractures Is Associated with Entry of Osx-Expressing, Blood Vessel-Associated Osteoblast Precursors
Given that the processes governing embryonic bone development are largely recapitulated during fracture healing, we wondered whether a similar coinvasion of osteoblast lineage cells and blood vessels may occur in the adult setting of bone repair. After confirming that the Osx/GFP+ cells in Osx-Cre:GFP mice showed a coinvasion with blood vessels into the developing primary ossification center similar to that observed with the Osx-CreERt mice (Figure 7A), we employed the Osx-Cre:GFP mouse line to allow direct visualization of the predominantly nuclear GFP signal by confocal microscopy of thick bone sections.
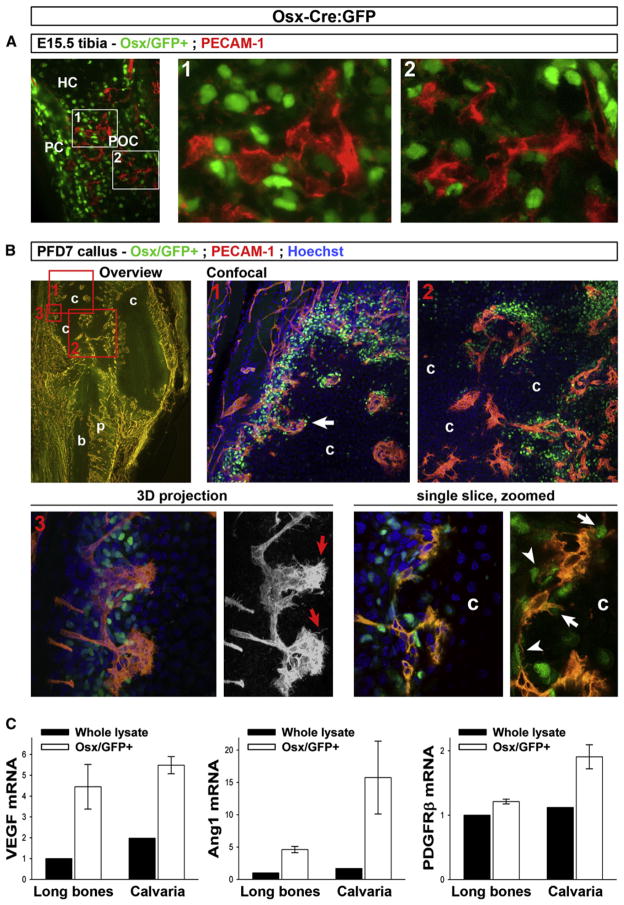
Figure 7. Simultaneous Osteoblastic and Angiogenic Invasion of Cartilage in Healing Fractures.
(A) PECAM-1 IHC on frozen E15.5 Osx-Cre:GFP tibia sections revealing Osx/GFP+ cells (green, note predominant nuclear localization of the Cre:GFP fusion protein) and endothelium (red) during the initial invasion of the hypertrophic cartilage (HC) and primary ossification center (POC) formation. PC, perichondrium. Right, magnified views of indicated areas.
(B) Thick section analysis of fractured tibia of Osx-Cre:GFP mice at postfracture day (PFD) 7, stained with PECAM-1 antibodies (red) and Hoechst (blue). Top row, overview of the callus at 5× by standard microscopy (left; yellow, vasculature; dark areas, cartilage [c] and cortical bone [b]; p, periosteum) indicating the areas (boxes 1–3) analyzed by confocal microscopy. Right panels, areas 1 and 2, showing projections of sets of stacked images taken with a 20× objective (also see Movies S1–S3). Bottom left, area 3, showing 3D projection of 60× confocal images (also see Movie S4). The 3D red channel (PECAM-1 signal) is also shown separately (black/white view), revealing the endothelial filopodia at the invasion front (red arrows). Bottom right, single optical slice extracted from the 60× stack, and enlarged detail omitting the blue channel signal for optimal appreciation of the Osx/GFP+ nuclei. Note abundant Osx/GFP+ cells immediately juxtaposing the endothelial lining (arrowheads) and tips at the invasion front (arrows).
