Abstract
The diagnosis of myoepithelial (ME) tumors outside salivary glands remains challenging, especially in unusual clinical presentations, such as bone or visceral locations. A few reports have indicated EWSR1 gene rearrangement in soft tissue ME tumors, and, in one case each, the fusion partner was identified as either PBX1 or ZNF444. However, larger studies to investigate if these genetic abnormalities are recurrent or restricted to tumors in soft tissue locations are lacking. Sixty-six ME tumors mainly from soft tissue (71%), but also from skin, bone and visceral locations, characterized by classic morphologic features and supporting immunoprofile were studied. Gene rearrangements in EWSR1, FUS, PBX1 and ZNF444 were investigated by FISH. EWSR1 gene rearrangement was detected in 45% of the cases. A EWSR1-POU5F1 fusion was identified in a pediatric soft tissue tumor by 3’RACE and subsequently confirmed in four additional soft tissue tumors in children and young adults. An EWSR1-PBX1 fusion was seen in five cases, while EWSR1-ZNF444 and FUS gene rearrangement was noted in one pulmonary tumor each. In conclusion, EWSR1 gene rearrangement is a common event in ME tumors arising outside salivary glands, irrespective of anatomic location. EWSR1 negative tumors were more often benign, superficially located, and showed ductal differentiation, suggesting the possibility of genetically distinct groups. A subset of soft tissue ME tumors with clear cell morphology harbor an EWSR1-POU5F1 fusion, which can be used as a molecular diagnostic test in difficult cases. These findings do not support a pathogenetic relationship between soft tissue ME tumors and their salivary gland counterparts.
Keywords: myoepithelial tumor, EWSR1, POU5F1, PBX1, ZNF444
INTRODUCTION
The spectrum of myoepithelial (ME) tumors represents a family of lesions with variable terminology, based on anatomic location: such as pleomorphic adenoma in the salivary gland, benign mixed tumor in the skin, and myoepithelial tumor/parachordoma in the soft tissue. Although genetic studies in pleomorphic adenoma have shown frequent rearrangements of PLAG1 and HMGA2 (Kas et al., 1997; Martins et al., 2005; Persson et al., 2009), similar abnormalities have not been identified in myoepithelial lesions from other tissues (Hallor et al., 2008). Thus it remains unclear if the myoepithelioma subsets described above share a common pathogenesis.
The genetic hallmark of soft tissue ME tumors is still under investigation, with only two cases analyzed cytogenetically, reporting disparate chromosomal translocations. A t(1;22)(q23;q12) resulting in an EWSR1-PBX1 fusion was first described as a sole cytogenetic event in a soft tissue ME tumor, arising in the foot of a 59-year old female (Brandal et al., 2008), while a second case, of an occipital soft tissue ME carcinoma, arising in a 40-year old female, showed a t(19;22)(q13;q12), resulting in an EWSR1-ZNF444 fusion (Brandal et al., 2009). In addition, EWSR1 gene rearrangement by FISH has been reported in two ME tumors in the pediatric age group (Gleason and Fletcher, 2007).
In this study we undertook a systematic molecular analysis of a large spectrum of ME tumors, including lesions from a variety of anatomic locations, age groups and lesions with differing biologic potential. Our hypothesis is that a better understanding of the molecular signature of ME tumors may help to refine the present classification based on immunophenotype alone, as well as providing insight concerning any pathogenetic relationship with the salivary gland counterpart and other related or morphologically similar entities.
MATERIALS AND METHODS
Histopathologic Diagnosis in Experimental and Control Groups
Seventy-one myoepithelial tumors with available tissue for molecular analysis were retrieved from the surgical pathology and consultation files of the authors. Slides were re-reviewed in corroboration with the immunohistochemical panel in all cases. Minimum criteria for confirming the morphologic diagnosis included the co-reactivity for EMA +/− cytokeratin and S100+/− GFAP. Five cases were excluded as failing FISH analysis: four due to decalcification and one due to hybridization failure.
The tumors were assessed morphologically for pattern of growth, duct formation, clear cell component, epithelioid versus spindle cell composition, nuclear pleomorphism, mitotic activity, and necrosis. The tumor location was recorded, including the anatomic structures involved, and lesions were subclassified into three subgroups, cutaneous, superficial (subcutaneous), and deep (below the fascial plane).
In order to investigate a potential relationship between soft tissue ME tumors and their salivary gland counterpart, we included five salivary gland ME carcinomas (ex-pleomorphic adenoma) for the genetic analysis. A group of five salivary mucoepidermoid carcinomas and five cutaneous eccrine hidradenomas were also tested, as recent evidence suggested similar genetic abnormalities (see Discussion section). As chordoma periphericum has been considered a potentially related tumor, we have included two such examples occurring in the bone, confirmed based on their immunopositivity for t-Brachyury and a similar microscopic appearance with their axial counterpart (Nielsen et al., 2001; Scolyer et al., 2004; Tirabosco et al., 2008). In addition, three ossifying fibromyxoid tumors (OFT) were included in the analysis, as this tumor often shares morphologic and immunophenotypic features with ME tumors.
Fluorescence in Situ Hybridization (FISH) and Chromosome Banding Analysis
FISH on interphase nuclei from paraffin embedded 4-micron sections was performed applying custom probes using bacterial artificial chromosomes (BAC), covering and flanking EWSR1 in 22q12, FUS in 16p11, PBX1 in 1q23, ZNF444 in 19q13 and POU5F1 in 6p21 (Fig. 1). BAC clones were chosen according to USCS genome browser (http://genome.uscs.edu). The BAC clones were obtained from BACPAC sources of Children's Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described (Agaram et al., 2008). The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems). A positive score was interpreted when at least 20% of the nuclei showed a break-apart signal. Nuclei with incomplete set of signals were omitted from the score.
Fig. 1.
