Abstract
Patient outcomes to drugs vary, but physicians currently have little data about individual responses. We designed a comprehensive system to organize and integrate patient outcomes utilizing semantic analysis, which groups large collections of personal comments into a series of topics. A prototype implementation was built to extract situational evidences by filtering and digesting user comments provided by patients. Our methods do not require extensive training or dictionaries, while categorizing comments based on expert opinions from standard source, or patient-specified categories. This system has been tested with sample health messages from our unique dataset from Yahoo! Groups, containing 12M personal messages from 27K public groups in Health and Wellness. We have performed an extensive evaluation of the clustering results with medical students. Evaluated results show high quality of labeled clustering, promising an effective automatic system for discovering patient outcomes from large volumes of health information.
Introduction
Online discussions now play an essential role in people’s lives. When a person plans to buy an electronic product, she would like to view other customers’ reviews from shopping websites. When a person considers trading on some specific stock, she would like to know other traders’ comments from stock discussion board. Web forums in specific area provide valuable information in support of users’ product judgment by exposing them to others’ recent experiences. Though subjective, they reflect comprehensive first hand opinions from actual people using actual products.
When it comes to the area of healthcare, the situation is similar: online bulletin boards and chat groups, such as Yahoo! Groups1 and WebMD2, offer patients and physicians a good platform to discuss health problems, e.g., diseases and drugs, diagnoses and treatments. These online user discussions also provide rich material for textual analysis to extract patient outcomes related to drug regimes for individual persons. Such analysis could enable physicians to better know the side effects of particular drugs, and patients to better know the experiences of similar patients, related to whether the drug is effective, under what conditions.
Online medical discussions have limitations hindering users’ effective knowledge of information. If a user searches for a specific drug, there are usually thousands of comments or reviews returned, many of which are useless. The user often has difficulty in digesting and understanding the information quickly – she has to select the useful posts from the pool and read each one by one. Therefore, the outcomes need to be organized and integrated in a targeted fashion. To address such problem, we propose a prototype system for digesting health messages.
Unlike product reviews, medical discussion messages are unstructured, i.e., each comment talks about several topics in one piece of plain text. These messages must be partitioned into parts, and these parts must be grouped together according to what topic category they each belong to. By doing this we will have a coherent view on different aspects of the medical issue based on all the information available from our source. Our purpose is to re-construct and integrate a large number of unstructured online messages, into meaningful groups according to the topics, in order to aid users navigate through the vast information pool and satisfy their information need.
We designed a prototype model for clustering patient outcomes by effectively digesting large volumes of personal health messages. First, the useful user comments are retained, while news and advertisements which are noise for our purpose, are filtered out by an Support Vector Machine (SVM)-based classifier. In the main step, similar topics are grouped from sentences appearing in different messages by Probabilistic Latent Semantic Analysis (PLSA) topic model, where the topic categories can be guided by standard outcome descriptions from expert sources. In addition to identifying the sentences which are similar to (agree with) expert opinion into the corresponding topic, the model also clusters the sentences which are opposite to (disagree with) expert opinion into the same topic. In other words, for each outcome provided by experts, the system automatically identifies the sentences which provide positive support for the expert opinion and those provide negative support. The process organizes and integrates all the messages from online medical discussions in a practical way, relevant to particular persons in particular situations.
We have implemented a prototype interactive system for text mining of health messages. This system has been tested with sample messages from our unique dataset from Yahoo! Groups, which contains 12M personal messages from 27K public groups in Health and Wellness. This outcome research utilizes deeper processing of natural language, such as SVM and PLSA, than our previous studies on drug reactions with the same dataset[1,2]. Our methods do not require extensive training nor dictionaries. In addition, they allow users to specify their own topics for digesting. Therefore, our methods provide general and powerful solutions to mine health messages.
We have evaluated the prototype system with a sample set of drugs using a sample cohort of medical students. 5000 sentences relevant to 10 representative drugs were randomly selected, and automatically clustered into topics extracted from PubMed Health database 3, a well known expert source for drug information. The accuracy of these clustering results was evaluated by medical students in the College of Medicine at the University of Illinois in Urbana. By comparing the automatically generated clustering results to the ones generated by these professional annotators, it is shown that our topic clustering methods produce highly accurate results. We also statistically prove that all judges were consistent in classifying the sentences and thus have produced a valid gold standard for our evaluation.
Guided by the standard expert opinions extracted from PubMed Health, our topic clustering provides robust automatic classification of patient-reported drug outcomes. That is, our system can automatically classify patient outcomes, which describe patients’ experience and result of using a particular drug, often using layman language, into standard categories derived from PubMed Health, with high accuracy. Drugs used for our evaluation can be divided into two classes: specialized and generalized. The first class treats a particular medical condition (e.g., Metformin), while the second class includes over-the-counter drugs (e.g., Ibuprofen) and commonly-prescribed-drugs (e.g., Heparin). The results show the accuracy of clustering specialized drugs is higher than that of generalized drugs. This is reasonable since specialized drugs often have a focused range of treatments and side effects, which makes patients’ outcome description more specific and consistent. In addition, we also observe that the clustering methods work better for more common drugs, possibly because users are likely to be more knowledgeable about drugs they encounter often.
We also show that our system can explore outcomes not included in the standard expert source. In this particular experiment, we have computed an additional cluster that groups together sentences not closely associated with any of the standard clusters. By examining this additional cluster, we discover some patient comments concerning serious side-effects or other treatments, but not discussed in the standard outcome description on PubMed Health. By referring to the medical literature, we are able to confirm many of these patient-provided outcomes have been recorded as possible results of using the particular drug. Patient-reported outcomes can be an important supplementary source of information, even when automatically extracted from health messages.
In summary, our outcomes system is accurate for clustering standard outcomes and effective for discovering novel outcomes, while fully automatic with text processing. Thus by using this system as the core engine, a national surveillance system is feasible to automatically extract drug outcomes from patient messages.
