Abstract
The Substitutable Medical Apps, Reusable Technologies (SMART) project provides a framework of core services to facilitate the use of substitutable health-related web applications. The platform offers a common interface used to “SMART-ready” health IT systems allowing any SMART application to be able to interact with those systems. At Partners Healthcare, we have SMART-enabled the Informatics for Integrating Biology and the Bedside (i2b2) open source analytical platform, enabling the use of SMART applications directly within the i2b2 web client. In i2b2, viewing the patient in an EMR-like view enables a natural-feeling medical review process for each patient.
Introduction
In April 2010, as part of the Strategic Health IT Advanced Research Projects (SHARP) program from the Office of the National Coordinator for Health Information Technology (ONC-HIT), Harvard Medical School began to lay the necessary groundwork to create a flexible HIT environment that would drive innovation in an otherwise outdated one-size-fits-most systems trend1. The team recognized that such an open environment characterized by substitutable applications (apps) constructed around shared core components would drive down healthcare technology costs, support standards evolution, foster competition in the market, and accelerate innovation2. Therefore, development commenced on the Substitutable Medical Apps, Reusable Technologies (SMART) platform architecture which allows substitutability of medical web apps in a SMART-ready HIT system.
An initial candidate and existing HIT system that would quickly benefit from being SMART-enabled was the Informatics for Integrating Biology and the Bedside (i2b2) analytics framework, developed at Partners Healthcare. This is because making SMART available in i2b2 would allow an investigator to review patient data on a per-patient basis in a familiar EMR-like view and directly inside the i2b2 web client—a feature that i2b2 currently lacks. The SMART EMR-like view also features a customizable dashboard allowing the investigator to select the best-of-breed available SMART app and substitute in apps that are relevant to their research, for example, adding a clinical trial SMART app on-the-fly that determines eligibility based on a patient’s demographics and diagnoses from the i2b2 data repository. Thus, a project goal was formulated to enable the i2b2 software, among other HIT platforms, to become SMART-ready which would allow for SMART apps to easily interact with medical data stored in i2b2’s Clinical Research Chart (CRC) data repository. This approach would introduce an i2b2 user to enhanced analyses and display capabilities in an EMR-like individual patient view.
Background
Informatics for Integrating Biology and the Bedside (i2b2) is a National Institute of Health (NIH) funded National Center for Biomedical Computing (NCBC) based at Partners Healthcare, which has developed a scalable informatics framework enabling clinical researchers to use existing clinical data for discovery research. This open source platform, known as the i2b2 “hive” software, is composed of core modules called “cells” that communicate through web services and a common i2b2 XML messaging format called the Patient Data Object (PDO). These cells, which manage users, patient data, projects, and workflows, together create a system that enables a variety of functions required for clinical research including Natural Language Processing (NLP) based querying for phenotype, de-identification that is HIPAA compliant, visualization, access to patient samples and analytic pipelines.3
As of early 2012, i2b2 is deployed and being used at more than 80 health institutions internationally.4
Substitutable Medical Apps, Reusable Technologies (SMART) is a project funded by The Office of the National Coordinator for Health Information Technology (ONC-HIT) through the Strategic Health IT Advanced Research Projects (SHARP) program. SMART is developing a platform to support a flexible health IT environment with low entry barriers by which the grassroots of the HIT community—patients, physicians, and small agile software vendors—can continuously drive innovation.5 The platform allows for substitutable medical apps within SMART-ready HIT systems, similar to successful implementations of the substitutability model in other IT sectors.6 For instance, Apple’s products such as the iPad and iPhone utilizes a software platform with a published interface for apps to access core shared components on the device. Based on those interfaces, developers have created over a half-million apps generating a competitive Apple app store ecosystem, enabling consumers to select and substitute the best apps onto their devices.7 In a similar approach, the SMART platform strives to capture the same type of app innovation and competitive app ecosystem within the health care sector, as apps are run in a substitutable fashion on top of existing HIT platforms.
