Abstract
Acronyms and abbreviations within electronic clinical texts are widespread and often associated with multiple senses. Automated acronym sense disambiguation (WSD), a task of assigning the context-appropriate sense to ambiguous clinical acronyms and abbreviations, represents an active problem for medical natural language processing (NLP) systems. In this paper, fifty clinical acronyms and abbreviations with 500 samples each were studied using supervised machine-learning techniques (Support Vector Machines (SVM), Naïve Bayes (NB), and Decision Trees (DT)) to optimize the window size and orientation and determine the minimum training sample size needed for optimal performance. Our analysis of window size and orientation showed best performance using a larger left-sided and smaller right-sided window. To achieve an accuracy of over 90%, the minimum required training sample size was approximately 125 samples for SVM classifiers with inverted cross-validation. These findings support future work in clinical acronym and abbreviation WSD and require validation with other clinical texts.
1. INTRODUCTION
Acronyms and abbreviations within clinical texts are widespread, and their use continues to increase1–3. Several reasons for this ongoing growth include adoption of electronic health record (EHR) systems with increased volume of electronic clinical notes accompanied by the wide usage of acronyms and abbreviations2, the time-constrained nature of clinical medicine encouraging the use of shortened word forms, and a longstanding tradition of commonly using acronyms and abbreviations in clinical documentation1. The process of understanding the precise meaning of a given acronym or abbreviation in texts is one of several key functions of automated medical natural language processing (NLP) systems4 and is a special case of word sense disambiguation (WSD)5. Automatic meaning discrimination by a machine is a complex task that is critical to accessing information encoded in clinical texts6, 7. Improved acronym and abbreviation WSD methods can therefore enhance automated utilization of clinical texts to support diverse applications that rely on NLP.
Acronyms and abbreviations each have a short form (the acronym or abbreviation) and a long form (the expansion of the acronym or abbreviation). In clinical documents, the expanded long form is rarely proximal to the short form of the acronym or abbreviation2, 8 because clinical texts rarely conform to the formalism of enclosing the long form in parentheses after the first mention of the abbreviation, as is customary in scientific literature9. This lack of the formalism is one of the significant barriers associated with using clinical texts for NLP research, which has resulted in limited data resources for research. Because of this informality and the shortage of the available resources/research, while researchers have explored the use of supervised machine learning (ML) approaches for acronym and abbreviation WSD3, 5, 10, some of the related issues with optimal window size and orientation and with training sample size minimization to reduce the associated cost and time to manually annotate training corpora remain open3, 10.
In this paper, we have three objectives: (1) to understand and validate the relative value of different features to automatically disambiguate senses of 50 clinical acronyms and abbreviations; (2) to determine the optimal window size and orientation for obtaining features for acronym and abbreviation sense disambiguation; and (3) to estimate minimum sufficient training sample size for good performance in the inverted cross-validation settings using supervised learning approaches.
2. Background
2. 1. Broad classes of features for WSD
Types of predictive features from clinical notes can be grouped into domain knowledge-based, linguistic, statistical, and general document features. These features utilize techniques developed in the biomedical NLP and computational linguistics domains. Optimal feature selection for WSD therefore requires a comprehensive understanding of the strengths and weaknesses of each feature type to maximize valuable information used for feature sets as input into ML algorithms.
