Abstract
OBJECTIVE:
To assess whether errors can be found in LOINC by changing its representation to OWL DL and comparing its classification to that of SNOMED CT.
METHODS:
We created Description Logic definitions for LOINC concepts in OWL and merged the ontology with SNOMED CT to enrich the relatively flat hierarchy of LOINC parts. LOINC - SNOMED CT mappings were acquired through UMLS. The resulting ontology was classified with the ConDOR reasoner.
RESULTS:
Transformation into DL helped to identify 427 sets of logically equivalent LOINC codes, 676 sets of logically equivalent LOINC parts, and 239 inconsistencies in LOINC multiaxial hierarchy. Automatic classification of LOINC and SNOMED CT combined increased the connectivity within LOINC hierarchy and increased its coverage by an additional 9,006 LOINC codes.
CONCLUSIONS:
LOINC is a well-maintained terminology. While only a relatively small number of logical inconsistencies were found, we identified a number of areas where LOINC could benefit from the application of Description Logic.
Introduction
There has been major progress both in Description Logic and ontology design since LOINC was originally developed in 19941. The emergence of the standard Web Ontology Language OWL, combined with the increase in computing power removed many of the limitations that hindered early application of DL in large clinical terminologies. Terminologies developed more recently have taken advantage of DL, for example the NCI Thesaurus2 and SNOMED CT3.
Comprehensive clinical terminologies such as SNOMED CT tend to overlap with specialised terminologies such as LOINC, and terminological systems such as the Unified Medical Language System (UMLS) can be used to bridge between them. A relatively flat hierarchy of LOINC terms can thus be augmented with the richness of SNOMED CT relations, which can provide novel insights into the original resource4.
Auditing clinical terminologies is an important step in assuring that they are fit for their purpose5. The objective is not to find errors, as any sufficiently large corpus is bound to have, but rather to identify areas for improvement. Graph-based approaches were proposed as one way to achieve this6;7, and more recently Description Logic was suggested as a method for quality assurance in the context of SNOMED CT8;9.
Background
LOINC is a universal standard for identifying laboratory observations. It can be considered the lingua franca of clinical observation exchange as it has more than 15,000 users in 145 countries10. It is recommended as part of the Meaningful Use and endorsed by American Clinical Laboratory Association and College of American Pathologists. A fully specified test result or clinical observation can be described formally with the following syntax: <Analyte/component>:<kind of property of observation or measurement>:<time aspect>:<system (sample)>:<scale>:-<method>11.
Description Logic and OWL are a family of knowledge representation languages and OWL is specifically aimed at authoring ontologies12. It is endorsed by the World Wide Web Consortium (W3C) and latest specification of the language (OWL2) was released in 2009. OWL is increasingly gaining traction as a standard for implementing clinical ontologies13.
The validation of an ontology by a DL-based classifier serves to ensure compliance with certain rules of classification, e.g., absence of cycles3. In general, it can be expected that the reasoner will identify two types of errors: duplicates and missing hierarchical relations14.
The integration of LOINC and SNOMED CT through DL has been explored since 199815;16;17. However, prior work focused primarily on achieving the most accurate mappings between the two terminologies, which is inherently difficult due to different scopes4. The specific contribution of this work is the application of DL specifically to quality assurance of LOINC and to our knowledge this is the first work describing LOINC audit of any kind.
Methods
Converting LOINC into OWL
LOINC 2.36 files were created by the Regenstrief Institute. This included two additional files that contain parts names and part links to full LOINC codes that are not part of a standard LOINC distribution. We expected they would contribute significantly to our OWL version, because the links between parts and codes play an important role in determining part links among observables.
Each LOINC code was defined using relations provided by the part links file that would form a logical conjunction defining the concept. LOINC part types were translated into respective OWL object properties, e.g., CHALLENGE part becomes has_challenge object property. We also created one additional relation (data property) has_index to model the fourth subpart of LOINC component axis. Example of a fully defined LOINC code in DL in a human-readable OWL Manchester syntax is provided in Figure 1.
Figure 1:
DL definition of LOINC code 15076-3 in OWL Manchester syntax.