The SMART platform enables applications built against the SMART API to be able to be embedded within any SMART “container”. A SMART container can be a variety of HIT platforms, such as an Electronic Medical Record (EMR) system, a Personal Health Record (PHR), or a data analytics framework such as i2b2, as described herein. The SMART API and data model specifications define a variety of common requirements and features, from data retrieval methods to medical coding systems to be used by SMART containers and apps. For instance, the SMART API outlines a number of RESTful (Representational State Transfer) web service calls which provide access to individual resources such as medications, fulfillment events, prescription events, problems, demographics, allergies, vital signs, and laboratory results from a SMART-enabled system. Additionally, medication, diagnoses, and lab data returned in a SMART message need to be coded in RxNorm, SNOMED-CT, and LOINC, respectively.
As of early 2012, the SMART team has published a set of API functions used to access patient data from a SMART-enabled HIT system—for instance, functions are available to GET all or individual medications, fulfillments, demographics, problems, allergies, vital signs, or laboratory records for a single patient.10 The SMART team has also made available an extensive set of JavaScript, Python, and Java libraries for SMART apps to easily invoke these API functions. Additionally, a data model for various types of medical data represented in Resource Descriptive Framework (RDF)/XML format is also published. Finally, a set of online developer tools—for instance, a SMART reference EMR sandbox containing sample patient data—are available to assist in the understanding and testing of SMART components from both perspectives of a SMART-enabler of an HIT system and a SMART app developer.10
Goals
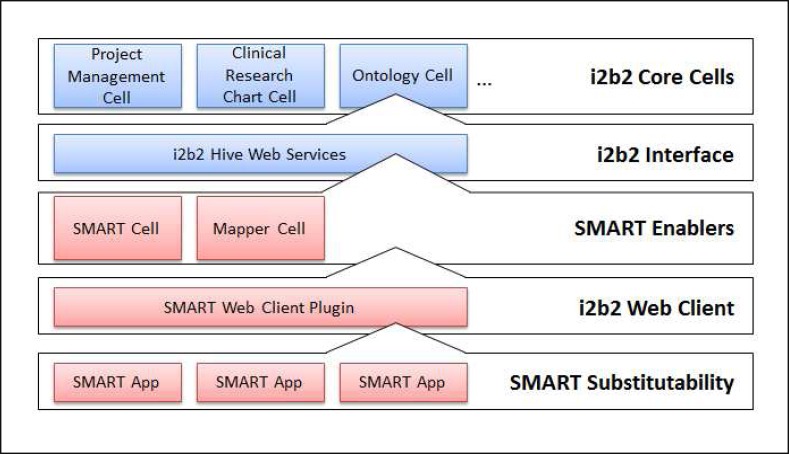
Our goal was to build upon the success of the i2b2 modular framework and add SMART support into i2b2 in an autonomous fashion. To accomplish this, we would develop a new i2b2 cell to expose the published SMART API functions as RESTful URLs defined by SMART, and create a new i2b2 web client plugin for capturing frontend interactions and displaying SMART apps (Figure 1). Specifically, we would target an initial subset of SMART API calls to retrieve medications, problems, demographics, allergies, fulfillments, and immunizations on a single patient. These independent components would allow for a straightforward implementation of SMART into new and existing i2b2 instances being used at many health institutions.
Figure 1.
A modular diagram that represents the new autonomous components that together, creates a SMART-ready i2b2 framework stack
To consider the technical implementation to be successful, we had to develop a system that allowed an external SMART app to be embedded directly into the i2b2 web client and furthermore, that SMART app would be authorized to interact with patient data directly from i2b2’s clinical data repository. Furthermore, the new i2b2 cell would work with existing core i2b2 cells including the Project Management cell which manages user roles and authorizations, and the CRC cell which contains patient data, to expose the SMART API functions and ultimately be able to communicate in the SMART defined RDF/XML common message type allowing any SMART app to be substituted into the i2b2 web client.