Because clinical notes are based upon medical knowledge, biomedical and clinical domain resources can serve as the knowledge base to enhance clinical WSD algorithms. In particular, the Unified Medical Language System (UMLS)11 and the Systematized Nomenclature of Medicine-Clinical Terms (SNOMED CT)12 are terminology resources in the biomedical and clinical domains respectively. These resources are used by the medical NLP community not only because they provide knowledge sources for identification of medical terms, but also because they offer semantic information and ontological relationships3, 4 that may be used to compute semantic similarity measures between concepts that can subsequently serve as features for ML13. On the other hand, while medical terminologies have face-validity of concept coverage, the curation and quality of these resources are variable for different subject domains and must be considered in any error analysis involving the use of these resources14. Automatic tools for the UMLS have been used with success in biomedical WSD research. For example, MetaMap automatically maps terms in texts to biomedical concepts of the UMLS15. McInnes et al.16 showed that Concept Unique Identifiers (CUIs) generated by MetaMap to biomedical concepts of the UMLS to be good features for general WSD using supervised ML algorithms in the biomedical domain. Leroy and Rindflesch17, 18 examined semantic types and groups with MetaMap. These ontology features produced high variability in accuracy of supervised ML algorithms, because these features rely on complicated hierarchical semantic knowledge representation and have low granularity19.
Linguistic features are based upon patterns of human natural languages and are applicable to clinical notes that result from human communication in the clinical domain. These features represent characteristics of language in general and reflect structural properties of a particular language that are independent of the medical domain. Automatic tools, like the Stanford part-of-speech (POS) tagger20 or MedPOST21, have been trained on general English and biomedical discourse, respectively. However, these tools may not always perform well with clinical texts due to frequent deviations from the standard English sentence structure22. The most common linguistic feature set used in WSD is POS information, which indicates the syntactic category of a given word as it is used within a sentence. Mohammad and Pedersen23 utilized lexical and syntactic features to improve performance of supervised classifiers for general English WSD.
Statistical features utilize distribution and co-occurrence of features of a given corpus. Because humans often describe ideas with similar words, these features are powerful and supported through well-established statistical theories, technologies, and tools. However, one of the weaknesses of these approaches is the difficulty of detecting rare cases or minor senses. In contrast, parameters of statistical models can increase bias through overfitting. Bag-of-words (BoW) is the simplest example of using the frequency of lexical items surrounding the ambiguous word as a predictive statistical feature24. Despite its apparent simplicity and a number of limitations, BoW approach has been demonstrated in previous studies to provide high quality information for many WSD tasks3, 10. Joshi et al.10 explored BoW and term frequency applying supervised approaches to improve accuracy of ML algorithms. Liu et al.25 investigated diverse feature sets including BoW with 15 biomedical abbreviations with supervised ML algorithms. In this later study, the authors show that BoW or BoW with a few word-based features (corresponding orientation within a three word windows with three nearest two-word collocations) produce the best performance for abbreviation disambiguation.
Finally, general document features include information related to the global discourse structure (e.g., document title or section headings). Document characteristics may be indicative of a type of the medical document or clinical sub-specialty and may help narrow down a particular rule set for a particular NLP task26. Discourse information therefore incorporates idiosyncrasies of clinical documentation into predictive features for WSD. For instance, Xu et al.8 used in part section information to build 12 sense inventories from a repository of admission notes through semi-supervised ML methods. One limitation of this class of features is that clinical notes do not always use the same structural format for the same note type, even within the same hospital system or same EHR system. This set of features may also require significant domain knowledge and development of specific rules based upon context, also resulting in a large overhead and lower scalability26.
2.2. Feature selection considerations
Even though researchers have used diverse approaches for WSD, limited studies in the clinical domain make optimal feature sets, optimal window size and orientation, and training sample size optimization an open question3, 10. Major findings in the literature include the following:
Harmonic feature combinations without overfitting results in high performance of supervised ML algorithms3, 10.
BoW has good performance for disambiguation and simple implementation compared to other single features10.
Wider window sizes (entire abstract) surrounding the ambiguous target word provide better performance for WSD within biomedical text10, 16 compared to general English text25.
UMLS CUI as a feature has better accuracy than UMLS semantic type information16.
To obtain optimal “learning”, supervised ML algorithms are required to have enough training samples. Liu et al.25 found supervised classifiers require at least “a few dozens of instances” for each sense. Xu et al.27 scrutinized “required sense size,” and found that increasing the training sample size tends to diminish the error rate if senses are well separated semantically. They also found that a well-separated sense distribution did not affect performance and error rate corresponds to the similarity of senses, and the major classifier performs competitively if the distribution of the majority sense is more than 90%.