LOINC has a multiaxial hierarchy that integrates LOINC codes and parts into a single hierarchy, e.g., LOINC code 35212-0:Glucose [Mass or Moles/volume] in Urine is child of LOINC part LP43854-6:Glucose | Urine. Because codes and parts are different in nature, it makes sense from an ontological perspective to separate them into two different hierarchies: one for LOINC codes and abstract observations grouping them, and a separate one for LOINC parts. In practice, we want to avoid situations where a substance, e.g., glucose subsumes the observation in which this substance is also an analyte.
Defining LOINC MULTIAXIAL parts logically
LOINC multiaxial hierarchy includes both parts and codes in the same graph. Most of the parts in this hierarchy are multiaxial, and can be defined in the context of their primitives, e.g., LP:43854-6:Glucose | Urine is a combination of COMPONENT Glucose and SYSTEM Urine parts. However, the composition of the multiaxial parts is not provided in LOINC explicitly.
We lexically matched the multiaxial parts to their primitive counterparts and created DL definitions for every part in the multiaxial hierarchy. An example is provided in Figure 2. When a lexical match was ambiguous and pointed to multiple LOINC parts, it was disambiguated based on the parts defining the underlying LOINC codes. 39,256 individual lexical matches were made in total. In 113 cases it was impossible to disambiguate the composing parts and multiple matches were accepted as valid. This did not affect subsequent results.
Figure 2:
DL definition in OWL Manchester syntax of a novel observation representing all measurements of glucose in urine. It was derived from a multiaxial LP43854-6:Glucose | Urine LOINC part.
We found that the string Bld-Ser-Plas occurring in 7,403 multiaxial part labels had no equivalent in primitive parts. It was modelled as a logical disjunction of three system parts, i.e., has_system some LP7057-5:Blood or has_system some LP7567-3:Serum or has_system some LP7479-1:Plasma.
Converting SNOMED CT into OWL
We followed the process described in the SNOMED CT Technical Implementation Guide18. SNOMED CT (July 2011 version) was converted into OWL by running the standard Perl transform script bundled with the distribution.
Acquiring LOINC to SNOMED CT mappings
LOINC parts were mapped to SNOMED CT terms using owl:equivalentClass statements. The n-to-n mappings were derived from UMLS 2011 AB release by parsing the Concept Names and Sources File19. In practice, we considered a LOINC concept equivalent to a SNOMED CT concept if both concepts were asserted under the same concept identifier in the UMLS. This provided 7,377 LOINC parts mappings to 8,161 SNOMED CT concepts. For example, LP16699-8:Erythrocyte and SCT_41898006:Erythrocyte share the same UMLS identifier C0014792 and an equivalence axiom was asserted in the ontology accordingly.
Additionally, equivalent relations (OWL object properties) in the two terminologies were mapped with owl:equivalent-Property statements. It was challenging because has_system relation in LOINC does not have an equivalent direct relation in SNOMED CT, but rather it can be represented by a combination of relationships; linking the laboratory test first to a specimen (has specimen), and then linking the specimen to a substance (specimen substance)4. This became possible to model in DL with the introduction of owl:propertyChainAxiom construct in OWL2. Thus, relation hasSystem in LOINC was asserted as equivalent to a property chain of relations has specimen o specimen substance in SNOMED CT.
Merging of the ontologies and classifying
The hierarchy and all the logical restrictions of SNOMED CT were preserved by merging LOINC and SNOMED CT into a single OWL ontology. The ontology was then classified using ConDOR reasoner, which was chosen because of a dramatic improvement in speed over existing ontology reasoners20.
Computing environment
Code for parsing and serialising LOINC into OWL was written in Java7. It depends on the Java CSV library (http://opencsv.sourceforge.net) and OWL API21 and is available under GNU lesser GPL license from: https://code.google.com/p/loinc-sem-web
Computations were performed on a dedicated server running Red Hat Enterprise Linux Server release 5.8 (Tikanga) with eight processors (Intel® Xeon™ CPU 3.20GHz) and 32GB of memory.
Results
1. LOINC and SNOMED CT overview
The final ontology consisted of 468,572 concepts in total and had over a million asserted axioms (1,577,861). 413,050 additional axioms were inferred by the reasoner (see Table 1).