(C) qRT-PCR analysis of VEGF, Ang1, and PDGFRβ in Osx/GFP+ sorted cell populations (mean ± SEM; n = 4) derived from collagenase-digested calvaria or long bones from newborn Osx-Cre:GFP mice, as compared with control digests of Osx-Cre:GFP-negative littermates (“whole lysate”). Video files (Movies S1–S4) of the stacked confocal images and data on other time points during the fracture healing process are provided in Figure S7.
Adult (3-month-old) Osx-Cre:GFP mice showed the presence of Osx/GFP-expressing cells in the thin periosteal layer overlaying the cortical bone surface of uninjured bones (postfracture day [PFD] 0) (Figure S7A). We next monitored the relationship between Osx/GFP+ cells and PECAM-1+ endothelium (red fluorescent IHC) during the subsequent stages of the endochondral healing process in a semistabilized fracture model applied to Osx-Cre:GFP mice (schematically depicted in Figure S7B). As observed previously, the induction of a bone defect is associated with an early proliferative response in the periosteum of the disrupted cortices (Maes et al., 2006). Osx-GFP+ cells were particularly abundant in this thickened periosteum at PFD 4 (Figure S7C). Further dynamic studies indicated that by PFD 14, Osx/GFP+ cells contributed massively to the woven bone callus, localizing within the regenerating bone regions as well as along the blood vessels interspersing the woven bone (Figure S7D).
Interestingly, during the endochondral ossification process occurring in the interim time, the infiltration of the cartilaginous callus tissues by invading endothelium was closely associated with coinvasion of Osx-GFP+ cells (PFD 7, Figure 7B). Adult Osx/GFP+ cells were markedly abundant in the vascularized peripheral portion of the callus bordering the cartilage and along the invasion fronts. Confocal microscopy revealed pericyte-like Osx/GFP+ cells surrounding the endothelium of the extending vascular plexus (Figure 7B; Movies S1–S4). These data suggest that similar mechanisms might govern the coinvasion of osteoblast precursors and blood vessels in bone development and fracture healing.
To address the relationship between osteoblast precursors and the vasculature at the molecular level, we performed qRT-PCR analysis on FACS-sorted Osx-expressing cells as compared with cell preparations without selection for Osx expression (see Supplemental Experimental Procedures). Osx/GFP+ cell digests showed 3- to 10-fold increased expression levels of VEGF and angiopoietin-1 (Ang1), factors secreted by pericytes that may mediate their recruitment of endothelial cells (Figure 7C). Moreover, the expression of the PDGF receptor PDGF-Rβ, a crucial receptor for recruitment of pericytes to endothelium, was also detected in Osx/GFP+ cells, suggestive of a molecular cross-talk between the colocalizing cell types (Figure 7C).
DISCUSSION
The Perichondrial Origin of Trabecular Osteoblasts
The origin of the osteogenic stroma and the osteoblasts that occupy the primary ossification center to form trabecular bone has been uncertain. Hypotheses have suggested that these cells derive from the perichondrium/periosteum, from chondrocytes, from delivery of circulating progenitors through the blood stream, and/or from pericytes (Colnot et al., 2004; Galotto et al., 1994; Kronenberg, 2007; Modder and Khosla, 2008; Roach and Erenpreisa, 1996). Here, we assessed the fate of osteoblast lineage cells from the embryonic perichondrium. Our results confirm and extend previous ex vivo findings by Colnot et al. (2004), who showed that perichondrial cells of unidentified genetic signature contribute osteoblasts to developing bones planted under the renal capsule. By using the mouse Osx- and Col1-promoters to drive the expression of the inducible CreERt transgene in stage-selective subsets of perichondrial osteoblast-lineage cells, we now find that osteoblast precursors, as opposed to mature perichondrial osteoblasts, have the capacity to enter the developing bone and differentiate there into trabecular osteoblasts (see Figure 8). This does not necessarily imply that the sole fate of all the labeled Osx/LacZ+ cells is to become mature osteoblasts. In fact, our data indicate that a subset of them contributes to the metaphyseal stroma, displaying a rather undifferentiated phenotype (see Figures 5A–5C). It is unclear whether such cells constitute reserve precursors that can be activated later to become osteoblasts. Alternatively, and not mutually exclusively, they may represent a subpopulation with different/independent fates and functions, such as hematopoietic support (Wu et al., 2008).