Related Work
Many applications have utilized formal medical literature to extract useful information, such as generating text summaries[3] and topic modeling[4]. We instead use informal medical messages which are generated by large numbers of online users. Compared with the formal literature, our dataset from web posted personal medical messages is more unstructured and noisy, which challenges the information extraction.
Instead of formal and structured literature, some research papers apply natural language processing techniques on unstructured clinical notes, such as abbreviation analysis[5] and social-history information detection[6]. Compared with these, our dataset from personal medical messages is more informal and contains more noisy information, which challenges the text processing. Meanwhile, the topic diversity of our dataset reflects various responses and opinions from various physicians and patients, which are rare in clinical notes.
There have been only a few studies using informal medical sources: Crain et al.[7] worked on consumer medical search by using Yahoo! Answer messages, while Yang et al. did a solid query log analysis[8] based on the Electronic Medical Record Search Engine (EMERSE)[9]. We have published several papers based on Yahoo! Groups messages, such as tracking users’ sentiments[1] and predicting adverse drug events[2]. Using the same dataset, we perform a comprehensive information extraction task and apply a series of text mining techniques to extract patient outcomes.
Recently semantic clustering represents a more effective approach for reorganizing texts in support of human understanding. For example. Lin and Demner-Fushman[10] proposed a hierarchical agglomerative algorithm to cluster Medline abstracts. Similar to their work, we try to cluster sentences of message into meaningful clusters, but in a much finer-grained way. Therefore, advanced clustering approach like topic model will be considered.
Topic models such as Probabilistic Latent Semantic Analysis (PLSA)[11] and Latent Dirichlet Allocation (LDA) [12] have been applied to text mining problems with good results. Lu et al.[13] applied PLSA model to integrate product aspects from online product reviews, while Kandulaweb et al.[14] utilized LDA model to discover diabetic-related medical materials. In the above work, limited evaluations upon a small number of manual post-labeling were performed. After performing a semi-supervised PLSA model, our work performs a comprehensive evaluation strategy to evaluate the effectiveness of the model, based upon gold standard values produced by medical professionals.
To the best of our knowledge, our work is the first to integrate and analyze patient drug outcomes from online personal health messages, with a comprehensive evaluation framework.
Problem Definition
For one particular drug, we collect all the related health messages from Yahoo! Groups, denoted as M, in which N is the set of all the news, C is the set of all the user comments (our target), and S is the set of all the spam such as advertisements. Clearly, we have M = N ∪ C ∪ S.
After successfully extracting C, we split it into a set of meaningful sentences, denoted as D. Each sentence d ∈ D is called a comment unit, which would potentially present one side of outcomes. Our target goal is to group all the comments into m meaningful outcome clustersO1, O2, ...Om, given the collection D.
Here are several key concepts to be introduced:
– Expert commentei: To better cluster the outcomes, semi-supervised PLSA model[11] is applied. Expert comments aim to offer the prior knowledge for PLSA and guide the topic of each Oi. For each Oi we have one expert comment ei. Compared with the user comments, the expert comments are more well-written, professional and semantically discriminative to each other. We collect the set of expert comments E (formed by e1, e2, ...em–1) for each drug, from the PubMed Health database of U.S. National Library of Medicine, Drug and Supplement Category4, which includes the detailed description of each drug. The reason why we choose m – 1 expert comments is that we want to create some groups of opinions with prior expert knowledge (O1, O2, ...Om–1) as well as another group of opinions whose topics are beyond the expert’s (Om).
– Similar opinionOi_sim and Opposite opinionOi_opp: Each outcome Oi (1 ≤ i ≤ m – 1) consists of a group of comment units Di and is associated with one expert comment ei. Some of the comments represent similar or relevant opinion with ei, which form Oi_sim. Others reflect different or opposite opinions from ei, though they still talk about the same topic. We call such collection Oi_opp.
System Architecture
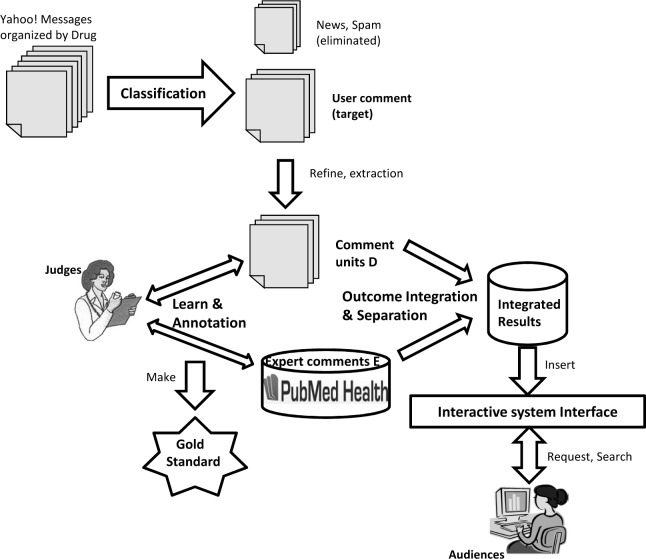
Figure 1 illustrates the whole design and implementation process of our system.
Figure 1:
The architecture of our system
Messages from Yahoo! Groups are firstly organized by drug and then classified into three categories: N, C, S. N and S are eliminated while the the collection of comment units (D) is extracted from user comments (C), together with the expert comments (E) as input. On the one hand, such comment units (D) will be re-organized and integrated into several meaningful outcomes O1, O2, ...Om by our topic model, and presented to audiences via our system interface. On the other hand, judges will annotate the extracted data (D and E). Their annotation results will become a gold standard to evaluate the performance of our model. In the following sections we will introduce the key components of our system in details.
Methodology
As we introduced before, there are several steps to complete the whole task.
Data Pre-processing (step 1): The input is the collection M. We will separate it into three categories: News (N), Comment (C) and Spam (S). C is our target while N, S will be filtered out. C is then split into a set of comment units D.