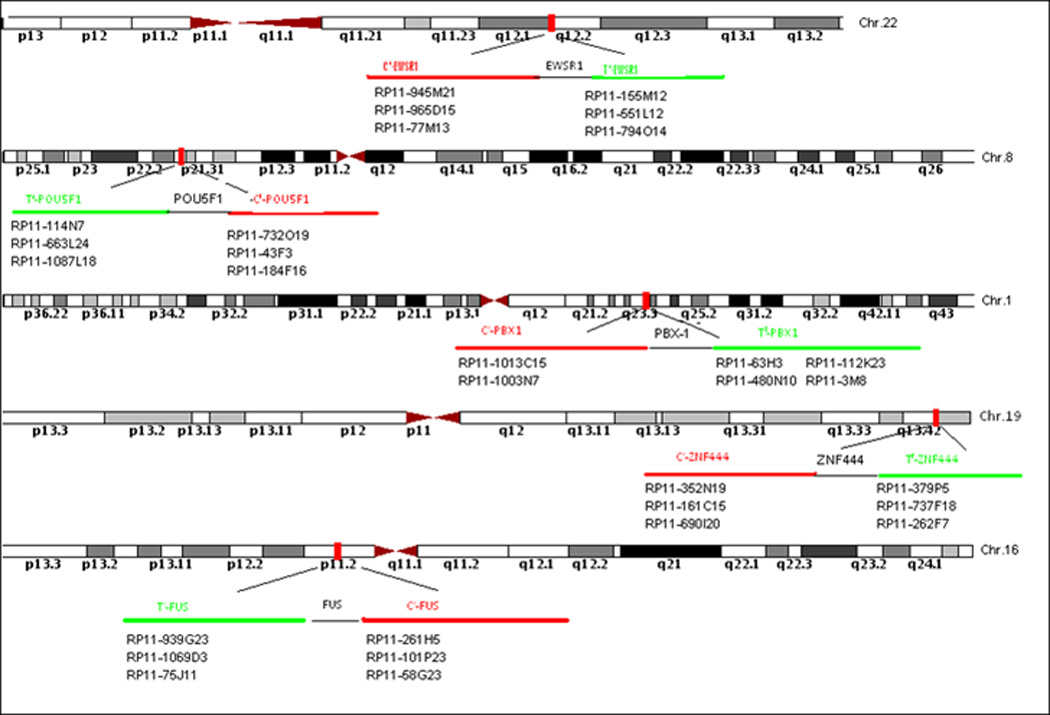
BAC probe sets used to determine breakpoints by FISH for EWSR1, FUS, POU5F1, PBX1, and ZNF444.
All cases were first tested with an EWSR1 probe. The EWSR1-rearranged tumors were then evaluated for break-apart signals using probes for PBX1, ZNF444, and POU5F1. The EWSR1 negative tumors were then tested for FUS break-apart, since FUS may substitute for the EWSR1 gene in certain translocation-associated sarcomas. In selective cases, two-color FISH was applied using probe-sets centromerically flanking one gene and telomerically flanking the partner gene, in order to confirm the fusion between EWSR1 and the partner genes. In one case a G-banded karyotype was obtained after short term culture.
3’RACE, RT-PCR and Sequence Analysis
RNA extraction from paraffin-embedded tissue was attempted in 4 cases in which both gene partners were identified as rearranged by FISH. However, the quality of RNA was suboptimal for further analysis by RT-PCR using the PGK housekeeping gene primers. Total RNA was extracted from one case with available frozen tissue, using Trizol reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). One microgram of total RNA was used for cDNA preparation, followed by a 3’-RACE, using the GeneRacer™ kit (Invitrogen, Carlsbad, CA). Reverse transcribing mRNA was initiated at the poly(A) tail of mRNA with appropriate GeneRacer™ Oligo dT Primers in a 20 µL reaction volume according to the manufacturer's protocol. First round polymerase chain reaction (PCR) was done by Clontech Advantage 2 PCR kit with the GeneRacer™ 3’Primer and EWSR1 forward primer EWSR1EX5.1 (5’-AGCCTGTCCAGGGGTATGGCACT-3’). Nested PCR was performed with the GeneRacer™ Nested 3’Primer and EWSR1 forward primer EWS22.14 (5’-GCACCTCCATCCTACCCTCC-3’). The PCR products were analyzed by electrophoresis. Confirmatory RT-PCR was performed using primers EWSR1EX5.1 and POU5F1 (5’- TCCCAAATAGAACCCCCAGG-3’) and the amplified product was sequenced using the Sanger’s method.
RESULTS
A total of 66 patients with confirmed diagnosis of ME tumor and adequate tissue for molecular analysis were included in the experimental group (Tables 1, 2 and Supplementary Table 3). There were 36 females and 30 males, with a wide age distribution (1–75 years old). More than one third of patients were younger than the age of 30: 15 (23%) being children (<18 years old) and 11 (17%) young adults (≥18, <30 years old). The anatomic distribution included: 47 in soft tissue, 7 cutaneous, 6 osseous and 6 visceral (lung).
Table 1.
Pathologic and Molecular Findings of ME tumors with EWSR1-POU5F1, EWSR1-PBX1 or EWSR1-ZNF444 fusion
| Case # | Age/Sex | Location | Morphology | Malignant Potential |
IHC | Fusion Transcript |
|---|---|---|---|---|---|---|
| 1 | 9/M | Arm/deep | Clear cells, nested | Benign | S100+/EMA+ | EWSR1-POU5F1 |
| 2 | 31/F | Thigh/deep | Clear cells, nested | Malignant | S100+/EMA+/CK+ | EWSR1-POU5F1 |
| 3 | 34/F | Wrist/deep | Clear cells, nested | Malignant | S100+/EMA+/CK+ | EWSR1-POU5F1 |
| 4 | 7/F | Hip/deep | Clear cells, nested | Malignant | S100+/EMA+ CK+/GFAP+ |
EWSR1-POU5F1 |
| 5 | 26/M | Thigh/deep | Clear cells | Malignant | S100+/EMA+ CK+/GFAP+ |
EWSR1-POU5F1 |
| 6* | 37/M | Hip/deep | Sclerosing | Benign | S100+/EMA+ | EWSR1-PBX1 |
| 7 | 75/F | Lung | Spindle/clear | Malignant | S100+/EMA+ | EWSR1-PBX1 |
| 8 | 59/F | Foot/deep | Sclerosing | Benign | S100+/EMA+ | EWSR1-PBX1 |
| 9 | 11/F | Forearm/deep | Epithelioid/clear | Malignant | S100+/EMA+ | EWSR1-PBX1 |
| 10 | 49/M | Iliac/Bone | Sclerosing | Benign | S100+/EMA+ | EWSR1-PBX1 |
| 11 | 64/F | Lung | Epithelioid/clear/ focal spindle |
Malignant | S100+/EMA- /CK+/GFAP+ |
EWSR1-ZNF444 |
M, male; F, female; IHC, immunohistochemistry;
Positive for t(1;22) by karyotyping.
Table 2.