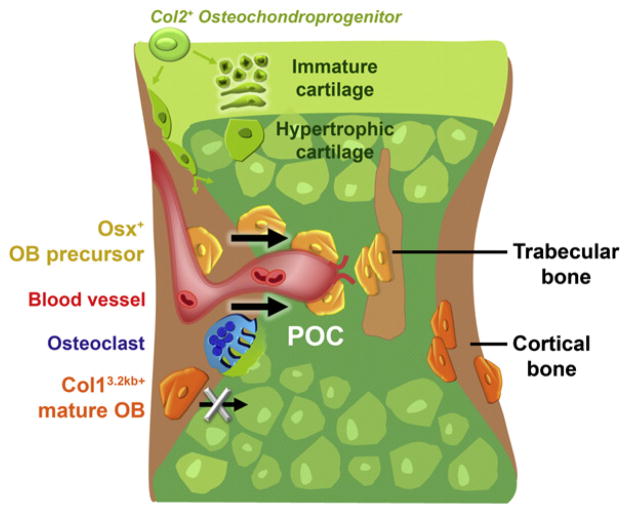
Figure 8. Model Summarizing Our Findings.
Schematic outline of the events taking place during the initial invasion of the cartilaginous bone model and its transformation into the primary ossification center (POC). Col2-expressing chondro-perichondrial progenitors (green) give rise to cells in the central growth cartilage and to transversally oriented cells at the periphery with presumed perichondrial/osteoblastic fates. The first committed osteoblast lineage cells appear in the perichondrium surrounding the middiaphyseal hypertrophic cartilage. The cells display differential destinies in the developing bone depending on their stage of maturation. Early cells of the lineage, represented by the Osx-expressing osteoblast precursors (Osx/LacZ+, yellow), move into the developing POC and populate it as stromal cells or differentiate further to become bone-forming trabecular osteoblasts. The entrance of the osteoblast precursors into the POC coincides with the initial invasion by blood vessels (red) and osteoclasts (blue), and is associated with a pericytic localization of a subset of the precursors onto the endothelium. In contrast, cells that differentiated to the stage of Col1 (3.2 kb) expression within the perichondrium/periosteum (Col1/LacZ+ mature osteoblasts, orange) are not found in vessel-covering positions and do not have the capacity to translocate into the POC. These osteoblasts are retained on and within the cortical bone surfaces.
Differentiated Chondrocytes Are Not a Major Source of Trabecular Osteoblasts
Since the initially labeled Col1/LacZ+ cells were confined to the perichondrium, whereas Osx/LacZ+ cells additionally included differentiating chondrocytes, we assessed whether the latter may have constituted or contributed to the trabecular osteoblast pool. Our data using an inducible Col2-CreERt confirm that early in development, Col2-expressing cells include bipotent progenitors contributing to the chondrocyte and osteoblast pools in endochondral bones (Hilton et al., 2007; Nakamura et al., 2006; Sakai et al., 2001) (Figure S3). Even later on, cells at the periphery of the growth cartilage particularly, exhibiting a longitudinal arrangement along the cartilage-perichondrium interface (“borderline chondrocytes”) (Bianco et al., 1998), may become incorporated in the perichondrium and/or give rise to osteoblasts at the endosteum and inside the bone. In contrast, our data outlined in Figure 3 indicate that Col2/LacZ+ cells that join the bulk of the more central growth cartilage, where they follow the classical chondrocyte differentiation program toward hypertrophy, do not become trabecular osteoblasts in the central metaphyseal regions (away from the endosteal surfaces) in the time span of our studies.