3. METHODS
3.1. Data sets
Clinical notes from Fairview Health Services 2004 to 2008 from four metropolitan hospitals in the Twin Cities were used from our research repository. These 604,944 notes were created primarily from voice dictation and transcription with the option of manual editing and included admission notes, inpatient consult notes, operative notes, and discharge summaries.
The 440 most frequently used clinical acronyms and abbreviations were identified using a hybrid heuristic rule-based and statistically-based technique. Potential acronyms and abbreviations were chosen if they consisted of capital letters with or without numbers and symbols (periods, comma, colon, or semicolon) and occurred over 500 times in the corpus. For each acronym or abbreviation, 500 random occurrences of the acronym and abbreviation were selected within the corpus, along with the surrounding previous and subsequent 12 word tokens and presented to two physicians to manually annotate for the senses of the potential acronyms or abbreviations. These 500 occurrences could potentially be extracted from the same discourse if the target acronym or abbreviation was repeated within the discourse. We selected 24 surrounding words as a conservative set of surrounding text, since previous work has demonstrated that humans can properly comprehend meaning given approximately five words including an acronym or abbreviation in the center position7. The inter-annotator agreement of the annotated sense was reported as Kappa with an overlap of 11,000 instances. Percentage agreement was 92.40% and Kappa statistic was 0.84 overall indicating a reasonable inter-rater agreement. These manual annotations were used as the gold standard.
Among 440 data sets, 50 acronyms and abbreviations were used for this study. We considered those acronyms and abbreviations with a majority sense less than or equal to 95%, then selected the same number of sets according to their majority sense ratio. Table 1 shows the 50 acronyms and abbreviations according to their major dominant sense rates. Table 2 summarizes the senses of acronyms and abbreviations and their coverage in the 500 samples. For example, ‘CVA’ has two different senses “cerebrovascular accident” (278 samples, 55.6% - majority sense) and “costovertebral angle” (222 samples, 44.4%).
Table 1.
Distributions of annotated senses of selected clinical acronyms and abbreviations
| Proportion of majority sense | Number of senses | Acronyms and Abbreviations |
|---|---|---|
| 90 ∼ 95% | 5 | BAL, CVS, DIP, IM, OTC |
| 85 ∼ 90% | 5 | C&S, CEA, CVP, ER, FISH |
| 80 ∼ 85% | 5 | ASA, MSSA, PE, SBP, T4 |
| 75 ∼ 80% | 6 | AVR, CA, CTA, IR, NAD, RA |
| 70 ∼ 75% | 4 | AV, PDA, SA, SMA |
| 65 ∼ 70% | 5 | AB, BK, DT, LE, RT |
| 60 ∼ 65% | 3 | IVF, MR, OP |
| 55 ∼ 60% | 5 | CVA, DC, DM, PCP, VBG |
| 50 ∼ 55% | 5 | C4, CDI, PAC, PR, T3 |
| 45 ∼ 50% | 2 | C3, T2 |
| Less than 45% | 5 | AC, IT, MP, PA, T1 |
Table 2.