Methods
Creating the SMART cell
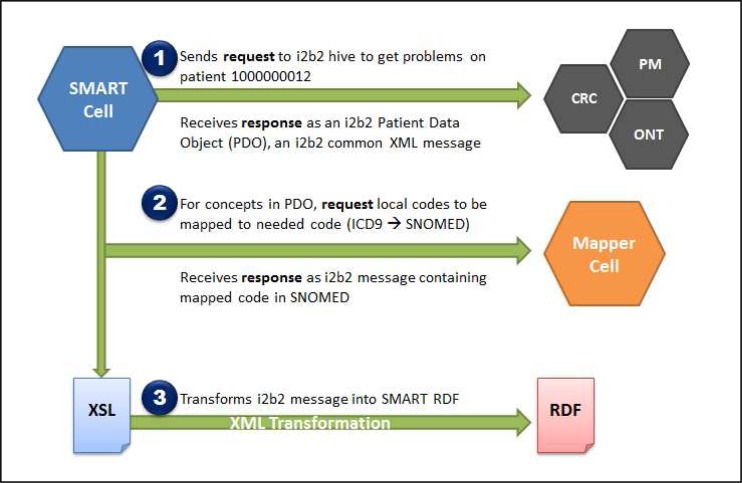
The i2b2 SMART cell is written in Java and follows the methodologies of developing an optional i2b2 cell that interacts with the core i2b2 cells in a JBoss/Tomcat Application Server environment.3 In short, the purpose of the SMART cell is to present medical data stored in i2b2 as a SMART RDF/XML message in order for SMART apps to be able to consume that data. At length, the SMART cell must work with the Project Management cell to properly authenticate the investigator’s i2b2 user session at the time SMART apps are being launched, authorize and assign OAuth tokens to each individual SMART app at runtime, communicate with the Clinical Research Chart (CRC) cell to retrieve the correct patient data based on the requested medical concepts, if necessary send those concepts to the Mapper cell to map local codes to the defined coding system, and finally, transform the result into a SMART RDF/XML message format based on the medical concept requested. OAuth is an open protocol to allow secure API authorization in a standard method in which the requesting application, i.e. a SMART app, does not ever get hold of the user’s account credentials.8 The described workflow occurs, in a timely fashion, on each and every API call made from a SMART app (Figure 2).
Figure 2.
An example workflow—via the i2b2 SMART cell—that occurs when a SMART application requests to get “All problems on a patient with an ID of 1000000012”
Each SMART API function is expressed as RESTful web service resources with specific formatted URLs, which are understood by all SMART apps. For example, to retrieve all medications for an i2b2 patient number ‘1000000012’, the entry point to the SMART cell would be an HTTP GET request to the following URL:
By setting a special “rewrite” configuration in JBoss, which allows for the creation of an alias for long and complicated web addresses, we were able to conform to the web address format described above. This technique allowed us to mask the true URL for that particular API call, which looks like:
The SMART cell also needs to know about SMART apps that are ‘installed’ or allowed to run in a SMART-enabled i2b2 instance. This is handled by placing a SMART app’s descriptor file, or a configuration file containing information about the SMART app, into a specified directory. The descriptor file looks like:
{
“name” : “Problems 2.0”,
“description” : “Display problems in a table view”,
“detailed” : “...”,
“author” : “Nich Wattanasin”,
“id” : “problems2@apps.smarti2b2.org”,
“version” : “2.0”,
“mode” : “ui”,
“scope” : “record”,
“index” : “http://localhost/problems2/index.html”,
“icon” : “http://localhost/problems2/icon.png”
}
Developing the EMR-like View
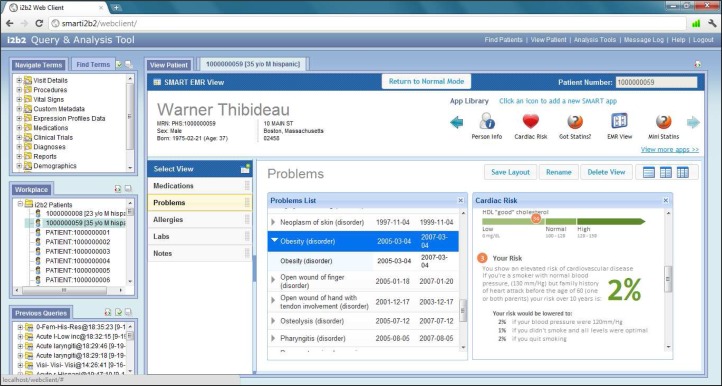
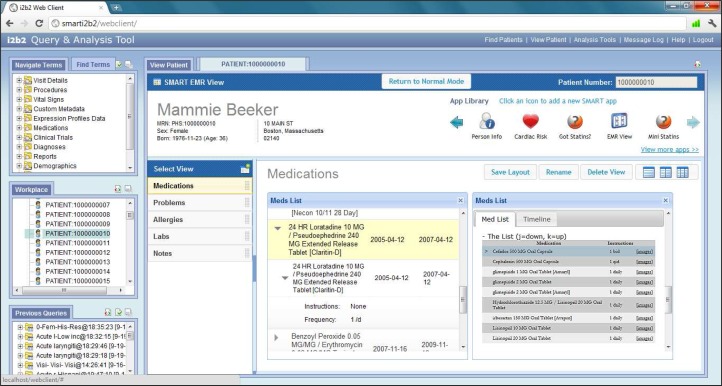
The i2b2 SMART web client plugin and EMR-like view is written in HTML/JavaScript and makes use of JQuery and SMART JavaScript libraries. The plugin facilitates the embedding of SMART apps directly in the i2b2 web client, and handles intricate asynchronous web service calls to the i2b2 SMART cell on behalf of the apps. Additionally, the EMR-like view can display multiple SMART apps running simultaneously on-screen and allow the i2b2 user to customize and save the layout of their dashboard (Figure 3). This allows for a fluid user experience, in which the user interacts with the i2b2 web client to query for a patient cohort based on various criteria as usual, but then is able to drag and drop a specific patient into the SMART web client plugin to view that patient’s medical data in an EMR-like view—without ever leaving, popping-up a new browser window, or changing web pages. The user can also choose to view a patient from their previously ran i2b2 queries.
Figure 3.
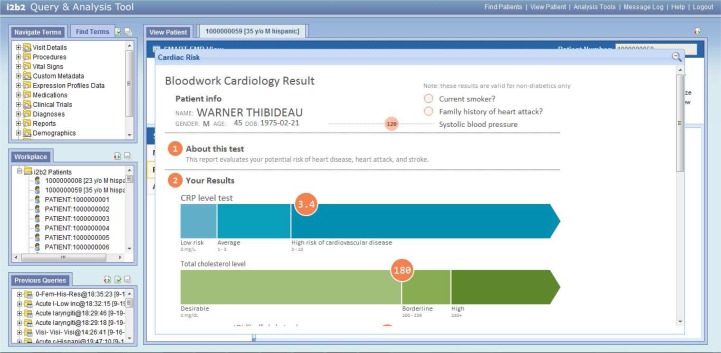
The SMART EMR-like view plugin running inside the i2b2 web client, displaying a sample i2b2 patient; Two SMART apps (Problems List and Cardiac Risk apps) can be seen running simultaneously.
The SMART web client plugin also communicates with the Project Management cell via AJAX to store user preferences as an ‘i2b2 user parameter’ allowing the user to save their customized views, dashboard layouts, and selected SMART apps. When the user starts a new session in the i2b2 web client or selects another patient to view, their previously saved layout is loaded and presented in real-time.