Coinvasion of Cartilage by Osteoblast Precursors and Blood Vessels
Several prior experiments have probed the interdependence of vascular and osteoblastic invasion of bone. Blocking the initial vascular invasion of the cartilage template, either physically, chemically, or genetically, inhibited the formation of the primary ossification center (Colnot et al., 2004; Maes et al., 2002). On the other hand, several genetic models involving an early block in perichondrial osteoblastogenesis through inactivation of Runx2, Osx, β-catenin, or Indian hedgehog also showed a near-complete absence of vascular invasion and primary ossification (Komori et al., 1997; Otto et al., 1997; Colnot et al., 2005; St. Jacques et al., 1999; Nakashima et al., 2002; Hill et al., 2005). Collectively, these studies suggest that perichondrial osteoblast differentiation, at least up to the stage of Runx2/Osx-expressing precursors, is required for vascular invasion of cartilage. In contrast, blockade of osteoclast formation genetically or through inhibitors of osteoclastogenesis (Figure S6) does not block invasion of either blood vessels or osteoblasts.
In our study, the two processes could be analyzed closely in a cell-specific way by combining osteoblast lineage tracing with monitoring of the vascular endothelium. Throughout the translocation of osteoblast precursors from the perichondrium into the nascent primary ossification center, several osteoblast lineage cells were seen to intimately associate with the invading blood vessels, while others comigrate near vessels, suggesting a coupled movement process that appears to be recapitulated during bone regeneration of fractures (see Figures 6D, 7A, and 7B).
Based on our data and current literature, three hypotheses for the coupling can be formulated. First, osteoblast precursors and endothelial cells may be attracted to the same or simultaneously expressed chemoattractant(s) produced by the hypertrophic cartilage, thereby moving in concert into its core. However, mere independent comigration would not explain the pericytic localization that we recognized for a portion of the osteoblast precursors during the movement process.
Therefore, a second mechanism may be that some osteoblast precursors use the blood vessels as a conduit to support their entry into the bone, as reported for melanoma tumor cells that migrate along the abluminal surface of the endothelium in a process termed “extravascular migratory metastasis” (Lugassy et al., 2004). Our studies indicated that Osx-expressing cells strongly express angiogenic stimuli as well as molecules involved in pericyte-endothelial interactions (VEGF, Ang1, PDGRβ). Of note, it is uncertain whether the perivascular osteoblast lineage cells detected in our study have functional characteristics generally attributed to pericytes. In fact, different from most other tissues, the blood vessels of bone lacked coverage by supporting cells expressing classical pericyte markers such as αSMA (see Figures S5F–S5H). This observation may be related to the presence in bone marrow of thin-walled and fenestrated sinusoids, in which trans-endothelial migration of hematopoietic cells is thought to take place (Kopp et al., 2005).
A third option is that osteoblast precursors themselves trigger the initial vascular invasion, by pulling vessels into the avascular cartilage. This may involve “looping angiogenesis,” a recently proposed mechanism describing the vascularization of previously avascular, contracting granulation tissue of healing wounds through biomechanical translocation of intact vessels as loops pulled by myofibroblastic contraction (Kilarski et al., 2009). Our EM analyses revealed that capillaries invade the cartilage as morphologically intact vessels that are presumably connected to the circulation, as evidenced by the abundance of RBCs in their lumens, with osteoblast precursors locating to the leading edges as well as along the lumen linings (see Figure 6C). The expansion of the vascular plexus most likely involves sprouting angiogenesis, as suggested by the abundant filopodia extending from the endothelial cells at the tips of the invading vascular sprouts, observed by EM and (confocal) histology (indicated by red arrows in Figure 6C1 during initial cartilage invasion in embryogenesis and in Figure 7B3 and Movie S4, showing a 3D projection of callus neovascularization during endochondral bone healing). Endothelial tip cells are known to display characteristically high expression of PDGF-B and VEGFR-2 (Gerhardt et al., 2003), possibly implicating a crosstalk with the PDGFRβ- and VEGF-enriched osteoblast lineage cells.