Annotated senses for selected acronyms and abbreviations in clinical corpus
| Abbr | Sense | Rate% |
|---|---|---|
| AB | abortion | 69.0 |
| blood group in ABO system | 27.4 | |
| other 10 senses | 3.6 | |
| AC | acromioclavicular | 31.8 |
| adriamycin cyclophosphamide | 23.6 | |
| (drug) AC | 21.8 | |
| before meals | 18.8 | |
| other 6 senses | 4.0 | |
| ASA | acetylsalicylic acid | 80.8 |
| American Society of Anesthesiologists | 18.6 | |
| aminosalicylic acid | 0.6 | |
| AV | atrioventricular | 74.8 |
| arteriovenous | 23.2 | |
| other 2 senses | 2.0 | |
| AVR | aortic valve replacement | 76.2 |
| augmented voltage right arm | 20.6 | |
| other 5 senses | 3.2 | |
| BAL | bronchoalveolar lavage | 91.2 |
| blood alcohol level | 8.6 | |
| unsure sense | 0.2 | |
| BK | BK (virus) | 68.6 |
| below knee | 31.4 | |
| C&S | conjunctivae and sclerae | 86.8 |
| culture and sensitivity | 9.4 | |
| other 3 senses | 3.8 | |
| C3 | cervical 3 | 49.8 |
| component 3 | 48.6 | |
| other 2 senses | 1.6 | |
| C4 | cervical 4 | 52.2 |
| component 4 | 46.2 | |
| other 3 senses | 1.6 | |
| CA | cancer | 78.2 |
| carbohydrate antigen | 21.0 | |
| other 2 senses | 0.8 | |
| CDI | Children’s Depression Inventory | 54.0 |
| center for diagnostic imaging | 45.0 | |
| other 2 senses | 1.0 | |
| CEA | carcinoembryonic antigen | 88.6 |
| carotid endarterectomy | 10.6 | |
| other 3 senses | 0.8 | |
| CTA | clear to auscultation | 79.2 |
| computed tomographic angiography | 20.0 | |
| other 3 senses | 0.8 | |
| CVA | cerebrovascular accident | 55.6 |
| costovertebral angle | 44.4 | |
| CVP | central venous pressure | 87.2 |
| cyclophosphamide, vincristine, prednisone | 12.4 | |
| cardiovascular pulmonary | 0.4 | |
| CVS | chorionic villus sampling | 91.4 |
| cardiovascular system | 8.2 | |
| customer, value, service | 0.4 | |
| DC | discontinue | 56.4 |
| direct current | 30.4 | |
| discharge | 6.2 | |
| District of Columbia | 6.2 | |
| other 3 senses | 0.8 | |
| DIP | distal interphalangeal | 92.4 |
| desquamative interstitial pneumonia | 7.2 | |
| dipropionate | 0.4 | |
| DM | dextromethorphan | 57.2 |
| diabetes mellitus | 41.8 | |
| other 2 senses | 1.0 | |
| DT | diphtheria-tetanus | 67.2 |
| delirium tremens | 25.8 | |
| dorsalis pedis:DP* | 4.4 | |
| other 4 senses | 2.6 | |
| ER | emergency room | 89.6 |
| extended release | 6.8 | |
| estrogen receptor | 3.6 | |
| FISH | fluorescent in situ hybridization | 89.8 |
| General English (‘fish’) | 10.2 | |
| IM | intramuscular | 92.2 |
| intramedullary | 7.6 | |
| unsure sense | 0.2 | |
| IR | interventional radiology | 78.8 |
| immediate-release | 20.4 | |
| other 3 senses | 0.8 | |
| IT | General English | 45.0 |
| information technology | 20.6 | |
| intrathecal | 11.6 | |
| ischial tuberosity | 9.6 | |
| iliotibial | 7.0 | |
| intertrochanteric | 2.8 | |
| other 4 senses | 3.4 | |
| IVF | in vitro fertilization | 61.6 |
| intravenous fluid | 37.2 | |
| unsure senses | 1.2 | |
| LE | leukocyte esterase | 68.4 |
| lower extremity | 26.8 | |
| other 7 senses | 4.8 | |
| MP | metacarpophalangeal | 35.4 |
| mercaptopurine | 21.4 | |
| metatarsophalangeal/metacarp ophalangeal | 21.0 | |
| metatarsophalangeal | 10.8 | |
| unsure senses | 6.8 | |
| other 4 senses | 4.6 | |
| MR | magnetic resonance | 62.8 |
| mitral regurgitation | 35.2 | |