Pathologic findings in EWSR1 positively rearranged ME tumors, without an identified fusion partner
| Case # |
Age/Sex | Location | Morphology | Malignant Potential |
IHC |
|---|---|---|---|---|---|
| 12 | 45/F | L1 vertebral body/bone |
Epithelioid, reticular, myxoid |
Benign | S100+/EMA-/CK+ |
| 13 | 20/F | Pelvic/peri- rectal/deep |
Epithelioid/spindle | Malignant | S100+/EMA+ |
| 14 | 13/F | Peri-ocular/deep | Undifferentiated/small blue cells |
Malignant | S100+/EMA+ |
| 15 | 32/F | Lung | Epithelioid, myxoid | Malignant | S100+/EMA-/Ker+ |
| 16 | 7/F | Peri-orbital/deep | Undifferentiated/small blue cells |
Malignant | S100+/EMA+/CK+ |
| 17 | 19/M | Sino-nasal/ deep | Plump spindle, focal epithelioid |
Malignant | S100+/EMA-/CK+ |
| 18 | 58/F | Calf/superficial | Epithelioid, reticular, myxoid |
Benign | S100+/EMA+/CK+ |
| 19 | 21/F | Shoulder/deep | Undifferentiated/small blue cells |
Malignant | S100+/EMA+/GFAP+ |
| 20 | 16/F | Fibula/bone | Undifferentiated/small blue cells |
Benign | S100+/EMA+ |
| 21 | 27/F | Lung | Small blue cells, epithelioid, clear |
Benign | S100+/EMA+ |
| 22 | 23/M | Humerus/bone | Epithelioid, clear cells | Benign | S100+/EMA+ |
| 23 | 1/M | Head &neck/ deep | Epithelioid, spindle | Malignant | S100+/EMA+/CK+ |
| 24 | 2/F | Mediastinum/ deep | Epithelioid, rhabdoid cells | Malignant | S100+/EMA+/GFAP+ |
| 25 | 53/F | Index finger/ superficial |
Undifferentiated/small blue cells |
Malignant | S100+/EMA+/CK+ |
| 26 | 58/F | Chest wall/ superficial |
Epithelioid, rhabdoid, myxoid |
Benign | S100- /EMA+/GFAP+ |
| 27 | 20/M | Foot/ superficial | Spindle, epithelioid, clear cells |
Benign | S100+/EMA-/CK+ |
| 28 | 32/M | Ankle/ superficial | Sclerotic, spindle, epithelioid |
Benign | S100+/EMA+/CK+ |
| 29 | 26/M | Thigh/ cutaneous | Epithelioid, histiocytoid | Benign | S100+/EMA+ |
| 30 | 46/M | Flank/ cutaneous | Epithelioid, spindle, nested | Benign | S100+/EMA-/CK+ |
EWSR1 Rearrangement is a Common Event in Deep-Seated Soft tissue and Bone ME Tumors
Thirty (45%) cases showed the presence of an EWSR1 gene rearrangement by FISH (Fig. 2F, Tables 1 & 2). There were 18 females and 12 males, with a mean age at diagnosis of 30 (range 1–75, median 26 years). More than half of the cases (n=16) occurred in children and young adults (less than 30 years of age). The most common presentation was in the soft tissues of the extremities (n=16), 10 deep-seated and 6 superficial to the fascia. An additional 4 cases occurred in the head and neck, including 3 peri-orbital soft tissue and one sino-nasal. Five of the six osseous ME tumors showed an EWSR1 rearrangement, and in one case it showed in addition high level amplification of the 3’end of EWSR1 gene (Fig. 2H). Four of the six lung ME tumors were positive, while only two of the six cutaneous tumors showed a break-apart signal with EWSR1 by FISH.
Fig. 2.
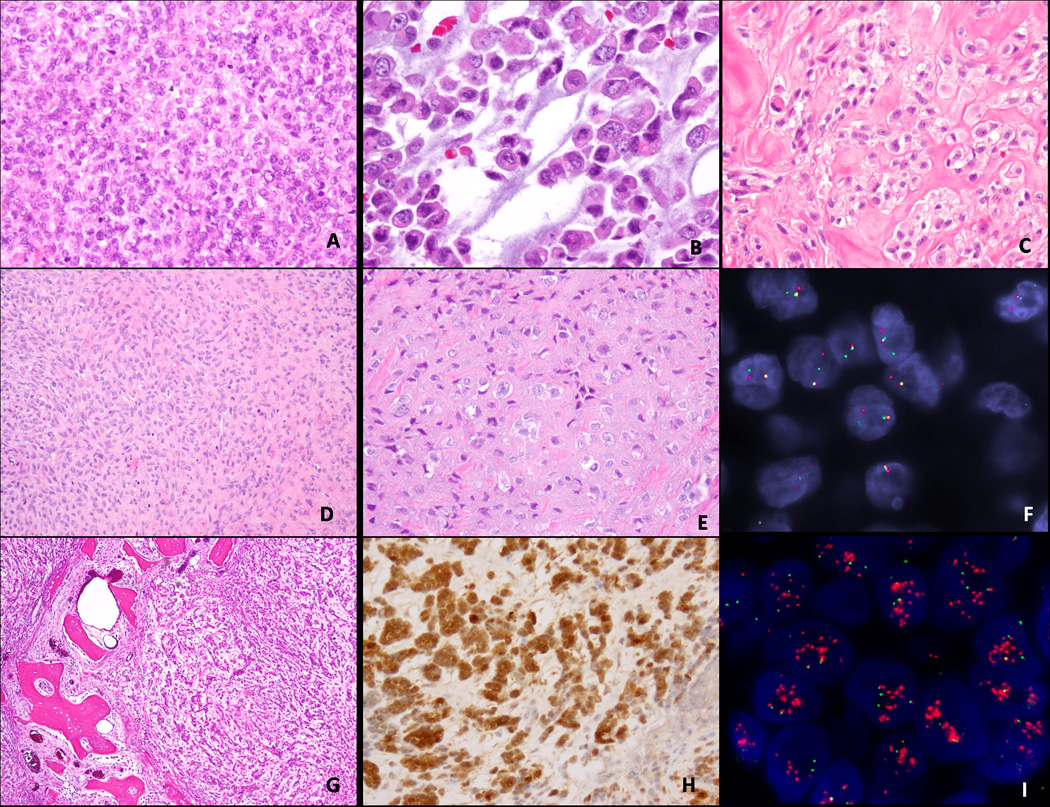
Morphologic spectrum of soft tissue ME tumors showing an EWSR1 gene rearrangement, composed of: (A) undifferentiated small blue cells with ill-defined cell borders (100x; 7 year-old female, peri-orbital mass); (B) rhabdoid morphology with eosinophilic cytoplasm (400x; 2 year-old female, mediastinum); (C) epithelioid cells with clear cytoplasm arranged in nests in a sclerotic background (200x, 20year-old male, subcutaneous foot); (D) bland spindle cell proliferation in a sclerotic background (100x, fibula, 16-year old); (E) fibrotic lesion with hypocellular epithelioid cells (200x; 32 year-old male, subcutaneous ankle); (F) EWSR1 gene rearrangement by FISH showing break-apart signal; (G) nested epithelioid morphology of an intra-osseous ME tumor (40x, humerus, 23 year-old male), showing diffuse and strong S100 protein (H) and by FISH in addition to EWSR1 gene rearrangement, amplification of the 3’end of the EWSR1 gene (I).