These three hypotheses are not mutually exclusive and may converge in the following model (see Figure 8): Osteoblast precursors developing in the perichondrium invade the future marrow space along with blood vessels, both while in association with the endothelium in pericytic fashion and by migrating along with the front of sprouting vessels and osteoclasts excavating the cartilage. Osteoblast precursor interactions with the endothelium may contribute to the translocation into the late-hypertrophic cartilage, possibly through combined mechanisms of looping and sprouting angiogenesis, that may be directionally steered by the high concentration and release of stimulatory signals such as VEGF. Osteoblasts that had differentiated into mature, matrix-producing cells in the perichondrium do not have pericyte properties, nor do they move into the bone and become trabecular osteoblasts. Instead, these cells are destined to generate the cortical bone.
EXPERIMENTAL PROCEDURES
Animal Studies
The generation of Osx-CreERt2 and Col1(3.2 kb)-CreERt mice is outlined in the Supplemental Experimental Procedures. Osx-Cre:GFP mice have been described (Rodda and McMahon, 2006). Experiments were conducted in compliance with the Guide for the Care and Use of Laboratory Animals and approved by Massachusetts General Hospital’s Center for Comparative Medicine and by the K.U. Leuven. Male CreERt(Tg/−);Rosa26R(R/R) mice were timed mated to CD1 females overnight and separated in the morning, designated E0.5. Pregnant mice received 2 mg 4OHTam i.p. (~50 mg/kg), dissolved stepwise in 20 μl dimethylfluoride and to a total volume of 200 μl in sunflower seed oil. At sacrifice, the embryos were weighed and dissected clear of skin and internal organs. Tail or liver biopsies were collected for genotyping by PCR (see Supplemental Experimental Procedures). Skeletal muscles were removed from the limbs from E15.5 onward. Selected embryos were stained with Alizarin red using the standard protocol. Fracture healing was studied in 3-month-old Osx-Cre:GFP mice using a semistabilized tibia fracture model described in detail before (Maes et al., 2006). The tibia was fixed overnight in 2% PFA and decalcified in 0.5M EDTA for 2 weeks.
X-Gal Staining of LacZ Activity
The embryo proper as well as the organs and separated limbs were fixed for 1 hr at RT in 0.2% glutaraldehyde and 1.5% formaldehyde, followed by overnight X-gal staining at 37°C according to the standard protocol, and 1 day postfixation in 2% PFA at 4°C.
Histology
X-gal-stained samples were paraffin embedded and sectioned at 4 μm. When necessary, bones were decalcified for 1–3 days prior to the embedding. Sections were dewaxed in xylenes, immersed stepwise in 100% and 95% ethanol, and stained with 1% eosinY in 95% ethanol for 5 min followed by rinsing, dehydrating, and mounting. Von Kossa staining was done according to standard methods and followed by eosin staining. Additional sections were subjected to staining for TRAP activity or to IHC using antibodies for PECAM-1 (CD31) (BD Biosciences) or BrdU (Harlan Sera-Lab) as described (Maes et al., 2002). Detailed information on IHC and ISH procedures for osteoblast and pericyte markers is provided in the Supplemental Experimental Procedures. Fracture calluses were studied using thick sections preserving the Osx/GFP+ signal, by embedding the bones in 5% low melting seaplaque agarose (Lonza Rockland) and sectioning at 100 μm on a Microm Vibratome. Sections were stained with PECAM-1 antibodies (Cy3 red fluorescent detection) and Hoechst dye (blue nuclear signal).