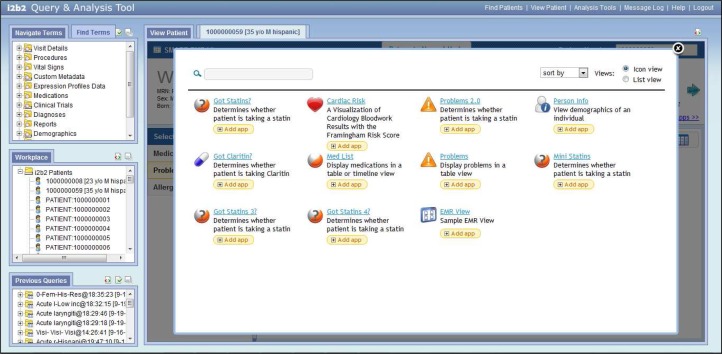
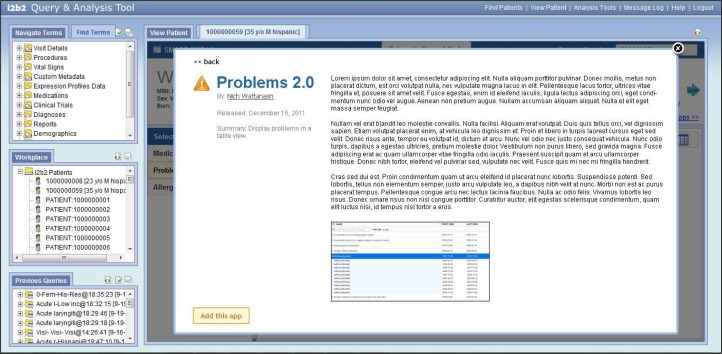
The SMART EMR-like view also features a navigable carousel of available SMART apps that allow a user to select to use a SMART app. The carousel (Figure 4) and expanded detail view (Figure 5) are populated based on the contents of the app descriptor files previously described. These selector tools assist the user to locate and choose the desired SMART apps to run in the EMR-like view if they choose to tailor additional custom layouts in addition to the available default views.
Figure 4.
The web client plugin comes with a SMART app carousel that allows a user to add apps to their views.
Figure 5.
The user is able to retrieve additional detailed information about a SMART app before adding it into their custom views.
Furthermore, the user has the ability to ‘zoom-in’ on a running SMART app directly inside the EMR-like view. This allows SMART apps that can recognize the width that it is currently running in to expand to a larger view (Figure 6). Most SMART apps are developed with widths of percent ratios rather than specific pixels allowing the natural and automatic stretch of apps in the larger view mode.
Figure 6.
The Cardiac Risk SMART app recognizes the width it is running in and re-draws its graphs in real-time to conform to a larger viewport.
Security and Authentication
In the i2b2 platform, users with specified data-viewing roles would login using their username and password— authentication can be managed by i2b2 or an organization’s enterprise services via LDAP or Active Directory—and the Project Management cell creates an automatically-expiring i2b2 session key valid for that user’s session only. To ensure that a SMART app is authorized to receive data from i2b2, every single app needs to get authorization from the SMART cell to be able to access patient data. This is handled by the SMART cell generating a set of valid OAuth access tokens scoped to an i2b2 user’s session, specific i2b2 project containing patient data, and to the specific and known SMART app that is making the request. As described previously, OAuth is an open protocol to allow secure API authorization in a standard method in which the requesting application, i.e. a SMART app, does not ever get hold of the user’s account credentials.8 OAuth tokens are generated every time a SMART app launches and expires automatically when the session ends. Therefore, each time a SMART app communicates with i2b2, it sends OAuth-signed requests which are authorized in real-time.
Presenting data in the SMART format
In the comprehensive demo dataset of sample patients packaged with the i2b2 software, medications utilize the National Drug Code (NDC) coding system and diagnoses utilize the International Statistical Classification of Diseases and Related Health Problems version 9 (ICD-9) coding system. Likewise, an institution may utilize and store i2b2 data in other coding systems including their own local coding system. Therefore, when the SMART cell receives a request, the cell will communicate with an i2b2 Mapper cell if necessary, which will map codes to the required coding systems.