Data Clustering (step 2): Given D and m–1 expert comments e1, e2, ...em–1, we will generate m outcome clusters O1, O2, ..., Om. Each cluster Oi refers to one meaningful drug outcome, either guided by expert opinion ei (1 ≤ i ≤ m – 1), or contributing to “other opinions” (i = m).
Data Post-processing (step 3): For each cluster with prior expert knowledge Oi, we will split it into Oi_sim – expressing the similar opinion to ei, as well as Oi_opp, which shows the opposite opinion.
A. Pre-processing: Filtering the messages
After extensively observing the messages on this online forum, we find that there are three types of messages:
News (N): The content of a news article is mainly about the FDA approval or scientific discovery of drugs. It is usually so long that useful information is difficult to extract. Therefore we will eliminate this group of messages.
User comment (C): User comments are the most informative part in the whole collection M. They are of proper length and provide good amount of useful information. This group of messages is our target.
Spam (S): Most advertisements[15], posted by human or robots, should be eliminated for our purpose. They are often short, and appear repeatedly, which makes it easy to identify them automatically.
Table 1 shows a snippet of each type of message, which are extracted from the real data.
Table 1:
Three Types of Messages
| (a). News |
| ...FDA officials could not maintain their iron grip in an effort to suppress evidence of far reaching lethal effects of Vioxx when their actions were in full public view. FDA officials were forced to lift the roadblocks they had put in Dr. David Graham’s way... |
| (b). User Comment |
| ...Taking Aspirin gives me bloodshot looking eyes (just enough to look horrible) yet I forgot to take it for 2 days and my eyes were very clear and bright. I don’t want to take aspirin either but my family has a strong history of heart attacks and migraine with aura... |
| (c). Spam |
| ...Everything is 70% off for this week only! Get for mens health Buy Valium for CHEAP Get Xanax for Anti Anxiety. Buy Meridia online for weight loss... |
To distinguish the messages, we apply SVM classification[16] to split all the messages into these three classes (N, C, S). Support vector machines (SVM) are a set of related supervised learning methods used for classification by analyzing data and recognizing patterns. Compared with other grouping approaches like manually labeling, rule-based parsing, SVM has relatively higher accuracy and can handle high-dimensional data automatically. It is a good solution to distinguish text messages and filter out the useless ones.
To apply SVM classification, we need to label some training data, select the proper features, transfer messages into feature vectors, train an SVM classifier on the training data and test it. Among them, the most essential step is the feature selection. In our previous work[17], we find the following three types of features can be considered: Term-appearance feature, i.e., word distribution; Lexical feature, such as the number of terms, the average length of sentences, etc; Semantic feature, such as the percentage of drug/treatment names, the percentage of positive/negative sentences, etc. We train a tri-class SVM classifier[18] and test on the real data. The results show a high accuracy.
B. Clustering: Outcome selection and integration
To achieve our core step of the system: grouping the comments into reasonable and discriminative clusters, where each cluster represent one main outcome of the drug, semi-supervised PLSA model[11] is applied. We would introduce the model first and then describe the integration process.
PLSA model
In PLSA model, we consider each comment unit d ∈ D is generated from a mixture of m+1 multinomial component models. One component model is the background model θB that absorbs non-discriminative (i.e., meaningless) words and the rest are m latent theme topic models (saying Θ = {θ1, θ2, ... θm}) via the Expectation Maximization (EM) algorithm[19]. Also, we we enroll some prior knowledge by extending the basic PLSA to incorporate a conjugate prior based on expert comments e1, e2, ..., em–1 (i.e., semi-supervised). For each outcome cluster Oi (1 ≤ i ≤ m–1), since we have already acquired the expert comment ei, we can build a unigram language model {p(w|ei)}, and estimate the language model for each cluster j: p(w|θj) by Formula 1 as below 5:
| (1) |
In Formula 1, c(w, d) refers to the frequency of term w appearing in comment unit d. p(zd,w,j) indicates the probability that the word w in comment unit d is generated using topic j. While μ can be interpreted as “equivalent sample size”, which means that the impact of adding the prior. Note, for cluster Om, there is no prior knowledge since we expect to discover some additional opinions rather than the experts’.
Integration Progress
We build m meaningful clusters by appling semi-supervised PLSA model, there are several steps described below:
Build the prior knowledge. For each cluster Oi (1 ≤ i ≤ m – 1), we have already acquired an expert comment ei from PubMed Health database of U.S. National Library of Medicine. Based on it, we estimate {p(w|ei)} by Maximum Likelihood as the prior estimator. Here only adjectives, adverbs, verbs and nouns are considered in the estimator since they are the terms which express the opinions.
Given such prior knowledge and the set of the comment units D, we could estimate the topic models {θ1, θ2, ... θm} by the formulas above.
- For each comment unit d ∈ D, we assign it to the most suitable cluster by the following formula:
(2) For each opinion Oi, we generate a topic model θj as well as a bunch of corresponding comment units Di. The terms which has high probability p(w|θj) in θj as well as featured comment unites can represent such topic.
C. Post-processing: Separating similar and opposite opinions
Now in each cluster Oi, there are a couple of assigned comment units Di. For the cluster which has a prior expert comment, we will split it into Similar opinion Oi_sim and Opposite opinion Oi_opp, by applying semi-supervised PLSA model (creating two clusters with ei as one Oi_sim’s prior knowledge while Oi_opp has no prior knowledge).
To create such two clusters, the straightforward approach is to build one cluster’s prior estimator {p(w|ei)} by the typical way: all the adjectives, adverbs, verbs and nouns are considered in the estimator since they are the terms which express the opinions, and leave another cluster’s prior as empty.