Table 1:
An overview of concepts and axioms in LOINC with and without SNOMED CT.
| LOINC | LOINC+SNOMED CT | |
|---|---|---|
|
| ||
| Number of classes | 173,091 | 468,572 |
| Number of asserted axioms: | 677,023 | 1,577,861 |
| Number of inferred axioms | 126,020 | 413,050 |
| LOINC codes | 65,003 | |
| LOINC parts | 82,102 | |
| LOINC multiaxial hierarchy | ||
| LOINC codes | 47,405 | |
| LOINC parts | 25,982 | |
Inferred equivalent LOINC parts
The reasoner identified 676 equivalent sets of LOINC parts comprising 1,549 LOINC parts. For example, LP7536-8:RBC, LP14304-7:Erythrocytes, and LP16699-8:Erythrocyte were classified by the reasoner to be equivalent to one another because they were asserted to be equivalent to the same SNOMED CT concept SCT_41898006:Erythrocyte via UMLS. Determining equivalent parts is an important step in the classification process for subsequent identification of equivalent LOINC concepts.
Inferred equivalent LOINC codes (intrinsic)
There were 325 sets of LOINC codes with the same definition provided by the part links table. These are not actual duplicates, but rather their are mistakenly linked to the same parts, for example:
56897-2:Cells.CD3-CD56+/100 cells:NFr:Pt:CSF:Qn
51279-8:Cells.CD3+CD56+/100 cells:NFr:Pt:CSF:Qn
are both linked to LP19037-8:Cells.CD3+CD56+ and LP35646-6:Cells.CD3-CD56+. As is also the case with:
10132-9:T’ wave amplitude.lead AVR:Elpot:Pt:Heart:Qn:EKG
10144-4:T wave amplitude.lead AVR:Elpot:Pt:Heart:Qn:EKG
both sharing the LP31227-9:T wave amplitude.lead AVR and LP31243-6:T’ wave amplitude.lead AVR component parts. This is characteristic of a number of LOINC codes in the EKG.MEAS class.
Some concepts were found to be simply missing a distinguishing part link, for example the following two concepts were both linked to LP72988-6:Note:
64071-4:Progress note:Find:Pt:Hospital:Doc:Medical student.critical care
64072-2:Consultation note:Find:Pt:Hospital:Doc:Medical student.critical care
Finally, in a smaller subset no distinguishing feature could be identified and they may require additional curation:
46062-6:Treatments:-:Pt:^Patient:Set:
46064-2:Therapies:-:Pt:^Patient:Set:
36748-2:Views oblique:Find:Pt:Spine.cervical:Nar:XR
42164-4:Views & oblique:Find:Pt:Spine.cervical:Nar:XR
45424-9:Epilepsy:Find:Pt:^Patient:Ord:MDS
45662-4:Seizure disorder:Find:Pt:^Patient:Ord:MDS
This approach can be considered intrinsic to LOINC as the aforementioned results could be achieved without Description Logic by simply running a detailed database query against the LOINC codes table.
Inferred equivalent LOINC codes (extrinsic)
However, once LOINC is augmented with additional information from SNOMED CT (see Acquiring LOINC to SNOMED CT mappings) it becomes possible to validate LOINC assertions externally. This extrinsic approach allowed to identify additional 102 sets of equivalent LOINC codes. Examples in this category include:
10374-7:Helmet cells:ACnc:Pt:Bld:Ord:Microscopy.light
800-3:Schistocytes:ACnc:Pt:Bld:Ord:Microscopy.light
8703-1:Physical findings:Find:Pt:Extremities:Nom:Observed
32430-1:Physical findings:Find:Pt:Extremity:Nom:Observed
39037-7:Multisection^W contrast IV:Find:Pt:Upper extremity:Nar:MRI
36208-7:Multisection^W contrast IV:Find:Pt:Upper arm:Nar:MRI
This method identified codes differing only in grammatical number (Extremities vs. Extremity), codes with synonymous components (Helmet cells vs. Schistocytes), as well as parts that could be considered synonymous (Upper extremity vs. Upper arm) depending on the context. However, it did identify some codes incorrectly as equivalent:
9105-8:Fluid intake.total:VRat:8H:^Patient:Qn:
9259-3:Fluid output.total:VRat:8H:^Patient:Qn:
Complete data set is available as Supplementary Information at http://goo.gl/DhXxo.