Imaging
Whole-mount images were captured on a Leica MZ16 stereomicroscope equipped with Image-Pro Plus digital imaging module. Histological sections were imaged on Nikon Eclipse E800 and Zeiss Axioplan microscopes using 2×, 4×, 10×, 20×, 40×, and 100×objectives, and SPOT Advanced or Axio-vision software. Whole-image adjustments of brightness and contrast were done using Adobe Photoshop software. Thick vibratome sections were analyzed on a Nikon Eclipse Ti-2000 inverse confocal scanning microscope using 0.75 N.A. 20× and 1.4 N.A. 60× (oil) objectives to acquire image stacks and single confocal optical slices using NIS-Elements AR3.1 software. Contrast and brightness were adjusted and 3D projections from stacks were composed with the Image J program (version 1.42).
Quantification of LacZ-Positive Cells
Perichondrial labeling was quantified on E14.5 humerus sections by counting the number of LacZ+ and LacZ+/BrdU+ double-positive cells in a fixed area (spanning 345 μm longitudinally, centralized at the middiaphysis) comprising most of the perichondrium (shown in Figure 2A, left). Three to five sections were analyzed per bone (n = 3 per genotype), including only central-cut sections at least 40 μm apart. The proliferation index was calculated as the percentage LacZ+/BrdU+ cells of the total LacZ+ cell count. At E16.5, LacZ+ cells were counted in defined (210 × 210 μm) regions of the trabecular and cortical bone areas, positioned centrally along perpendicular axes through the bone, as indicated in Figure 2A (right, blue squares). Trabecular regions were taken 50 μm distal of the hypertrophic chondrocyte zone to exclude the junction. Three to five sections per bone of n = 2–9 embryos were analyzed, blindly with regard to genotype. Statistical analysis was done by two-sided, two-sample t tests. Results are represented as average ± SEM, with the symbols * for p < 0.05, **p < 0.01, and ***p < 0.001.
EM Analysis
After X-gal staining, bones were cleared completely of surrounding soft tissues, cut transversally at middiaphysis, fixed in EM fixative (2.5% glutaraldehyde, 2.0% PFA, 0.025% calcium chloride in a 0.1 M sodium cacodylate buffer [pH 7.4]), and processed for EM. The samples were embedded vertically (E13.5–E14.5) or horizontally (E16.5) in resin, to create transversal or longitudinal sections, respectively. Three sets of consecutive sections were prepared, as illustrated in Figure 4A: (1) 1 μm sections stained with toluidine blue for 15 s to visualize the tissue morphology, (2) 0.3 μm sections that were placed on top of grids for EM analysis, and 3) 1 μm sections that remained unstained to visualize the blue LacZ+ cells. Aligning the information from the respective sections allowed for a cell-specific analysis of the labeled cells as inspected by EM. The correct identification of each of the labeled cells via this procedure was confirmed by the recognition on EM of X-gal reaction product deposits in the cytoplasm of LacZ+ cells, as patchy distributed black amorphous material, in several of the analyzed samples. Samples were examined with a Phillips 301 transmission electron microscope and digital images were captured using an AMT (Advanced Microscopy Techniques) CCD camera at magnifications ranging from ×4500 to ×19,500. Quantifications were done by classifying the LacZ+ cells based on their morphology and spatial characteristics: pericytic cells, displaying immediate perivascular localization; stromal cells, displaying no connection to bone matrix and having little ER and few organelles; early osteoblast lineage cells, located in the proximity or onto collagen matrix but still containing limited ER, sparse plasma membrane convolutions and no or sparse evidence of secretion activity; mature osteoblasts, defined as cuboidal cells located on osteoid and/or mineralized bone and actively secreting matrix; osteocytes, cells immediately flanked by bone matrix on at least three sides.