This approach has one limitation that it does not address the sentiment meaning. Look at the following two sentences: “I took some Aspirin to treat the back pain and it works well”. “I took some Aspirin to treat the back pain, but bad effect.” They have nearly the same vocabularies to address the same topic but with opposite opinions. For such sentences with similar lexical structure, a better way to distinguish them is to detect the sentiment terms. i.e., “well” and “bad”, which are key points to express the positive opinion and negative opinion, respectively. Start with this observation, we propose another approach to build the prior estimator {p(w|ei)} where only positive/negative terms in ei are considered. Since all the sentences in Oi have already been considered to have the same topic with ei. It is more appropriate to focus on the sentiments while splitting Oi into Oi_sim and Oi_opp.
In the experiment, we will implement each approach respectively and compare their performances.
Experiments and results
A. Data and Setup
We utilize our unique dataset which is segmented from Yahoo! Groups with Health and Wellness data. The dataset consists of 27, 290 public groups and over 12, 519, 807 messages in total, spanning seven years and multiple topics. All the experiments run on a 4TB-disk, 4GB-RAM, and 10-core server.
We have trained an SVM-based tri-class classifier with an RBF kernel on the real data and tested it. Evaluation results[17] show that our classifier can achieve 90.15% overall accuracy as well as 93.31% accuracy of detecting user comments (C), which indicates that our approach could successfully distinguish messages’ categories, especially user comments.
Since we are targeting personal medical information, we choose to evaluate the system with outcomes of specific drugs. The evaluation system first checks the frequency of each drug from the complete list offered in our previous work[2], Only the drugs appearing more than 1000 times are considered, since their relatively high frequency of appearance may ensure that sufficient personal messages can be processed. Finally, we carefully choose 10 drugs judged to be representative and known by physicians. Half of the drugs are Prescription - Variety Medical Conditions(Prescript-VMC) drugs: Metformin, Clonidine, Gabapentin, Clonazepam and Oxaliplatin, and five are Pain Relief or Anti-coagulation(PRAC) medications: Aspirin, Heparin, Ibuprofen, Hydrocodone and Naproxen. For each drug, we collect all the messages containing its name or synonyms, process them by our classifier and get the user comments C, split C and collect sentences D which either contain the drug name, or are next to the sentences containing the drug. These sentences D potentially represent users’ diverse opinions on the drug.
We were aware that a more straightforward approach is to compare Prescription drugs with over-the-counter (OTC) drugs. However, OTC drugs tend to contain more noisy data and we noticed that, after pre-processing, many OTC drugs simply do not have enough information on this forum of proper length and diversity for our evaluation purpose. In this case, we set two groups as Prescript-VMC drugs with different specific treatments, and PRAC with more general treatments, such as some OTC drugs (Aspirin, Naproxen, Ibuprofen) as well as drugs with similar treatments (Hydrocodone and Heparin).
Due to the uniqueness of our data source – Yahoo! Groups, it is difficult to apply the traditional evaluation approaches by comparing to a gold standard like TREC medical informatics[20]. Thus, a comprehensive evaluation system which contains a large-scale of professional-labeled sentences as our gold standard should be built and applied.
B. Annotation Framework
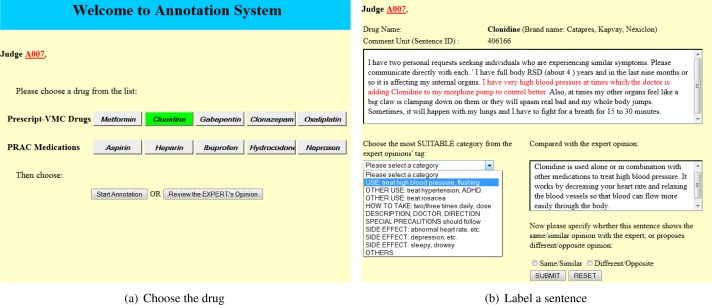
We have built an interactive web-based database system to support the evaluation process. 500 comment units (i.e., sentences) are randomly generated for each drug and stored in the database (Note, for some drugs such as Naproxen, the total number of available comment units is slightly more than 500). For a specific drug, the professional evaluation judge needs to become familiar with the pre-defined expert comments (8–10 per drug), each of which is associated with a given tag, and then enter the actual annotation. Each time one comment unit d is given together with its context in the actual personal message. The judge needs to understand the meaning of d and assign it to the most suitable cluster (recognized by the tag of the corresponding expert opinion) it belongs to, or to “other” cluster if no prior expert opinion matches. After that, the judge is also asked to determine whether d shows similar or opposite opinion with the chosen expert’s. The annotation of one comment unit is then finished and the result will be stored in the database. Figure 2 simplifies the interface for the annotation process.
Figure 2:
Annotation Interface
Similar to Blake’s work[21], we design a two-step annotation process. The purpose of the first step, a pilot study, is to validate the design of annotation process, including the instruction and defined categories, is easy to understand and unambiguous for human annotators. If the pilot study shows there is no large variance of understanding about the annotation process among the annotators, we will proceed to the main study to complete the actual annotation.
Pilot study: In this experiment, three graduate students (majors are computer science, nutrition and bioinformatics) and one medical school student were enrolled. We assigned each of them 100 identical sentences (50 for Prescript-VMC and 50 for PRAC, randomly generated, covering all common clusters) and they labeled the sentences independently following the instructions. Then we collect their annotations and test the inter-rater agreement by using Fleiss’ kappa[22]. Fleiss’ kappa is widely used for assessing the reliability of agreement between a fixed number of judges when assigning categorical ratings to a number of items or classifying items.
From computation, the four judges’ kappa-value reaches 0.84. According to the interpretation of the kappa statistic[23], this result shows almost-perfect agreement among the judges, which proves that our annotation process is designed with minimum ambiguity.
Main study: To reach our initial goal, a gold standard should be made by professional annotators upon all the sentences. In the main study, we invited 10 medical school students who are experienced, and familiar with information about drugs and treatments, among other qualifications to develop our gold standard. For time and quality concern, it is impossible to ask one judge to label all the sentences so we randomly split the whole annotation task to 10 judges. To test the inter-judge reliability, we repeated the process of pilot study – assigning them 50 identical sentences, which are randomly selected from the sentence pool and cover all common clusters. The Fleiss’ kappa value reaches 0.81, which is considered almost perfect agreement.