Inferred multiaxial hierarchy
There are 73,387 concepts in the multiaxial hierarchy, which covers 47,461 (73%) LOINC codes terms and 26,834 (32%) LOINC parts. 17,598 LOINC codes are outside of the hierarchy (see Table 1).
The reasoner starting from a list of newly defined abstract observations and LOINC codes created a hierarchy with 56,411 LOINC codes and essentially enhanced the original hierarchy by 9,006 additional codes that were otherwise placed outside this hierarchy. General characteristics of the two graphs: original multiaxial and inferred are presented in Table 2.
Table 2:
Network analysis comparing LOINC multiaxial and inferred hierarchies.
| LOINC | Inferred | |
|---|---|---|
|
| ||
| Number of connected nodes | 73,387 | 82,350 |
| Network diameter | 15 | 13 |
| Connected components | 8 | 513 |
| Shortest paths | 425,976 | 1,119,232 |
| Characteristic path length | 3.39 | 3.81 |
| Average number of neighbours | 2.01 | 3.40 |
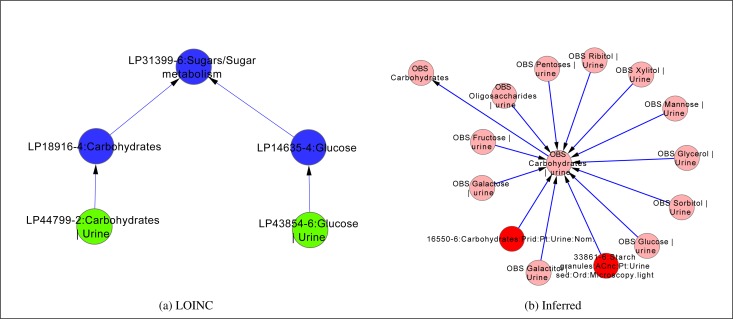
Figure 3b demonstrates two important aspects of the inferred hierarchy. There is a new access point to Glucose | Urine codes via Carbohydrates | Urine. Figure 3a shows that this path does not exist in the original LOINC hierarchy, where the two abstract observations are sharing the same parent Sugar metabolism, but otherwise are not in direct relationship to one another. Furthermore, some codes are now subsumed as more specific, e.g., 22705-8:Glucose:SCnc:Pt:Urine:Qn:Test strip became a child of a more general 15076-3:Glucose:SCnC:pt:Urine:Qn: test.
Figure 3:
Paths between Carbohydrates | Urine and Glucose | Urine observations in LOINC multiaxial and inferred hierarchies. LOINC codes are shown in red, abstract observations in pink, MULTIAXIAL parts in green, and COMPONENT parts in blue. Edges represent is_a (subClassOf ) relations. [created in Cytoscape22]
Inconsistencies in LOINC multiaxial hierarchy
The reasoner identified 198 sets of equivalent multiaxial parts. No concepts already asserted in the multiaxial hierarchy were excluded by the reasoner. 122 discouraged LOINC codes already in the hierarchy were omitted from the analysis. 239 LOINC codes were found to be incorrectly asserted in respect to their original LOINC hierarchy. In majority of cases the assertions were correct lexically, but the codes lacked a link to more specific part. These codes predominantly (183 concepts) were of scale type Document, e.g., 28626-0:History and physical note:Find:Pt:Setting:Doc:Physician was asserted under History and physical note, but the reasoner placed it under a more general observation Note (Document Ontology branch), because 28626-0 was defined as has_component some Note. Another class of findings were chemical compounds, e.g., 38639-1:Boron trifluoride:MCnc:Pt:Air:Qn: was asserted under Boron trifluoride | air, but the reasoner inferred it under Boron as it was missing the more specific Boron trifluoride connection.