In order to facilitate the institution’s exercise of mapping codes to the required SMART coding systems of RxNorm, SNOMED-CT, and LOINC, we have released a set of ‘top terms’ that an institution can map to in order to cover 85% of most used terms. This extensive work was based off of another Partners Healthcare more than decade-long project called the Research Patient Data Registry (RPDR), a centralized clinical data warehouse for Partners investigators with more than 5 million patients and 1 billion diagnoses, medications, labs, procedures, demographics, and visit entries.9 Additionally, the Mapper cell is released with a mapper tool, an i2b2 Eclipse plug-in, that allows a point-and-click mapping of various ontologies to the set of top terms. 11
Lastly, to produce the output message of SMART RDF/XML that all SMART apps understand, the i2b2 SMART cell takes a mapped PDO message and transforms it using Extensible Stylesheet Language Transformations (XSLT).
Components together
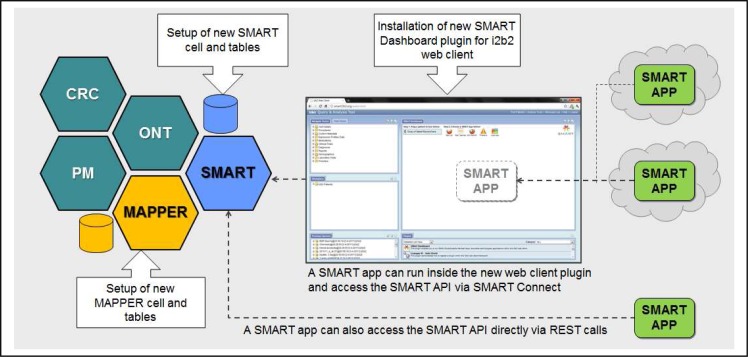
The architecture of the SMART and i2b2 integration consists of a new i2b2 SMART cell, Mapper cell, and web client plugin as previously described (Figure 7). These components were deployed on all of the supported i2b2 operating systems—Windows and Linux—and database management systems—Oracle and SQL Server. Additionally, the components including the i2b2 released demo set of patients is hosted as a public demo on a virtual machine in Amazon’s Elastic Compute Cloud (EC2) platform. 12 By doing so, we created an all-inclusive standalone and publicly available SMART-enabled i2b2 test bed that is used to demonstrate a working live example of embedded external SMART applications. Functionally, the test bed also helps facilitate SMART application development among both the SMART and i2b2 communities.
Figure 7.
A high-level architecture diagram highlighting the components required to SMART-enable an existing i2b2 instance. Shown in teal color are core i2b2 cells (CRC, Ontology, and Project Management cells).
Results
We have released as open source, version 1.0 of the SMART cell, Mapper cell, and web client EMR-like view plugin fully achieving our goals described herein. The SMART cell is exposing SMART API web service calls for the following SMART data types from the i2b2 clinical data repository: medications, problems, demographics, vital signs, lab results, allergies, and fulfillment events. In order to allow new and existing i2b2 users to test SMART functionality in i2b2, we have setup a publicly facing i2b2 instance that has been SMART-enabled at http://www.smarti2b2.org. Our web site also contains architecture, technical, and installation documentation that will allow an institution running an i2b2 installation to SMART-enable their instance with little intrusion to their existing configuration.
With the EMR-like view plugin, allowing a user to create and customize new dashboard views with SMART apps did actually strive towards the SMART project’s goal of creating a competitive app ecosystem. This was realized when two ‘Medication List’ SMART apps—one developed by us and the other by the SMART project team—were both added to our SMART-enabled i2b2 instance. When both medication list apps were displayed side-by-side (Figure 8), the user decidedly chose what they thought was the better app—the SMART app that had the features they needed. Specifically, the medication list app on the right-hand side in Figure 8 only displayed a non-sortable list of medications with no display of dates. However, the more advanced medication list app on the left-hand side displayed a sortable and searchable list of medications including dates when a medication was first and last observed to be taken.