The above results indicate that there is no significant variance among all the annotators, and prove that, by comprehending the task instruction, our judges are well-trained enough to provide generally consistent gold standard. Therefore, we could confidently apply the gold standard to evaluate our clustering results. The whole annotation process lasted half a month and a random follow-up check was executed by two other medical school students afterward. The output of such well-designed and professional-enrolled annotation framework is good enough to become the gold standard of our integrated system.
C. Clustering Results and Analysis
Now we have the set of comment units D for each drug, as well as the expert comments E extracted from PubMed Health database of U.S. National Library of Medicine. The number of expert comments for each drug is based on the content of the description in the database. Thus it varies from 8 to 10 for different drugs. According to the semi-supervised PLSA model introduced before, we assign each comment unit to the suitable cluster with prior expert knowledge or “other outcome” cluster without prior knowledge and compare the results to the gold standard data.
To measure the quality of our clustering results, we utilize the following measurements: accuracy, precision, recall and F-score, which are defined by:
| (3) |
| (4) |
| (5) |
| (6) |
In Formula 4 and 5, “expert sentence” means the sentence which is assigned into a cluster associated with one expert opinion (O1, ...Om–1), regardless of annotated or auto-retrieved. In other words, the measurements precision, recall and F-score ignore the potential effect by “other opinion” cluster (Om). Note, our measurements only evaluate the correctness of clusters, not the correctness of the sentences though “incorrect” user comments do exist.
Table 2 shows the performance of the clustering result by each drug, each category (prescript-VMC or PRAC) and overall. we can observe the following facts:
– Our semi-supervised PLSA model can achieve a relatively high performance of clustering. i.e., the overall accuracy is 0.657 and the overall F-score is 0.670, considering the large number of clusters (9 to 11 per drug).
– Our result also shows that F-score is higher than the corresponding accuracy in all cases. The paired t-test[24] between F-score and accuracy for the 10 drugs is also significant (p-value < 0.05). It indicates that cluster with prior knowledge could effectively improve the performance compared to that of no prior knowledge.
– Compared with PRAC medications, Prescript-VMC drugs perform better in all cases. i.e., accuracy: 0.694 v.s. 0.620, F-score: 0.707 v.s. 0.632, etc. We also conduct t-tests between PRAC and Prescript-VMC for each of the measurements. It shows, compared to PRAC, the recall and F-score of Prescript-VMC are significantly higher(p-value < 0.05), and the accuracy(p-value = 0.06) and recall(p-value = 0.10) are marginally significantly higher. The results further confirm our conclusion that the clustering on Prescript-VMC outperforms that of PRAC. This makes sense since people can relatively easily describe the outcome of Prescript-VMC drugs since they may have more specific treatments, more strict usage and easier-described side effects.
– Some interesting phenomena: among all the Prescript-VMC drugs, Oxaliplatin, a cancer chemotherapy drug, is probably the most uncommon one since patients are not likely to know it unless they are facing colorectal cancer. In contrast, among all the PRAC, Aspirin is the most popular one since it is a well-known pain-relief that most people have encountered or heard of. Our results show that the performance of Oxaliplatin is the worst among all the Prescript-VMC while Aspirin is the best among all the PRAC. It reveals that: the more people get familiar with a drug, the more accurately that people can describe its outcomes, thus the better the model achieves the performance.
Table 2:
The performance of the clustering result for all the drugs
| Drug | Accuracy | Precision | Recall | F-score |
|---|---|---|---|---|
| Metformin | 0.675 | 0.706 | 0.661 | 0.683 |
| Clonidine | 0.705 | 0.783 | 0.696 | 0.737 |
| Gabapentin | 0.665 | 0.669 | 0.663 | 0.680 |
| Clonazepam | 0.770 | 0.766 | 0.740 | 0.753 |
| Oxaliplatin | 0.655 | 0.709 | 0.653 | 0.680 |
| Prescript-VMC(standard deviation) | 0.694(0.05) | 0.726(0.05) | 0.683(0.04) | 0.707(0.04) |
| Aspirin | 0.725 | 0.768 | 0.708 | 0.737 |
| Heparin | 0.580 | 0.635 | 0.563 | 0.597 |
| Ibuprofen | 0.600 | 0.634 | 0.586 | 0.609 |
| Hydrocodone | 0.620 | 0.665 | 0.616 | 0.640 |
| Naproxen | 0.575 | 0.591 | 0.569 | 0.580 |
| PRAC(standard deviation) | 0.620(0.06) | 0.659(0.06) | 0.608(0.05) | 0.632(0.06) |
| Overall | 0.657 | 0.693 | 0.646 | 0.670 |
Also for all the correctly clustered “expert sentences”, we split each group Oi into two sub-groups Oi_sim – showing the similar opinion with the expert’s ei, and Oi_opp – showing the opposite opinion. Two different strategies to form the prior estimator {p(w|ei)} are applied and compared. Table 3 shows the accuracy of two approaches compared to the gold standard. Approach 1 refers to that all meaningful terms are considered while Approach 2 refers to that only sentiment terms are considered. We utilize the technique and open source introduced in Hu and Liu’s sentiment analysis work[25].
Table 3:
The performance of distinguishing Oi_sim and Oi_opp
| Accuracy | Approach 1 | Approach 2 | Change |
|---|---|---|---|
| Prescript-VMC | 0.820 | 0.831 | +1.4% |
| PRAC | 0.792 | 0.811 | +2.4% |
| Overall | 0.806 | 0.821 | +1.9% |
From Table 3 we observe that both of the approaches reach a high accuracy to determine the similar or opposite opinion (overall accuracies are above 0.80), which indicates that semi-supervised PLSA model can successfully solve such problem. Furthermore, compared to the traditional way to build estimator (Approach 1), the novel way where sentiment analysis is highly addressed (Approach 2) performs better across Prescript-VMC, PRAC, and overall case.