Only one class of true errors was identified. A number of codes for measurements of fatty acids, e.g., 44084-2:Fatty acids:Imp:Pt:Ser/Plas:Nom: were originally asserted under 7-hydroxyoctanoate | Urine (note that both component and system assertions were wrong), and the reasoner correctly inferred them elsewhere, in this particular case under Lipids | bld-ser-plas.
A full list is available as Supplementary Information at http://goo.gl/E97O4.
Discussion
LOINC is an extremely useful terminology and the results of this study should not be considered a criticism or even an evaluation of LOINC. Nevertheless, this study does show LOINC in a very favourable light as only a relatively small number of inconsistencies were identified. For example, 427 sets of equivalent LOINC codes were identified, which represents an occurrence rate of only 0.24%. The creation and maintenance of LOINC is a resource-intensive process, therefore any methods that can focus curators’ attention on potentially troublesome content would help maximize its effectiveness.
Significance
-
Error detection
-
Duplicates
We found that while the DL approach does in fact identify potential duplicates it is much more sensitive to insufficiencies in modelling.
-
Missing hierarchical relations
The reasoner was successful in identifying a significant number of missing connections between the LOINC codes. A comparison of the two networks (see Table 2) confirms that the inferred hierarchy is indeed richer as it has more connections.
-
Inconsistencies in hierarchy
When LOINC codes were identified to be incorrectly asserted in the hierarchy, it was most likely due to insufficient modelling rather than erroneous assertions. However, some errors might have been more difficult to detect due to inconsistencies in parts hierarchy. For example, LP51132-6:N-methyl valine | Bld-Ser-Plas is asserted directly under LP501109-5:Amino Acids | Amniotic fluid. When the reasoner takes this assertion at face value, it fails to identify the clash in systems (Bld-ser-Plas vs. Amniotic fluid) between the two parts.
-
-
Enhanced navigation
In the current multiaxial hierarchy, LOINC codes have at most one parent, though the abstract observations are interlinked more. The DL representation provides access to more LOINC codes and more paths between the codes in the hierarchy, which directly translates into more ways a particular code can be discovered. By adding new paths to the hierarchy it also enables queries that were otherwise not possible. Figure 3b demonstrates this on the example of Carbohydrates | Urine, which in the inferred hierarchy not only returns a set of Glucose | Urine tests, but also all other carbohydrate measurements in urine.
-
Enhanced subsumption
The new paths are not limited to abstract observations that group LOINC codes, but also LOINC codes themselves can be in direct relationships. This actually limits the need for creating abstract observations to group them.
-
Maintenance
A typical scenario where a new LOINC code is requested by an external user requires several manual and error-prone steps to add the code. Firstly, you need to confirm that the test or its close variant does not exist already in the terminology. This requires several queries and expert knowledge on the actual naming conventions in a particular field. Secondly, you need to identify the best place in the hierarchy to place the new term.
DL approach can to a large extent simplify both steps through automated classification. There is also now standard tooling to work and manipulate OWL ontologies, such as OWL API21 and Protégé23.
Intrinsic vs. extrinsic approach
It is important for logical assertions in the ontology to be externally validated as the resulting inference is only as good as the original assertions. An external resource can also shed some light on areas that were not modelled sufficiently.
Recommendations
-
Create logical definitions for codes
Description Logic has long been recognised as indispensable to achieving convergence of clinical terminologies24. Having logical definitions for codes enables the “lexically assign, logically refine” strategy followed by other clinical terminologies such as SNOMED CT9. Transition to DL could be considered a natural consequence as LOINC codes already have multiaxial composition.
-
Have an inferred hierarchy
The hierarchy of parts could be taken directly from SNOMED CT thus minimising the effort involved. A hierarchy of parts would mean that hierarchy of codes could be inferred automatically.
-
Parts vs. codes
Multiaxial codes represent abstract groupings of observations and as such logically are no longer parts. Parts in general should not be in hierarchical relation to codes, which is especially true if they are at the same time used to define them.
-
Alignment with SNOMED CT
Consider the example of SNOMED CT term 385421009:Site of distant metastasis and LOINC parts: LP73362-3:Sites of distant metastasis, LP73358-1:Site of distant metastasis, LP72485-3:Distant metastasis site, where a single SNOMED CT concept maps to several LOINC parts. What does it mean to have several parts in LOINC map to the same SNOMED CT concept? If indeed they are different concepts then their names should not represent the same entity.