Figure 8.
Two Medication List SMART apps are shown operating side-by-side in the EMR-like view dashboard, utilizing a sample patient’s data
Conclusion
When developing the components necessary to SMART-enable the i2b2 platform, we adhered to the principle of never modifying the core i2b2 code or existing schemas. In turn, we have successfully created an add-on i2b2 cell and web client plugin that have been packaged for deployment into existing i2b2 instances. Additionally, after conducting multiple tests and deployments in various i2b2 environments as previously described, the SMART-ready i2b2 framework demonstrated excellent performance with user interactions in the i2b2 web client. Furthermore, running the system on our publicly hosted Amazon virtual machine revealed that the user experience was not hindered during normal network latency.
During usability testing, we learned that the i2b2 SMART web client plugin only fully-operated on the latest major web browsers (Internet Explorer 7+, Firefox 3.5+, and Chrome.) This is an important aspect to keep in mind since some institutions may still be using outdated web browsers such as Internet Explorer 6.
Since releasing version 1.0 of our work as open source, we are currently working on extending the functionalities and features of the SMART cell, and further refining the EMR-like view web client plugin.
The future direction of implementing substitutability among HIT systems can be tremendously rewarding for the community. The work accomplished here to SMART-enable the i2b2 platform strategically demonstrates a single feasible workflow, among many more, that other existing HIT systems could potentially use to create their SMART-ready counterparts.
Footnotes
This work was funded by the SHARP Program Area Three award: 90TR0001/01
References
- 1.Mandl KD, Kohane IS. SHARP research area three overview. ONC-HIT web site. http://healthit.hhs.gov/portal/server.pt?open=512&mode=2&objID=3127&PageID=20707 (March 5, 2012)
- 2.Mandl KD, Kohane IS. No small change for the health information economy. N Engl J Med. 2009;360:1278. doi: 10.1056/NEJMp0900411. [DOI] [PubMed] [Google Scholar]
- 3.Murphy SN, Mendis M, Hackett K, et al. Architecture of the open-source clinical research chart from informatics for integrating biology and the bedside. Proceedings, Annual Fall Symposium of the American Medical Informatics Association; 2007. [PMC free article] [PubMed] [Google Scholar]
- 4.N.A. i2b2 Installations and Collaborations I2b2 web site. https://www.i2b2.org/work/i2b2_installations.html (March 5, 2012)
- 5.Mandl KD, Kohane IS. About page. Smart Platforms. http://www.smartplatforms.org/about (March 8, 2012)
- 6.Kibbe DC. Toward a modular EHR. Fam Pract Manag. 2009 Jul-Aug;16(4):8–9. PubMed. [PubMed] [Google Scholar]
- 7.Apple.com “Over 500,000 apps” http://www.apple.com/iphone/built-in-apps/app-store.html (March 8, 2012)
- 8.N.A. OAuth overview web site. http://www.oauth.net/ (March 10, 2012)
- 9.Murphy SN. Research Patient Data Registry (RPDR) overview. Partners Healthcare Research Computing web site. http://rc.partners.org/rpdr (March 10, 2012)
- 10.Mandl KD, Mandel JC, Murphy SN, et al. J Am Med Inform Assoc. 2012 doi: 10.1136/amiajnl-2011-000622. 3 of 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.N.A. Set of top terms released as part of SMART-i2b2 source code on i2b2 Community Wiki site. http://community.i2b2.org/wiki/display/SMArt/SMART+i2b2 (July 15, 2012)
- 12.N.A. Amazon Elastic Compute Cloud, EC2 web site. http://aws.amazon.com/ec2/ (July 15, 2012)