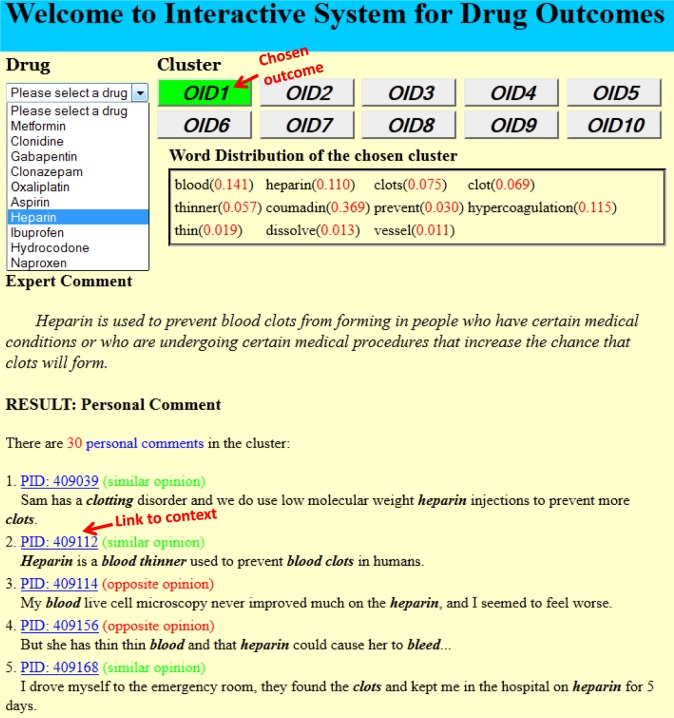
D. Clustering System for Drug Outcomes
Interface: Our prototype system is able to provide clustered and integrated drug information from personal health messages to enable examination through a user-friendly interface 6, whose composite format for the entire session is shown in Figure 3. Users can choose the specific drug and outcome that they are interested in, and the system will respond to the request by displaying the topic word distribution, the corresponding expert comment and all the comment units which belong to this outcome. Each comment unit is labeled by “similar” or “opposite” and users can also click the link of the corresponding PID to read its complete context.
Figure 3:
User interface
Example of Clonazepam: Table 5 shows the outcome integration results with expert comments, for the drug Clonazepam, which achieves the best performance out of 10 drugs. (accuracy: 0.770, F-score: 0.753). In the table, “Topic model” column shows the most common terms in this outcome as well as its probability. From this column we expect users could easily conceptualize the particular cluster at a glance. The third and fourth column show the number of “Similar Opinions” and “Opposite Opinions” for the corresponding outcome (denoted by s), respectively, as well as one sample personal sentence (for the space limit).
Table 5:
The outcome results for Clonazepam with expert comments
| OID | Topic model | Expert comment | Similar Opinions | Opposite Opinions |
|---|---|---|---|---|
| 1 | seizures(0.10), panic(0.09) attacks(0.07), seizure(0.06) brain(0.05), activity(0.02) |
Clonazepam is used alone or in combination with other medications to control certain types of seizures. It is also used to relieve panic attacks and works by decreasing abnormal electrical activity in the brain. | [s=59] She has only had a handful of seizures since then, Klonopin seems to control her seizures well | [s=16] Shy, Klonopin did not seem to contribute to my brain fog. |
| 2 | disorder(0.05), restless(0.04) plmd(0.03), dystonia(0.03) movement(0.02), mental(0.02) |
Clonazepam is also used to treat symptoms of akathisia that may occur as a side effect of treatment with anti psychotic medications and to treat PLMD, dystonia, and acute catatonic reactions) | [s=26] Klonopin works really well for Periodic Limb Movement Disorder, or any other med in the benzo class. | [s=12] Tried clonazepam for stress induced issues but it was too strong for me. |
| 3 | times(0.09), mg(0.08) daily(0.07), three(0.06) bedtime(0.04), tablet(0.03) |
Clonazepam comes as a tablet to take by mouth. It usually is taken one to three times a day with or without food. Take clonazepam at around the same time(s) every day. | [s=33] Zach has been on Klonopin (Clonazepam) .5mg three times a day for years | [s=9] Please do not take more than 8 Klonopin tablets a month |
| 4 | doctor(0.03), ask(0.02) dr(0.02), prescription(0.02) explain(0.01) pharmacist(0.01) |
Follow the directions on your prescription label carefully, and ask your doctor or pharmacist to explain any part you do not understand. | [s=18] Patricia,Klonopin is hands down the best medication to take, but you need to be monitored by a physician. | [s=6] I know they can’t prescribe the klonopin but a recomendation would be helpful. |
| 5 | allergic(0.07), pregnant(0.06) allergic(0.06), myoclonus(0.05) pregnancy(0.02) |
Before taking clonazepam, tell your doctor if you are allergic to clonazepam, tell your doctor if you are pregnant. | [s=24] Most anti-seizure meds aren’t allowed to be taken while you are pregnant, like Klonopin | [s=20] Sydnie has never had any allergy from the klonopin so we have been pretty pleased with it! |
| 6 | anxiety(0.21), anti(0.04) depression(0.03), mood(0.02) emotional(0.02), suicide(0.02) |
Report any new or worsening symptoms such as: mood or behavior changes, or if you feel agitated, irritable, hostile, aggressive, or have thoughts about suicide or hurting yourself. | [s=55] I have been worried about taking the Klonopin for the anxiety and sleeplessness because I have this history of depression, and it really brings you down. | [s=26] I just started taking Klonopin a couple of months ago for my anxiety. |
| 7 | tired(0.05), redness(0.04) rash(0.03), eye(0.02) breathing(0.02), liver(0.02) |
Call your doctor if you have a serious side effect such as: tiredness, shallow breathing; unusual eye movements; stomach problem, liver or kidney problem, redness, abnormal weight | [s=23] The klonopin just made me tired and that kind of made me feel more out of control. | [s=8] Only on the left side, and it is more like a rash than a redness |
| 8 | addiction(0.14), abuse(0.13) addictive(0.07), highly(0.02) pregnancy(0.02) |
Clonazepam may cause someone to succumb to drug abuse or addiction. | [s=31] Unfortunately, Klonopin is a very addictive drug. | [s=9] Klonopin is a little addictive, but does help when you need it |
From Table 5, we build 8 clusters for Clonazepam, where each of them focuses on one meaningful semantic outcome, guided by an expert comment. For example, Outcome ID 1 talks about the main treatment of Clonazepam – to treat seizures, which we know from the topic model and the expert comment e1. 59 comments express the similar/same opinion, while other 16 show different/opposite opinions on the same topic. From each row, we could easily understand the general user experience and how common this experience are among users regarding the particular outcome while taking Clonazepam: how do they feel about this drug? Do they agree or disagree with the experts’ opinion? Similarly, the rest outcomes such as side effects, dosage are shown in the following rows.