Morphologically, in the EWSR1 rearranged tumors there were several different patterns identified: (1) tumors composed mainly of small blue cells with scant cytoplasm, monotonous cytomorphology and ill-defined cell borders, arranged in solid sheets (Fig. 2A); (2) tumors with a predominantly epithelioid or rhabdoid appearance, with moderate to abundant eosinophilic or clear cytoplasm and eccentric nuclei (Fig. 2B,C,G); or (3) tumors composed of spindle or ovoid cells embedded in a prominent sclerotic stroma (Fig 2D,E). In EWSR1-positive tumors having a small cell/undifferentiated appearance, an alternative Ewing sarcoma/PNET diagnosis was excluded either by negativity for CD99 immunostaining or by RT-PCR for EWSR1-FLI1 and EWSR1-ERG. None of the EWSR1 positive tumors showed the presence of ductal or glandular differentiation or cartilage/bone matrix formation. Immunohistochemically, all tumors showed S100 protein staining, typically strong and diffuse (Fig. 2G), as well EMA and/or cytokeratin AE1/AE3.
Histologically, of the cases which showed EWSR1 rearrangement, 16 (53%) were deemed to be malignant, based on increased mitotic activity and/or prominent nucleoli or nuclear pleomorphism, while 14 (47%) were classified as benign. Most of the deep seated ME tumors in this groups appeared to have atypical histologic features in keeping with a malignant lesion, as were 3 of the 4 pulmonary lesions. In contrast, most of the superficially located tumors were benign, as were all the osseus lesions.
EWSR1-POU5F1 Fusion Identified in a Subset of Deep Soft Tissue ME Tumors of Children and Young Adults, with Distinctive Clear Cell Morphology
The 3’RACE applied in one of the EWSR1 rearranged tumors with available frozen tissue showed that a 24-bp fragment of exon 7 of EWSR1 was fused in-frame to the distal 10-bp fragment of intron 1, followed by exon 2 of POU5F1 (Fig. 3D). RT-PCR with primers flanking this break-point was performed and confirmed the fusion transcript by direct sequencing (Fig. 3F). Furthermore, BAC probes flanking and spanning POU5F1 were designed (Fig. 1) and FISH confirmed the break-apart signal (Fig. 3G). FISH analysis using the POU5F1 break-apart probe was then applied to all EWSR1 rearranged tumors, identifying 4 additional tumors to be positive.
Fig. 3.
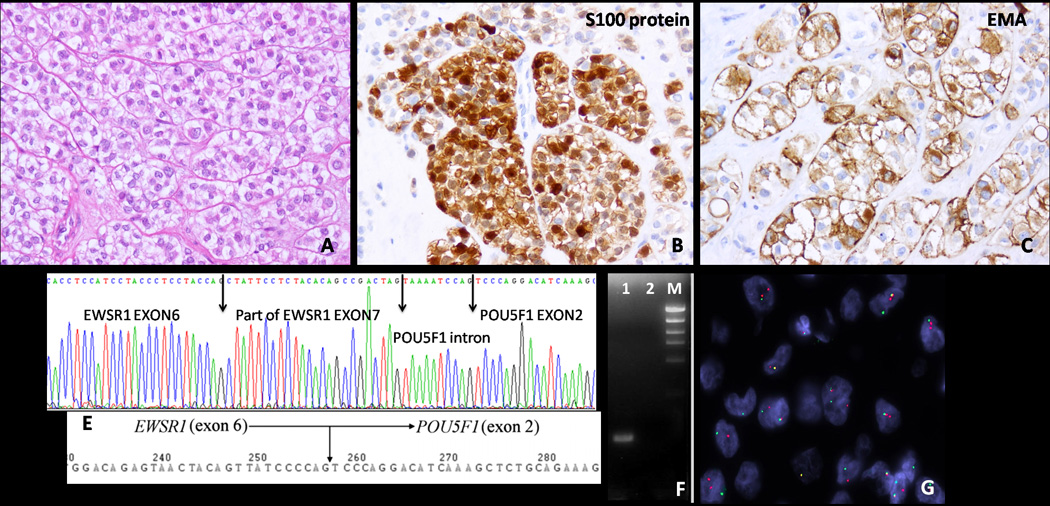
Pathologic and molecular findings in a deep-seated arm ME tumor from a 9-year old male, showing (A) epithelioid cells with abundant clear cytoplasm, arranged in a nested growth pattern, being immunoreactive for (B) S100 protein and (C) EMA, and by (D) 3’RACE showing fusion of exon 7 of EWSR1 with the last portion of POU5F1 intron 1; (E) this fusion transcript structure is similar with the one previously reported in eccrine hidradenoma/ mucoepidermoid carcinoma (Moller et al., 2008); the gene fusion results were further confirmed by (F) RT-PCR: showing an amplified product in lane 1; lane 2 is the negative control; M, size marker and by (G) FISH, with a break-apart signal for POU5F1 probe.
Thus a total of 5 (8%) patients had tumors positive for EWSR1-POUF1 fusion (Table 1). There were 3 females and 2 males, with a mean age of 21 years (range 7–34, median 26). All tumors were located in the deep soft tissues of the extremities. Morphologically they shared a predominantly solid or nested growth pattern, separated by thin fibrous septa. A clear cell phenotype was present in all 5 cases and in 4 cases the clear cell change was present in >90% of the tumor (Fig. 3A). All except one tumor had histologic features in keeping with malignancy, with brisk mitotic activity, hyperchromasia and at least mild nuclear pleomorphism. Immunohistochemically, all showed strong positivity for S100 protein and EMA (Fig. 3.B,C), with additional focal cytokeratin staining in 4 cases. However, all five tumors tested with an OCT4 (Ventana) antibody were completely negative.
EWSR1-PBX1 Fusion Present in a Small Subset of ME tumors Associated with Bland Sclerotic Appearance or Clear Cell Morphology
Cytogenetic analysis performed in one case identified the presence of a t(1;22)(q23;q12)(Fig. 4C) as a sole abnormality. Subsequent FISH analysis on this case using EWSR1 and PBX1 probes confirmed the presence of a break-apart signal in both genes (Fig. 4D,E). FISH for PBX1 rearrangement was then performed in all tumors showing an EWSR1 abnormality, with an additional 4 cases being identified as positive. Thus a total of 5 patients (Table 1), with an age range at diagnosis of 11–75 years (mean 46), had tumors positive for EWSR1-PBX1 fusion. Three tumors occurred in the deep soft tissue of the extremities, one in the pelvic bone of a 49-year old male and one in the lung of a 75-year old female smoker. S100 protein and EMA were diffusely positive in four and only focal in one tumor.