FACS Sorting, RNA Extraction, and Gene Expression Analysis by qRT-PCR
The preparation of calvaria and long bone digests from newborn Osx-Cre:GFP mice, their FACS sorting, and qRT-PCR analysis are described in detail in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Abhishek Gole, Jennifer Paruch, Greg Nachtrab, and Ingrid Stockmans for assistance with mouse breeding and genotyping. The Histology Core at the Endocrine Unit of Massachusetts General Hospital, Riet Van Looveren, and Karen Moermans (Legendo, K.U. Leuven) are acknowledged for excellent help with histology. For expert advice on ISH, FACS analysis, confocal microscopy, and image analysis, we thank Przemko Tylzanowski (Rheumatology, K.U. Leuven), Aernout Luttun (CMVB, K.U. Leuven), Sebastian Munck (CME and VIB, K.U. Leuven), Nick Van Gastel, Maarten Depypere, and An Vanden Bosch (Legendo, K.U. Leuven). We thank Andy McMahon (Harvard) for generously providing the Osx-Cre:GFP mouse. We are also grateful to Joy Wu, Ernestina Schipani, and the members of the Endocrine Unit for helpful discussions. This work was supported by NIH grant DK056246 to H.M.K., FWO grants G.0569.07 and G.0500.08 to G.C., and a postdoctoral fellowship of the Fund for Scientific Research Flanders (FWO) to C.M.
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four movies and can be found online at doi:10.1016/j.devcel.2010.07.010.
All authors declare no conflicts of interest.
References
- Bianco P, Cancedda FD, Riminucci M, Cancedda R. Bone formation via cartilage models: the “borderline” chondrocyte. Matrix Biol. 1998;17:185–192. doi: 10.1016/s0945-053x(98)90057-9. [DOI] [PubMed] [Google Scholar]
- Colnot C, Lu C, Hu D, Helms JA. Distinguishing the contributions of the perichondrium, cartilage, and vascular endothelium to skeletal development. Dev Biol. 2004;269:55–69. doi: 10.1016/j.ydbio.2004.01.011. [DOI] [PubMed] [Google Scholar]
- Colnot C, de la Fuente L, Huang S, Hu D, Lu C, St-Jacques B, Helms JA. Indian hedgehog synchronizes skeletal angiogenesis and perichondrial maturation with cartilage development. Development. 2005;132:1057–1067. doi: 10.1242/dev.01649. [DOI] [PubMed] [Google Scholar]
- Feil R, Brocard J, Mascrez B, LeMeur M, Metzger D, Chambon P. Ligand-activated site-specific recombination in mice. Proc Natl Acad Sci USA. 1996;93:10887–10890. doi: 10.1073/pnas.93.20.10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galotto M, Campanile G, Robino G, Cancedda FD, Bianco P, Cancedda R. Hypertrophic chondrocytes undergo further differentiation to osteoblast-like cells and participate in the initial bone formation in developing chick embryo. J Bone Miner Res. 1994;9:1239–1249. doi: 10.1002/jbmr.5650090814. [DOI] [PubMed] [Google Scholar]
- Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, Jeltsch M, Mitchell C, Alitalo K, Shima D, Betsholtz C. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 2003;161:1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TP, Spater D, Taketo MM, Birchmeier W, Hartmann C. Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev Cell. 2005;8:727–738. doi: 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Hilton MJ, Tu X, Cook J, Hu H, Long F. Ihh controls cartilage development by antagonizing Gli3, but requires additional effectors to regulate osteoblast and vascular development. Development. 2005;132:4339–4351. doi: 10.1242/dev.02025. [DOI] [PubMed] [Google Scholar]
- Hilton MJ, Tu X, Long F. Tamoxifen-inducible gene deletion reveals a distinct cell type associated with trabecular bone, and direct regulation of PTHrP expression and chondrocyte morphology by Ihh in growth region cartilage. Dev Biol. 2007;308:93–105. doi: 10.1016/j.ydbio.2007.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/s1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- Kilarski WW, Samolov B, Petersson L, Kvanta A, Gerwins P. Biomechanical regulation of blood vessel growth during tissue vascularization. Nat Med. 2009;15:657–664. doi: 10.1038/nm.1985. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Kopp HG, Avecilla ST, Hooper AT, Rafii S. The bone marrow vascular niche: home of HSC differentiation and mobilization. Physiology (Bethesda) 2005;20:349–356. doi: 10.1152/physiol.00025.2005. [DOI] [PubMed] [Google Scholar]