| other 4 senses | 2.0 | |
| MSSA | modified selective severity assessment | 83.6 |
| methicillin-susceptible Staphylococcus aureus | 16.4 | |
| NAD | no acute distress | 75.4 |
| nothing abnormal detected | 24.6 | |
| OP | oropharynx | 61.6 |
| oblique presentation/occiput posterior | 24.2 | |
| operative | 11.0 | |
| other 5 senses | 3.2 | |
| OTC | over the counter | 93.8 |
| ornithine transcarbamoylase | 6.2 | |
| PA | posterior-anterior | 42.4 |
| pulmonary artery | 27.6 | |
| physician associates | 16.6 | |
| physician assistant | 12.2 | |
| other 4 senses | 1.2 | |
| PAC | premature atrial contraction | 55.0 |
| physician assistant certification | 27.4 | |
| post anesthesia care | 9.2 | |
| picture archiving communication | 5.0 | |
| other 5 senses | 3.4 | |
| PCP | pneumocystis carinii pneumonia | 58.8 |
| primary care physician | 22.2 | |
| phencyclidine | 18.6 | |
| other 2 senses | 0.4 | |
| PDA | posterior descending artery | 72.2 |
| patent ductus arteriosus | 27.6 | |
| patient-controlled analgesia:PCA† | 0.2 | |
| PE | pulmonary embolus | 81.6 |
| pressure equalization | 17.8 | |
| other 2 senses | 0.6 | |
| PR | pr interval | 50.4 |
| per rectum | 28.2 | |
| progesterone receptor | 17.6 | |
| other 3 senses | 3.8 | |
| RA | right atrium | 78.8 |
| rheumatoid arthritis | 13.2 | |
| room air | 7.2 | |
| other 2 senses | 0.8 | |
| RT | radiation therapy | 67.2 |
| respiratory therapy | 29.6 | |
| other 5 senses | 3.2 | |
| SA | slow acting/sustained action | 74.0 |
| sinuatrial | 17.6 | |
| unsure senses | 6.6 | |
| other 4 senses | 1.8 | |
| SBP | spontaneous bacterial peritonitis | 83.4 |
| systolic blood pressure | 16.6 | |
| SMA | superior mesenteric artery | 70.6 |
| sequential multiple autoanalyzer | 16.8 | |
| spinal muscular atrophy | 11.2 | |
| other 3 senses | 1.4 | |
| T1 | tumor stage 1 | 39.6 |
| thoracic vertebra 1 | 38.8 | |
| T1 (MRI) | 20.6 | |
| other 2 senses | 1.0 | |
| T2 | T2 (MRI) | 45.4 |
| tumor stage 2 | 33.2 | |
| thoracic vertebra 2 | 19.4 | |
| other 3 senses | 2.0 | |
| T3 | triiodothyronine | 53.6 |
| tumor stage 3 | 31.2 | |
| thoracic vertebra 3 | 12.8 | |
| other 2 senses | 2.4 | |
| T4 | thyroxine | 84.8 |
| thoracic vertebra 4 | 8.2 | |
| tumor stage 4 | 7.0 | |
| VBG | vertical banded gastroplasty | 59.8 |
| venous blood gas | 40.2 |
DP (dorsalis pedis) should be used instead of DT
PCA (patient-controlled analgesia) should be used instead of PDA
3.2. Features
For this study, the following features were included and defined as follows:
Window size is the number of word tokens on each side of the given acronym or abbreviation. Window size was varied as follows: ±3, 5, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60 words, entire section, and entire document levels. The sentence level was not analyzed separately in our experiments because of the lack of formal sentence structure within clinical notes. On average, the number of word tokens per document section was 67.97 and for a given note was 391. The window size of 3 means that three previous word tokens, the given acronym or abbreviation, and three post word tokens were included in the window. Windows were also examined asymmetrically (e,g., more context on the left of the acronym than on the right) to understand the relative value of the left and right-sided information.