Fig. 4.
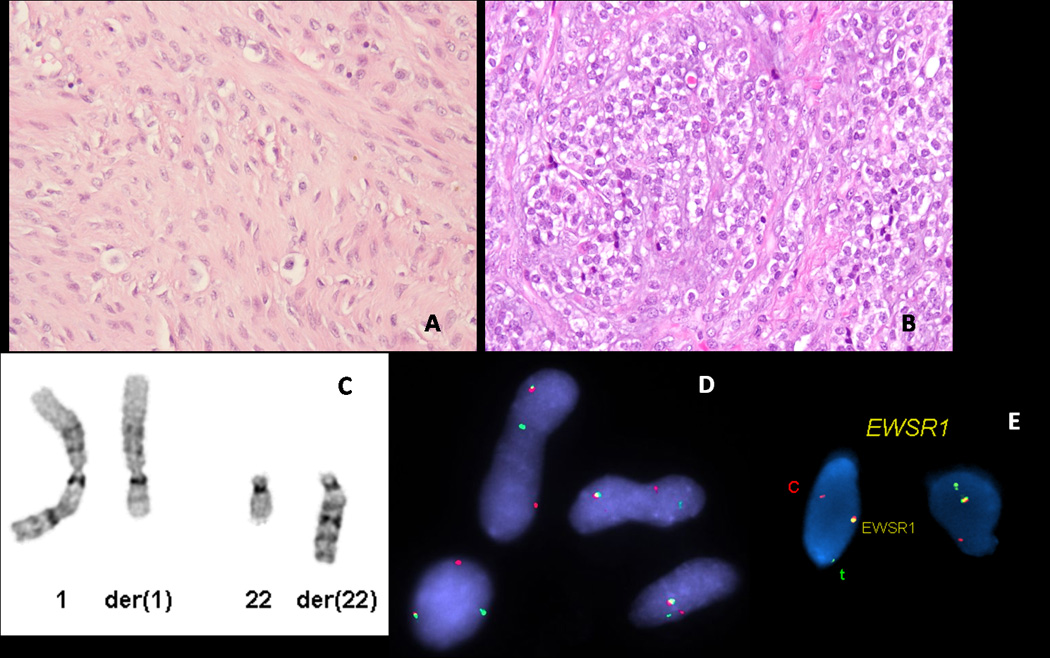
ME tumors characterized by EWSR1-PBX1 fusion: (A) 37-year old male with a left hip lesion, showing a bland spindle cell proliferation embedded in a fibrotic stroma (200x); (B) 6-year old female with a locally recurrent forearm mass showing bone invasion, composed of uniform epithelioid cells with scant clear cell cytoplasm (200x); (C) partial karyotype showing a t(1;22)(q23;q12), confirmed by FISH to have a rearrangement of (D) PBX1 and (E) EWSR1.
Morphologically, three of the tumors, located in the foot, hip and pelvic bone, showed a deceptively bland appearance, being composed mainly of spindle cells embedded in a fibrotic stroma, resembling in areas desmoid-type fibromatosis (Fig. 4A). The other two cases, located in the forearm and lung, were composed of epithelioid or ovoid uniform cells with abundant clear cytoplasm (Fig. 4B). The forearm tumor had alternating cellular areas, composed mainly of bland epithelioid cells with clear cytoplasm, with other areas displaying a more sclerotic background. This tumor occurred in a 6 year-old female, who developed three subsequent local recurrences with similar morphologic features, one showing direct invasion into underlying bone (Fig. 4B). The patient is alive with no evidence of disease, four years after the initial diagnosis.
EWSR1-ZNF444 Fusion is Rare in ME Tumors and May Also Occur in Visceral Sites
Only one tumor with EWSR1 rearrangement showed a break-apart signal with the ZNF444 probe by FISH (Fig. 5B), which occurred in the lung of a 64-year old female. The morphologic appearance was quite typical, with predominantly epithelioid cells with scant, clear cytoplasm, arranged in Indian-files or pseudo-rosettes separated by prominent sclerotic stroma (Fig. 5A). A focal spindle cell component was also noted. Tumor cells were positive for cytokeratin AE1:AE3 and S100 protein, but negative for EMA.
Fig. 5.
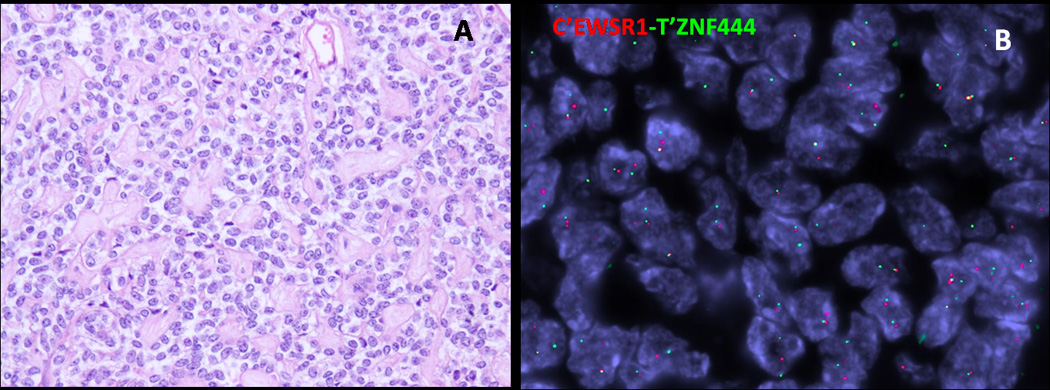
Pulmonary ME tumor from a 64-year old female showing (A) epithelioid cells with clear cytoplasm separated by a prominent collagenous stroma (200x), which had a (B) ZNF444 rearrangement by FISH.
ME Tumors Negative for EWSR1 Rearrangement are More Often Benign, Cutaneous or Superficially Located
The remaining 36 (56%) cases were negative for EWSR1 gene rearrangement (Supplementary Table 3). The patients in this group showed equal sex distribution (18 females, 18 males) and a mean age at presentation of 42 years (range 1–74, median 45 years). Most of the tumors were located in the soft tissue (78%), with predilection for the skin and subcutaneous tissue (17 cases - 61%), while 11 (39%) were deep-seated. In fact, four of the six cutaneous tumors and all of the tumors that showed ductal, glandular or chondroid/osseous matrix formation lacked rearrangement of the EWSR1 gene. Twenty tumors (55%) in this group were classified as benign. There was no predilection for any particular morphologic pattern that emerged within this group.