Acknowledgments
We would like to thank Dr. Clement McDonald for helpful comments and Regenstrief Institute for providing the necessary files. This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM) and the Oak Ridge Institute for Science and Education (ORISE) Training Program in Clinical Informatics managed for the U.S. Department of Energy (DOE) by Oak Ridge Associated Universities (ORAU).
Glossary
- OWL
W3C Web Ontology Language
- DL
Description Logic
- HL7
Health Level Seven
References
- [1].Forrey aW, McDonald CJ, DeMoor G, Huff SM, Leavelle D, Leland D, Fiers T, Charles L, Griffin B, Stalling F, Tullis A, Hutchins K, Baenziger J. Logical observation identifier names and codes (LOINC) database: a public use set of codes and names for electronic reporting of clinical laboratory test results. Clinical chemistry. 1996 Jan;42(1):81–90. URL http://www.ncbi.nlm.nih.gov/pubmed/8565239. [PubMed] [Google Scholar]
- [2].Hartel Frank W, de Coronado Sherri, Dionne Robert, Fragoso Gilberto, Golbeck Jennifer. Modeling a description logic vocabulary for cancer research. Journal of biomedical informatics. 2005 Apr;38(2):114–29. doi: 10.1016/j.jbi.2004.09.001. ISSN 1532-0464. URL http://www.ncbi.nlm.nih.gov/pubmed/15797001. [DOI] [PubMed] [Google Scholar]
- [3].Bodenreider Olivier, Smith Barry, Kumar Anand, Burgun Anita. Investigating subsumption in SNOMED CT: an exploration into large description logic-based biomedical terminologies. Artificial intelligence in medicine. 2007 Mar;39(3):183–95. doi: 10.1016/j.artmed.2006.12.003. ISSN 0933-3657. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2442845&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Bodenreider Olivier. Issues in mapping LOINC laboratory tests to SNOMED CT. AMIA ... Annual Symposium proceedings/AMIA Symposium. AMIA Symposium; January 2008; pp. 51–5. ISSN 1942-597X. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2655945&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- [5].Geller J, Perl Y, Halper M, Cornet R. Special issue on auditing of terminologies. Journal of biomedical informatics. 2009 Jun;42(3):407–11. doi: 10.1016/j.jbi.2009.04.006. ISSN 1532-0480. URL http://www.ncbi.nlm.nih.gov/pubmed/19465342. [DOI] [PubMed] [Google Scholar]
- [6].Peters Lee, Bodenreider Olivier. Using the RxNorm web services API for quality assurance purposes. AMIA ... Annual Symposium proceedings/AMIA Symposium. AMIA Symposium; January 2008; pp. 591–5. ISSN 1942-597X. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2656097&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- [7].Bodenreider Olivier, Peters Lee B. A graph-based approach to auditing RxNorm. Journal of biomedical informatics. 2009 Jun;42(3):558–70. doi: 10.1016/j.jbi.2009.04.004. ISSN 1532-0480. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2722378&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rector Alan L, Brandt Sam, Schneider Thomas. Getting the foot out of the pelvis: modeling problems affecting use of SNOMED CT hierarchies in practical applications. Journal of the American Medical Informatics Association : JAMIA. 2011;18(4):432–40. doi: 10.1136/amiajnl-2010-000045. ISSN 1527-974X. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3128394&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rector Alan, Iannone Luigi. Lexically suggest, logically define: Quality assurance of the use of qualifiers and expected results of post-coordination in SNOMED CT. Journal of biomedical informatics. 2011 Oct; doi: 10.1016/j.jbi.2011.10.002. ISSN 1532-0480. URL http://www.ncbi.nlm.nih.gov/pubmed/22024315. [DOI] [PubMed] [Google Scholar]
- [10].McDonald CJ. LOINC, a Universal Standard for Identifying Laboratory Observations: A 5-Year Update. Clinical Chemistry. 2003 Apr;49(4):624–633. doi: 10.1373/49.4.624. ISSN 0009-9147. URL http://www.clinchem.org/cgi/content/abstract/49/4/624. [DOI] [PubMed] [Google Scholar]