- Kronenberg HM. The role of the perichondrium in fetal bone development. Ann N Y Acad Sci. 2007;1116:59–64. doi: 10.1196/annals.1402.059. [DOI] [PubMed] [Google Scholar]
- Lugassy C, Kleinman HK, Engbring JA, Welch DR, Harms JF, Rufner R, Ghanem G, Patierno SR, Barnhill RL. Pericyte-like location of GFP-tagged melanoma cells: ex vivo and in vivo studies of extravascular migratory metastasis. Am J Pathol. 2004;164:1191–1198. doi: 10.1016/S0002-9440(10)63207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes C, Carmeliet P, Moermans K, Stockmans I, Smets N, Collen D, Bouillon R, Carmeliet G. Impaired angiogenesis and endochondral bone formation in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188. Mech Dev. 2002;111:61–73. doi: 10.1016/s0925-4773(01)00601-3. [DOI] [PubMed] [Google Scholar]
- Maes C, Coenegrachts L, Stockmans I, Daci E, Luttun A, Petryk A, Gopalakrishnan R, Moermans K, Smets N, Verfaillie CM, et al. Placental growth factor mediates mesenchymal cell development, cartilage turnover, and bone remodeling during fracture repair. J Clin Invest. 2006;116:1230–1242. doi: 10.1172/JCI26772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes C, Kobayashi T, Kronenberg HM. A novel transgenic mouse model to study the osteoblast lineage in vivo. Ann N Y Acad Sci. 2007;1116:149–164. doi: 10.1196/annals.1402.060. [DOI] [PubMed] [Google Scholar]
- Marie PJ. Transcription factors controlling osteoblastogenesis. Arch Biochem Biophys. 2008;473:98–105. doi: 10.1016/j.abb.2008.02.030. [DOI] [PubMed] [Google Scholar]
- Metzger D, Clifford J, Chiba H, Chambon P. Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc Natl Acad Sci USA. 1995;92:6991–6995. doi: 10.1073/pnas.92.15.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modder UI, Khosla S. Skeletal stem/osteoprogenitor cells: current concepts, alternate hypotheses, and relationship to the bone remodeling compartment. J Cell Biochem. 2008;103:393–400. doi: 10.1002/jcb.21423. [DOI] [PubMed] [Google Scholar]
- Nakamura E, Nguyen MT, Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev Dyn. 2006;235:2603–2612. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, de Crombrugghe B. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Roach HI, Erenpreisa J. The phenotypic switch from chondrocytes to bone-forming cells involves asymmetric cell division and apoptosis. Connect Tissue Res. 1996;35:85–91. doi: 10.3109/03008209609029178. [DOI] [PubMed] [Google Scholar]
- Rodda SJ, McMahon AP. Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development. 2006;133:3231–3244. doi: 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- Sakai K, Hiripi L, Glumoff V, Brandau O, Eerola R, Vuorio E, Bosze Z, Fassler R, Aszodi A. Stage-and tissue-specific expression of a Col2a1-Cre fusion gene in transgenic mice. Matrix Biol. 2001;19:761–767. doi: 10.1016/s0945-053x(00)00122-0. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- St Jacques B, Hammerschmidt M, McMahon AP. Indian hedgehog signaling regulates proliferation and differentiation of chondrocytes and is essential for bone formation. Genes Dev. 1999;13:2072–2086. doi: 10.1101/gad.13.16.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JY, Purton LE, Rodda SJ, Chen M, Weinstein LS, McMahon AP, Scadden DT, Kronenberg HM. Osteoblastic regulation of B lymphopoiesis is mediated by Gs{alpha}-dependent signaling pathways. Proc Natl Acad Sci USA. 2008;105:16976–16981. doi: 10.1073/pnas.0802898105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.