FUS Gene Rearrangement is a Rare Event in Soft Tissue ME tumors
Thirty ME tumors that lacked EWSR1 gene abnormalities were then screened for a FUS gene break-apart signal. Only one tumor showed the presence of a FUS gene rearrangement, which occurred in a pulmonary ME tumor from a 30-year old male. The tumor was composed of uniform epithelioid cells with scant cytoplasm, arranged in cords and reticular pattern, separated by an abundant hyalinized stroma.
Immunohistochemically, the tumor cells were diffusely positive for S100 and Cam5.2. FISH analysis for detecting a fusion partner was performed, but no rearrangements of POU5F1, PBX1 or ZNF444 genes were found.
Myoepithelial Carcinoma of the Salivary Gland and Other Related Tumors do not Show EWSR1 Gene Rearrangement
None of the tumors included in the control groups, including five salivary gland myoepithelial carcinomas, ex-pleomorphic adenoma; six salivary gland mucoepidermoid carcinoma; five cutaneous eccrine hidradenomas; three ossifying fibromyxoid tumors and two examples of chordoma periphericum, showed an abnormality of the EWSR1 gene.
DISCUSSION
Myoepithelial (ME) neoplasms represent a clinicopathologically heterogeneous group of tumors, ranging from benign to highly aggressive lesions, which based on their anatomic location are sometimes designated under different terminologies: soft tissue myoepithelioma, parachordoma, cutaneous mixed tumor, salivary gland pleomorphic adenomas, etc (Hornick and Fletcher, 2003; Kilpatrick and Limon, 2002). Although all these entities share morphologic and immunohistochemical features in keeping with ME differentiation, their potential genetic relationship has not been well established. PLAG1 and HMGA2 gene rearrangements have been associated only with salivary gland pleomorphic adenomas (Kas et al., 1997; Persson et al., 2009), and not with similar lesions at other locations (Hallor et al., 2008).
Before the present study, there have been only two reports of soft tissue ME tumors which have been successfully karyotyped (Brandal et al., 2008; Brandal et al., 2009). One tumor showed a t(1;22)(q23;q12), resulting in an EWSR1-PBX1 fusion (Brandal et al., 2008). This lesion occurred in the deep soft tissue of the foot of a 59-year old female, with a 10-year history of disease. Microscopically, the tumor was composed of a mixture of bland epithelioid and spindle cells, embedded in a fibrotic stroma, lacking any features of malignancy. Immunohistochemically, reactivity for both S100 and EMA was noted. Likewise, three of the five EWSR1-PBX1 positive tumors in our study showed very similar morphology, with a deceptively sclerotic appearance. In addition to the soft tissue location, seen in three of our cases, one tumor was located in the bone and the other one in the lung. PBX1 was previously implicated in another neoplasia-specific translocation, being fused to TCF3, resulting in a t(1;19), and seen mostly in pre-B acute lymphoblastic leukemia (Troussard et al., 1995). PBX1, like all the others EWSR1 partners described to date, is DNA binding.
The second soft tissue ME tumor cytogenetically investigated showed a t(19;22)(q13;q12), resulting in EWSR1-ZNF444 fusion (Brandal et al., 2009). It occurred in a 40-year old female, who presented with an occipital soft tissue mass, and then underwent three local recurrences before the tumor metastasized to lung. Microscopically, the tumor showed an epithelioid, nested appearance, similar to the findings noted in the only EWSR1-ZNF444 positive lesion in our study, which occurred in the lung of a 64-year old female. Clinical follow-up showed that our patient is alive with no evidence of disease after 36 months since diagnosis, suggesting that the pulmonary ME tumor is most likely the primary rather than a metastatic implant from an occult primary. Thus, EWSR1-ZNF444 fusion occurs only in a small subset of soft tissue and visceral ME tumors, accounting for <2% of cases.
A significant finding in this study was the identification of a novel EWSR1-POU5F1 fusion in soft tissue ME tumors. The five positive tumors had striking similarities, all of them presenting in the deep soft tissues of extremities, in children or young adults, and microscopically composed predominantly of epithelioid cells with abundant clear cytoplasm, arranged in a nested growth pattern. These tumors were consistently strongly and diffusely positive for both EMA and S100 protein, but lacked OCT4 expression, which seems surprising since this is the transcription factor encoded by POU5F1 (see below). Interestingly, a similar fusion between EWSR1 and POU5F1 has been reported previously in a case of undifferentiated bone tumor of the pelvis, carrying a t(6;22)(p21;q12) (Yamaguchi et al., 2005). The tumor was composed of both primitive round cells and short spindle cells and reportedly was immunopositive for S100 protein and focally for cytokeratin. These morphologic features raise the possibility of an intra-osseous ME tumor.
Subsequently, a similar EWSR1-POU5F1 fusion was identified in three hidradenomas of the skin and one case of mucoepidermoid carcinoma of salivary gland, which, in contrast to our findings, showed OCT4 expression immunohistochemically (Moller et al., 2008). Although not identical, the transcript composition of the EWSR1-POU5F1 reported in these cases was quite similar with the one identified in our ME tumor (Fig. 3D,E). These two epithelial tumor types have been previously shown to be molecularly related, both entities showing a CRTC1-MAML2 fusion in approximately half of the cases (Behboudi et al., 2006; Winnes et al., 2007). A FISH investigation of ten additional hidradenomas lacking the CRTC1-MAML2 fusion transcript identified two cases with EWSR1 gene rearrangement (Moller et al., 2008). Hidradenoma is a benign adnexal tumor composed of different cell types. When clear cell changes predominate the tumor is referred to as a ‘clear cell hidradenoma’. In a previous study on 20 cases of hidradenoma, CRTC1-MAML2 fusion transcript was particularly prevalent in this histologic subtype, all ten fusion-positive cases contained clear cells, while such features were absent or sparse in the fusion-negative hidradenomas (Winnes et al., 2007). Similar to hidradenoma, mucoepidermoid carcinoma of salivary gland is composed of multiple cell types, including epidermoid, mucinous and intermediate. The presence of a CRTC1-MAML2 fusion transcript in mucoepidermoid carcinoma has been associated with smaller and lower grade tumors having a favorable outcome (Behboudi et al., 2006; Okabe et al., 2006). However, none of the five eccrine hidradenomas and six salivary gland mucoepidermoid carcinoma in our study showed an EWSR1 rearrangement.