Bag-of-words (BoW) uses each unique word as a feature in a non-weighted vector, not considering word order. Taking into account frequency and form (i.e. stems) of words, Lexical Variant Generation (LVG)28 normalization tool distributed with MetaMap was used to normalize the 1,000 most frequent words. We limited normalization to the most frequent items in order to speed up the processing for a large number of experiments conducted in this study. We recognize, however, that normalization of lower frequency words may be of further but likely marginal benefit. We also experimented with BoW both with and without stop words29 to further reduce the feature space..
Concept Unique Identifiers (CUIs) were generated from MetaMap. Unique or multiple CUIs were obtained by putting the phrase of a given window size including the acronym or abbreviation into MetaMap. Metamap also generates a score for each potential mapped CUI with a maximum score of 1000 (high likelihood of a positive match). Score cutoffs were varied in our analysis as shown in the Results section.
Semantic types were generated from each of the CUI mappings. The feature set consisted of unique or multiple semantic types generated by putting the selected phrase within a given window size including the acronym or abbreviation into MetaMap. Semantic type groups were also used, aggregating into the pre-defined 15 groups proposed by McCray et al30.
Position information in clinical notes was defined as the relative position of the acronym at the section level and document level. Positions were calculated relatively as the location of the target abbreviations over total words of each level.
Section information from clinical notes is a local contextual feature. We extracted the relevant section information for the given sample as the closest previous section header to the target acronym or abbreviation. Four heuristic conditions were used to detect section information for the given acronym or abbreviation: (1) the previous line is an empty line or other line return symbol only; (2) the position of phrase starts at the beginning of a line; (3) the section indicator symbol “:”; and (4) words from the beginning to the section indicator symbol in the line are written in upper case characters. A physician merged sections tags manually because of the variability in expression for sections names in clinical notes.
Word level POS tags were generated using the Stanford POS tagger. POS tags were collected by putting the word chunk with a given word window size including the acronym or abbreviation into the Stanford POS tagger.
3.3. Algorithms and evaluation
Three fully supervised classification algorithms (Naïve Bayes, Support Vector Machines, and Decision Tree) were implemented with different window sizes using the 10-fold cross-validation setting in Weka (NaiveBayes, LibSVM, and J48 with the default settings), respectively. Window sizes and orientations were also varied to include different numbers of left or right word tokens to find optimal window orientation. Accuracy was reported for system performance with 10-fold cross-validation. Baseline performance was considered to be the majority sense, which helped in evaluating the performance of our ML algorithms. BoW without LVG or stopwords was used for these simulations as a representative baseline methodology.
To explore minimum training sample sizes for acronyms and abbreviations we used inverted cross-validation (ICV). With IVC, various size sub-sets of samples of the acronym or abbreviation were used one time for testing by ICV and the results for sub-sets were averaged to assess performance. ICV is a useful approach for estimating the minimum number of samples required to reach stable performance at a desired accuracy level. Because the average number of senses of selected acronyms and abbreviations was 4.72, we used ICV with 100 and lower number of iterations. Table 3 illustrates training and testing sample sizes with various ICV or cross-validation for each evaluation. For inverted cross-validation, the average accuracy of simulations was reported for the system performance.
Table 3.
Setting parameters of various cross-validation per acronym or abbreviation
| 100 ICV | 50 ICV | 25 ICV | 20 ICV | 10 ICV | 5 ICV | 4 ICV | 2 CV | 5CV | 10CV | |
|---|---|---|---|---|---|---|---|---|---|---|
| Number of training samples | 5 | 10 | 20 | 25 | 50 | 100 | 125 | 250 | 400 | 450 |
| Number of testing samples | 495 | 490 | 480 | 475 | 450 | 400 | 375 | 250 | 100 | 50 |
| Number of simulations | 100 | 50 | 25 | 20 | 10 | 5 | 4 | 2 | 5 | 10 |
4. RESULTS
When aggregating the performance, particularly overall accuracy for 50 acronyms and abbreviations, there were several general findings. From the perspective of classifiers, similar performance was achieved regardless of the classifier type with 10-fold cross-validation. However, SVM classifiers tended to show slightly better performance compared to NB classifiers, and NB classifiers tended to show better performances compared to DT classifiers. With respect to individual features, most features contain better information relative to the baseline majority sense. Among them, BoW features showed better but not statistically different performance compared to other features. As a second best feature, CUI demonstrated better performance than UMLS semantic type with grouping when using the threshold score 900 from MetaMap for a match compared to 1,000.