Such information is scattered in the huge amount of messages and impossible to integrate by hand. With our system, users can can effectively re-organize and integrate the drug-based information in a well-readable way.
New discovery of “other outcome”: Note for each drug, we also generate an additional cluster Om, i.e., “other outcome” which includes information mentioned by online users but not in standard description from expert comments. Table 4 shows some sentences (31 sentences in total, of which 8 relate to mouth burning) in Om for Clonazepam. We examine this cluster for each drug and find some interesting opinions. e.g., 34 comments report that Metformin is also used to treat obesity. For Clonazepam, 8 sentences show that Clonazepam may help to relief stomatodynia (burning mouth syndrome). For Aspirin, 5 sentences say that taking Aspirin causes eye problem, such as bursting eye vessels. For Heparin, 6 sentences show the concern that Heparin may cause severe bleeding to death. etc.
Table 4:
Sample results of Om for Clonazepam
| He said klonopin would help the burning sensation so I tried it and it did. The Klonopin is the only drug that helped me with the burning taste. We reintroduced the klonopin in a dropper full of water sublingually and eventually stabilized.... |
Although such “other outcomes” are not mentioned in expert comments, i.e., not recorded as standard outcome of the drug, they are discussed by actual users. In fact, formal medical literatures have mentioned each of the above additional outcomes – Metformin[26], Clonazepam[27], Aspirin[28] and Heparin[29]. Therefore, our system can effectively discover such “new outcome” from the clinical experiences as reported directly by the patients, which will provide supplemental information for the drug’s standard description.
E. Advantages and limitations of model
One advantage of our system over other simple statistic methods relies on its capacity of capturing the coherence of terms (e.g., appositive, synonym). The PLSA model is able to detect such connection between two comments which contain relevant, but not identical information since they share the similar contexts. Take one of the Clonazepam’s outcomes (OID 4) as an example, the expert comment is “Follow ... and ask your doctor or pharmacist to explain any part you do not understand.” The 18 similar sentences include not only “I would like to discuss with my Doctor...” which capture the word “doctor” exactly, but also “I first tried Klonapam prescribed by Dr. Cheney”, and “you need to be monitored by a physician”, which capture the words “dr”, “physician”.
Another advantage of the model is the flexibility of setting expert comments. Users can follow our example, i.e., extracting E from a professional drug database, or define and input the expert comments by themselves as prior knowledge of PLSA model. The clustering results will vary according to different expert comments. This customizable design could cater to various information needs of different users.
PLSA model requires to manually set a fixed number of clusters and lacks a way to dynamically determine the proper number of clusters. We will try to solve the problem in the future work. Furthermore, our model can analyze each independent drug effectively by embedding its specific expert comments. However, a unified expert-comment setting strategy should be designed and implemented while extending our system to arbitrary drugs or even treatments.
Conclusion and Future Work
In this paper, we describe a useful system we built to filter, organize and integrate drug-based medical information. SVM classifier, PLSA model and sentiment analysis techniques are applied in the system. We design a large-scale and professional-quality annotation framework, the output of which is good enough to be the gold standard to test the performance of the model. The experiment results with high accuracy and F-score show that our system can successfully organize the online medical information in a meaningful way. Users can request and search the well-organized personal opinions on different types of drugs via our prototype system, which could satisfy not only physicians’, but also patients’ information need.
In the future, we plan to explore and analyze different online healthcare resources, such as twitter7 – a more general social network where people discuss their health problem in a casual way, or MedHelp8 – a more professional medical forum providing well documented user demographic information. We expect to compare the up-to-date contents and language features across different online medical discussion platforms.
Recently, Comparative Effectiveness Research (CER) has become a new paradigm to analyze and compare different interventions and strategies in clinical trials, e.g., CER in chronic obstructive pulmonary disease[30]. Starting from our work, CER can be conducted to compare the harms or benefits of different drugs or treatments (e.g., Aspirin is more effective than Ibuprofen to treat migraine headache), by exploring the integrated personal messages.
From a system perspective, we will design and implement a flexible expert-comment setting strategy which offers the choice of unified standard from expert resource or patient-oriented prior knowledge. Building upon the clustering engine described in this paper, a wide range of drugs and treatments can be automatically analyzed from patient-specified personal health messages, and an effective interactive system can be built upon their integrated results.
Acknowledgments
Funding was provided by United States Department of Agriculture (USDA) National Research Initiative (NRI), grant 2009 – 35302 – 05285. Facilities were also provided by the Institute for Genomic Biology at UIUC.