In addition to hidradenoma, a cytogenetic report of porocarcinoma identified a translocation affecting the long arm of chromosome 22 (Jin et al., 1998), suggesting that the EWSR1 gene may be involved in additional subtypes of skin appendageal tumors. The relationship between these EWSR1 -fusion positive skin adnexal tumors (i.e. hidradenoma, poroma, porocarcinoma) and tumors reported in this series with features of superficial myoepithelioma and cutaneous benign mixed tumor requires additional investigation. However, most of the cutaneous ME tumors and the lesions displaying ductal/glandular differentiation in this study were negative for EWSR1 gene rearrangement, suggesting an alternative pathogenesis. Furthermore, the presence of EWSR1 gene rearrangement in only one of the three mucoepidermoid carcinoma studied by Moller and coworkers (Moller et al., 2008) and none of the five in the present series suggests that other mechanisms may be involved in the pathogenesis of that tumor type. In addition, none of the five myoepithelial carcinomas of salivary gland studied showed involvement of the EWSR1 gene, suggesting that at least a subset of myoepithelial tumors arising in salivary gland may not be related to their soft tissue counterparts.
POU5F1 expression is restricted to germ cells in mature adult tissue and in human tumors is typically present in germ cell tumors (Santagata et al., 2007). POU5F1 encodes a transcription factor which binds to the octamer motif (ATGCAAAT) present in the promoter or enhancer regions of target genes. Murine POU5F1 is expressed in embryonic stem cells (ESCs) and germ cells. The level and duration of POU5F1 expression has been found to be tightly regulated, a critical amount of POU5F1 sustains the stem cell phenotype, while up- or down- regulation of expression induces divergent developmental programs (Niwa et al., 2000). The multiple cell types seen in the composition of tumors positive for the EWSR1-POU5F1 fusion in our study may be indicative of a multi-phenotypic differentiation. ME tumor is a prototypical example of dual differentiation to both epithelial and mesenchymal lineages. POU5F1 expression as evidenced by OCT4 immunoreactivity was documented in the EWSR1-POU5F1 positive hidradenomas (Moller et al., 2008) – however, it was negative in all five ME tumors in the present study, using the same antibody. Furthermore, the mRNA expression of POU5F1 studied by expression profiling was not increased as compared to an EWSR1-ZNF444 positive ME tumor or other sarcoma types (data not shown). In this regard, transcriptional activation assays of the chimera suggested the possibility of a negative regulatory effect on target genes (Moller et al., 2008). Although being capable of transcriptional activation, the EWSR1-POU5F1 chimera activates transcription less efficiently than wild type EWSR1 (Moller et al., 2008). As such, translocation may represent a mechanism of POU5F1 activation by inappropriate expression of POU5F1 DNA binding sites.
The differential diagnosis of ME tumors is typically quite broad and may vary depending on patient age and anatomic location. Ossifying fibromyxoid tumor, an S100 protein-positive soft tissue tumor with an uncertain line of differentiation, shares significant morphologic overlap with ME tumors. Although only three OFMTs were included for genetic analysis, the lack of EWSR1 abnormalities suggests that this entity may not be related to ME tumors.
The distinction from extraskeletal myxoid chondrosarcoma (EMC) can often be quite difficult, especially in large, deep-seated soft tissue tumors associated with myxoid changes. The presence of EWSR1 gene rearrangement, used previously to support the diagnosis of EMC, is now demonstrated in both tumor entities, in a significant number of cases. In spite of the morphologic overlap and EWSR1 gene abnormality, we do not believe the two tumors are related. The consistently strong EMA and S100 protein co-reactivity, the common clear cell changes and nested growth pattern seen in ME tumors, but not in most EMC, should help in the distinction. Furthermore a predominant myxoid stroma throughout the tumor is rarely seen in ME lesions.
The presence of EWSR1 gene rearrangement in about half of ME tumors which can be readily detected by FISH analysis can serve as a powerful diagnostic tool in challenging cases. However, this finding adds ME tumors to an already growing family of EWSR1 gene-rearranged tumors, which are often considered in the differential diagnosis, especially in the pediatric age group (Romeo and Dei Tos, 2010). As illustrated in this study, ME tumors with EWSR1 gene rearrangement often have uniform rounded cell morphology and clear cytoplasm, presenting in the deep soft tissues of the extremities. These findings pose significant overlap both microscopically and clinically with other pediatric tumors, especially with Ewing sarcoma, also characterized by recurrent translocations involving the EWSR1 gene. In difficult cases which are positive for EWSR1 rearrangement, efforts to identify the translocation partner should be undertaken by RT-PCR methods for a definitive diagnosis and to avoid unnecessary systemic treatment.
These findings reinforce the fact that recurrent chromosomal translocations involving EWSR1 do not occur only in aggressive high grade sarcomas, but also in tumors with low or undetermined malignant potential, like ME tumors and so-called angiomatoid fibrous histiocytoma. Furthermore, these results provide solid evidence for a unifying concept of soft tissue ME with similar tumors arising in bone and at visceral locations. However, the data presented here do not support a pathogenetic relationship between soft tissue ME tumors and their salivary gland counterparts. Additional cases will need to be analyzed in order to further investigate the relationship of soft tissue ME lesions and cutaneous benign mixed tumors.