- [11].McDonald C, Huff SM, Suico J, Mercer K. Logical Observation Identifiers Names and Codes (LOINC R) users’ guide. Indianapolis: Regenstrief Institute. 2004 [Google Scholar]
- [12].Baader Franz, Calvanese Diego, McGuinness Deborah, Nardi Daniele, Patel-Schneider Peter., editors. The Description Logic Handbook: Theory, Implementation, and Applications. Cambridge University Press; 2003. [Google Scholar]
- [13].Staab Steffen, Studer Rudi. Handbook on Ontologies. Springer; 2009. [Google Scholar]
- [14].Wei Duo, Bodenreider Olivier. Using the abstraction network in complement to description logics for quality assurance in biomedical terminologies - a case study in SNOMED CT. Studies in health technology and informatics. 2010 Jan;160(Pt 2):1070–4. ISSN 0926-9630. URL http://www.ncbi.nlm.nih.gov/pubmed/20841848. [PMC free article] [PubMed] [Google Scholar]
- [15].Dolin RH, Huff SM, Rocha RA, Spackman KA, Campbell KE. Evaluation of a “lexically assign, logically refine” strategy for semi-automated integration of overlapping terminologies. Journal of the American Medical Informatics Association : JAMIA. 1998;5(2):203–13. doi: 10.1136/jamia.1998.0050203. ISSN 1067-5027. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=61291&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Spackman KA. Integrating sources for a clinical reference terminology: experience linking SNOMED to LOINC and drug vocabularies. Studies in health technology and informatics. 1998 Jan;52(Pt 1):600–3. ISSN 0926-9630. URL http://www.ncbi.nlm.nih.gov/pubmed/10384525. [PubMed] [Google Scholar]
- [17].Srinivasan Arunkumar, Kunapareddy Narendra, Mirhaji Parsa, Casscells S Ward. Semantic web representation of LOINC: an ontological perspective. AMIA ... Annual Symposium proceedings/AMIA Symposium. AMIA Symposium; Jan, 2006. p. 1107. ISSN 1942-597X. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1839278&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- [18].SNOMED CT ® Technical Implementation Guide January 2012 International Release (US English). URL http://ihtsdo.org/fileadmin/user_upload/doc/download/doc_TechnicalImplementationGuide_Current-en-US_INT_20110731.pdf
- [19].UMLS ® Reference Manual, September 2009. URL http://www.ncbi.nlm.nih.gov/books/NBK9676/
- [20].Kazakov Yevgeny. Consequence-Driven Reasoning for Horn SHIQ Ontologies. Proceedings of the 21st International Conference on Artificial Intelligence (IJCAI 2009); 2009. pp. 2040–2045. URL http://ijcai.org/papers09/Papers/IJCAI09-336.pdf. [Google Scholar]
- [21].The OWL API: A Java API for Working with OWL 2 Ontologies. OWLED 2009, 6th OWL Experienced and Directions Workshop, Chantilly; Virginia. October 2009; 2009. Matthew; Bechhofer Sean Horridge. [Google Scholar]
- [22].Shannon Paul, Markiel Andrew, Ozier Owen, Baliga Nitin S, Wang Jonathan T, Ramage Daniel, Amin Nada, Schwikowski Benno, Ideker Trey. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003 Nov;13(11):2498–504. doi: 10.1101/gr.1239303. ISSN 1088-9051. URL http://genome.cshlp.org/cgi/content/abstract/13/11/2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rubin Daniel L, Noy Natalya F, Musen Mark A. Protégé: a tool for managing and using terminology in radiology applications. Journal of digital imaging : the official journal of the Society for Computer Applications in Radiology. 2007 Nov;20(Suppl 1):34–46. doi: 10.1007/s10278-007-9065-0. ISSN 0897-1889. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2039856&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Spackman KA, Campbell KE. Compositional concept representation using SNOMED: towards further convergence of clinical terminologies. Proceedings/AMIA ... Annual Symposium. AMIA Symposium; Jan, 1998. pp. 740–4. ISSN 1531-605X. URL http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2232179&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]