Footnotes
For the space limit, other formulas can be seen in http://www.ideals.illinois.edu/bitstream/handle/2142/24194/Jiang_Yunliang.pdf
Currently the system only works on the 10 drugs used for the evaluation task
References
- 1.Chee B, Berlin R, Schatz B. Measuring Population Health Using Personal Health Messages. AMIA Annual Symposium Proceedings. 2009:92–96. [PMC free article] [PubMed] [Google Scholar]
- 2.Chee B, Berlin R, Schatz B. Predicting Adverse Drug Events from Personal Health Messages. AMIA Annual Symposium Proceedings. 2011:217–226. [PMC free article] [PubMed] [Google Scholar]
- 3.Agarwal S, Yu H. FigSum: Automatically Generating Structured Text Summaries for Figures in Biomedical Literature. AMIA Annual Symposium Proceedings. 2009:6–10. [PMC free article] [PubMed] [Google Scholar]
- 4.Arnold CW, El-Saden SM, Bui AA, Taira R. Clinical Case-based Retrieval Using Latent Topic Analysis. AMIA Annual Symposium Proceedings. 2010:26–30. [PMC free article] [PubMed] [Google Scholar]
- 5.Xu H, Stetson P, Friedman C. Methods for building sense inventories of abbreviations in clinical notes. Journal of the American Medical Informatics Association. 2009;15(1):87098. doi: 10.1197/jamia.M2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen ES, Manaktala S, Sarkar IN, Melton GB. A Multi-Site Content Analysis of Social History Information in Clinical Notes. AMIA Annual Symposium Proceedings. 2011:227–236. [PMC free article] [PubMed] [Google Scholar]
- 7.Crain SP, Yang S, Zha H, Jiao Y. Dialect Topic Modeling for Improved Consumer Medical Search. AMIA Annual Symposium Proceedings. 2010:132–136. [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, Mei Q, Zheng K, Hanauer DA. Query Log Analysis of an Electronic Health Record Search Engine. AMIA Annual Symposium Proceedings. 2011:915–924. [PMC free article] [PubMed] [Google Scholar]
- 9.Hanauer DA. EMERSE: the electronic medical record search engine. AMIA Annual Symposium Proceedings. 2006;941 [PMC free article] [PubMed] [Google Scholar]
- 10.Lin J, Demner-Fushman D. Semantic Clustering of Answers to Clinical Questions. AMIA Annual Symposium Proceedings. 2007:458–462. [PMC free article] [PubMed] [Google Scholar]
- 11.Hofmann T. Probabilistic latent semantic indexing. ACM SIGIR Conference on Research and Development in Information Retrieval. 1999:50–57. [Google Scholar]
- 12.Blei DM, Ng AY, Jordan MI. Latent Dirichlet Allocation. Journal of Machine Learning Research. 2003:993–1022. [Google Scholar]
- 13.Lu Y, Zhai C. Opinion Integration Through Semisupervised Topic Modeling. Proceedings of the 17th international conference on World wide web; 2008. pp. 121–130. [Google Scholar]
- 14.Kandula S, Curtis D, Hill B, Zeng-Treitler Q. Use of Topic Modeling for Recommending Relevant Education Material to Diabetic Patients. AMIA Annual Symposium Proceedings; 2011. pp. 674–682. [PMC free article] [PubMed] [Google Scholar]
- 15.Debarr D, Wechsler H. Social Network Analysis for Spam Detection; International Conf. on Social Computing, Behavioral Modeling, and Prediction; 2010. pp. 62–69. [Google Scholar]
- 16.Cortes C, Vapnik V. Support-Vector Networks. Machine Learning. 1995;20:273–297. [Google Scholar]
- 17.Jiang Y, Lin CX, Schatz B. Multi-class classification for online personal healthcare messages. The 2nd International Workshop on Web Science and Information Exchange in the Medical Web; 2011. [Google Scholar]
- 18.Songsiri P, Kijsirikul B, Phetkaew T. Information-based dichotomization: A method for multiclass Support Vector Machines. International Joint Conference on Neural Networks; 2008. pp. 3284–3291. [Google Scholar]
- 19.Dempster AP, Laird NM, Rubin DB. Maximum Likelihood from Incomplete Data via the EM Algorithm. Journal of the Royal Statistical Society Series B (Methodological) 1977;39(1):1–38. [Google Scholar]
- 20.Roberts PM, Cohen AM, Hersh WR. Tasks, topics and relevance judging for the TREC Genomics Track: five years of experience evaluating biomedical text information retrieval systems. Information Retrieval. 2009;12:81–97. [Google Scholar]
- 21.Blake C. Beyond genes, proteins, and abstracts: Identifying scientific claims from full-text biomedical articles. Journal of Biomedical Informatics. 2010;43:173–189. doi: 10.1016/j.jbi.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Fleiss JL. Measuring nominal scale agreement among many raters. Psychological Bulletin. 1971;76(5):378–382. [Google Scholar]
- 23.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 24.STUDENT(Gosset WS) The Probable Error of a Mean. Biometrika. 1908;6(1):1–25. [Google Scholar]
- 25.Hu M, Liu B. Mining and summarizing customer reviews. ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; 2004. pp. 168–177. [Google Scholar]
- 26.Lee A, Morley JE. Metformin decreases food consumption and induces weight loss in subjects with obesity with type II non-insulin-dependent diabetes. Obesity Research. 1998;6(1):47–53. doi: 10.1002/j.1550-8528.1998.tb00314.x. [DOI] [PubMed] [Google Scholar]
- 27.Woda A, Navez ML, Picard P, Gremeau C, Pichard-Leandri E. A possible therapeutic solution for stomatodynia (burning mouth syndrome) Journal of Orofacial Pain. 1998;12(4):272–278. [PubMed] [Google Scholar]
- 28.Lecuona K. Assessing and managing eye injuries. Community Eye Health. 2005;18(55):101–104. [PMC free article] [PubMed] [Google Scholar]
- 29.Jick J, Slone D, Borda IT, Shapiro S. Efficacy and Toxicity of Heparin in Relation to Age and Sex. N Engl J Med. 1968;279:284–286. doi: 10.1056/NEJM196808082790603. [DOI] [PubMed] [Google Scholar]
- 30.Krishnan JA, Mularski RA. Acting on comparative effectiveness research in COPD. Journal of the American Medical Association. 2010;304(14):1554–1556. doi: 10.1001/jama.2010.807. [DOI] [PubMed] [Google Scholar]