REFERENCES
- Agaram NP, Laquaglia MP, Ustun B, Guo T, Wong GC, Socci ND, Maki RG, DeMatteo RP, Besmer P, Antonescu CR. Molecular characterization of pediatric gastrointestinal stromal tumors. Clin Cancer Res. 2008;14:3204–3215. doi: 10.1158/1078-0432.CCR-07-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behboudi A, Enlund F, Winnes M, Andren Y, Nordkvist A, Leivo I, Flaberg E, Szekely L, Makitie A, Grenman R, Mark J, Stenman G. Molecular classification of mucoepidermoid carcinomas-prognostic significance of the MECT1-MAML2 fusion oncogene. Genes Chromosomes Cancer. 2006;45:470–481. doi: 10.1002/gcc.20306. [DOI] [PubMed] [Google Scholar]
- Brandal P, Panagopoulos I, Bjerkehagen B, Gorunova L, Skjeldal S, Micci F, Heim S. Detection of a t(1;22)(q23;q12) translocation leading to an EWSR1-PBX1 fusion gene in a myoepithelioma. Genes Chromosomes Cancer. 2008;47:558–564. doi: 10.1002/gcc.20559. [DOI] [PubMed] [Google Scholar]
- Brandal P, Panagopoulos I, Bjerkehagen B, Heim S. t(19;22)(q13;q12) Translocation leading to the novel fusion gene EWSR1-ZNF444 in soft tissue myoepithelial carcinoma. Genes Chromosomes Cancer. 2009 doi: 10.1002/gcc.20706. [DOI] [PubMed] [Google Scholar]
- Gleason BC, Fletcher CD. Myoepithelial carcinoma of soft tissue in children: an aggressive neoplasm analyzed in a series of 29 cases. Am J Surg Pathol. 2007;31:1813–1824. doi: 10.1097/PAS.0b013e31805f6775. [DOI] [PubMed] [Google Scholar]
- Hallor KH, Teixeira MR, Fletcher CD, Bizarro S, Staaf J, Domanski HA, von Steyern FV, Panagopoulos I, Mandahl N, Mertens F. Heterogeneous genetic profiles in soft tissue myoepitheliomas. Mod Pathol. 2008;21:1311–1319. doi: 10.1038/modpathol.2008.124. [DOI] [PubMed] [Google Scholar]
- Hornick JL, Fletcher CD. Myoepithelial tumors of soft tissue: a clinicopathologic and immunohistochemical study of 101 cases with evaluation of prognostic parameters. Am J Surg Pathol. 2003;27:1183–1196. doi: 10.1097/00000478-200309000-00001. [DOI] [PubMed] [Google Scholar]
- Jin Y, Jin C, Mertens F, Persson B, Jonsson N. Characterization of a malignant eccrine poroma by cytogenetic and fluorescence in situ hybridization techniques. Cancer Genet Cytogenet. 1998;102:100–103. doi: 10.1016/s0165-4608(97)00347-6. [DOI] [PubMed] [Google Scholar]
- Kas K, Voz ML, Roijer E, Astrom AK, Meyen E, Stenman G, Van de Ven WJ. Promoter swapping between the genes for a novel zinc finger protein and beta-catenin in pleiomorphic adenomas with t(3;8)(p21;q12) translocations. Nat Genet. 1997;15:170–174. doi: 10.1038/ng0297-170. [DOI] [PubMed] [Google Scholar]
- Kilpatrick SE, Limon J. Pathology and Genetics of Tumours of Soft Tissue and Bone. Lyon: IARC Press; 2002. pp. 198–199. [Google Scholar]
- Martins C, Fonseca I, Roque L, Pereira T, Ribeiro C, Bullerdiek J, Soares J. PLAG1 gene alterations in salivary gland pleomorphic adenoma and carcinoma ex-pleomorphic adenoma: a combined study using chromosome banding, in situ hybridization and immunocytochemistry. Mod Pathol. 2005;18:1048–1055. doi: 10.1038/modpathol.3800386. [DOI] [PubMed] [Google Scholar]
- Moller E, Stenman G, Mandahl N, Hamberg H, Molne L, van den Oord JJ, Brosjo O, Mertens F, Panagopoulos I. POU5F1, encoding a key regulator of stem cell pluripotency, is fused to EWSR1 in hidradenoma of the skin and mucoepidermoid carcinoma of the salivary glands. J Pathol. 2008;215:78–86. doi: 10.1002/path.2327. [DOI] [PubMed] [Google Scholar]
- Nielsen GP, Mangham DC, Grimer RJ, Rosenberg AE. Chordoma periphericum: a case report. Am J Surg Pathol. 2001;25:263–267. doi: 10.1097/00000478-200102000-00016. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Okabe M, Miyabe S, Nagatsuka H, Terada A, Hanai N, Yokoi M, Shimozato K, Eimoto T, Nakamura S, Nagai N, Hasegawa Y, Inagaki H. MECT1-MAML2 fusion transcript defines a favorable subset of mucoepidermoid carcinoma. Clin Cancer Res. 2006;12:3902–3907. doi: 10.1158/1078-0432.CCR-05-2376. [DOI] [PubMed] [Google Scholar]
- Persson F, Andren Y, Winnes M, Wedell B, Nordkvist A, Gudnadottir G, Dahlenfors R, Sjogren H, Mark J, Stenman G. High-resolution genomic profiling of adenomas and carcinomas of the salivary glands reveals amplification, rearrangement, and fusion of HMGA2. Genes Chromosomes Cancer. 2009;48:69–82. doi: 10.1002/gcc.20619. [DOI] [PubMed] [Google Scholar]
- Romeo S, Dei Tos AP. Soft tissue tumors associated with EWSR1 translocation. Virchows Arch. 2010;456:219–234. doi: 10.1007/s00428-009-0854-3. [DOI] [PubMed] [Google Scholar]
- Santagata S, Ligon KL, Hornick JL. Embryonic stem cell transcription factor signatures in the diagnosis of primary and metastatic germ cell tumors. Am J Surg Pathol. 2007;31:836–845. doi: 10.1097/PAS.0b013e31802e708a. [DOI] [PubMed] [Google Scholar]
- Scolyer RA, Bonar SF, Palmer AA, Barr EM, Wills EJ, Stalley P, Schatz J, Soper J, Li LX, McCarthy SW. Parachordoma is not distinguishable from axial chordoma using immunohistochemistry. Pathol Int. 2004;54:364–370. doi: 10.1111/j.1440-1827.2004.01633.x. [DOI] [PubMed] [Google Scholar]
- Tirabosco R, Mangham DC, Rosenberg AE, Vujovic S, Bousdras K, Pizzolitto S, De Maglio G, den Bakker MA, Di Francesco L, Kalil RK, Athanasou NA, O'Donnell P, McCarthy EF, Flanagan AM. Brachyury expression in extra-axial skeletal and soft tissue chordomas: a marker that distinguishes chordoma from mixed tumor/myoepithelioma/parachordoma in soft tissue. Am J Surg Pathol. 2008;32:572–580. doi: 10.1097/PAS.0b013e31815b693a. [DOI] [PubMed] [Google Scholar]
- Troussard X, Valensi F, Salomon-Nguyen F, Debert C, Flandrin G, MacIntyre E. Correlation of cytoplasmic Ig mu (C mu) and E2A-PBX1 fusion transcripts in t(1;19) B lineage ALL: discrepancy in C mu detection by slide immunofluorescence and flow cytometry. Leukemia. 1995;9:518–519. [PubMed] [Google Scholar]
- Winnes M, Molne L, Suurkula M, Andren Y, Persson F, Enlund F, Stenman G. Frequent fusion of the CRTC1 and MAML2 genes in clear cell variants of cutaneous hidradenomas. Genes Chromosomes Cancer. 2007;46:559–563. doi: 10.1002/gcc.20440. [DOI] [PubMed] [Google Scholar]
- Yamaguchi S, Yamazaki Y, Ishikawa Y, Kawaguchi N, Mukai H, Nakamura T. EWSR1 is fused to POU5F1 in a bone tumor with translocation t(6;22)(p21;q12) Genes Chromosomes Cancer. 2005;43:217–222. doi: 10.1002/gcc.20171. [DOI] [PubMed] [Google Scholar]