Increasing window size was found to have a tendency to improve performance at the lower but not the higher end of the window size range. Moreover, entire section and document-size windows showed further deterioration in performance. In contrast, larger window sizes for POS tag features tended to initially decrease performance at lower sizes and then increase performance at larger sizes. The best window size for classifier performance was found to vary with individual features and classifiers. Using SVM classifiers, the best window size with a symmetric window for BoW was 40 (left 40 and right 40 words) and for CUI features with MetaMap was 45 words. Taking out simple English stopwords resulted in better performance when the window size was larger than 20 words in our dataset using NB classifiers. However, removal of stopwords was not helpful for symmetric windows smaller than 20.
As a single feature, section information alone resulted only in an accuracy of 80%. However, it contributed additional information to other single or combined features. Compared to CUI or semantic type features, the combination of sections with CUI or semantic type features improved the ML performance. Although the combination of sections with BoW features did not perform significantly better than BoW features, this combination still gave enough information to make it the best combination of features from the feature types examined.
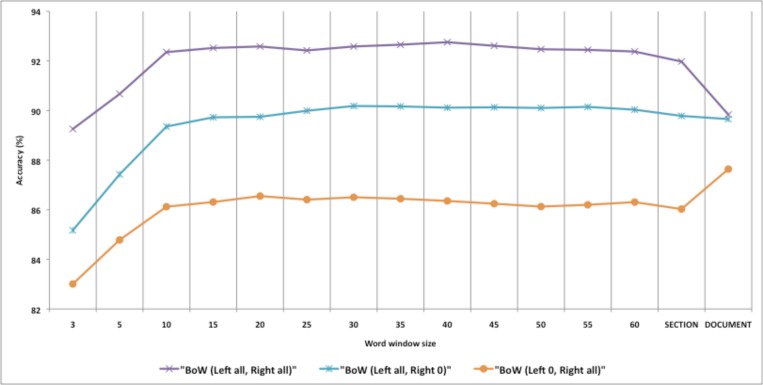
Because BoW resulted in best performance, we investigated information contained in each window of BoW using only one side of window. We utilized BoW along with the SVM machine learning algorithms.
Figure 1 contains a graphical representation of performance with a symmetric window and with windows containing only words on the right or left side. The figure shows that the left word window of the target acronym or abbreviation contains more information for WSD compared to the right word window. The use of both sides of word windows offers better discriminating information than the left side alone.
Figure 1.
Accuracy depending on different sides of word window for BoW with SVM classifiers
The summarized result using SVM classifiers with an expanding left window BoW is shown in Table 4 with acronyms and abbreviations separated by majority sense ratios. Table 4 illustrates a tendency of acronyms and abbreviations with low majority ratios to require a wider left window for best performance. However, if the majority ratio of acronyms and abbreviations is higher (over 80%), it paradoxically performed best with the entire document (left of the target acronym or abbreviation). When this was repeated with the right window, we observed that the maximum performance with the right window was achieved with the use of the entire right document window regardless of the majority sense ratio.
Table 4.
Depending on left word window, sub-aggregated accuracies of grouping by majority sense ratios of abbreviations
| Left BoW using SVM | 3 | 5 | 10 | 15 | 20 | 25 | 30 | 35 | 40 | 45 | 50 | 55 | 60 | Section | Document |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <0.50 (7 acronyms) | 71.486 | 73.514 | 77.171 | 77.086 | 76.514 | 76.943 | 77.000 | 77.057 | 76.943 | 77.086 | 76.886 | 77.429 | 76.943 | 76.914 | 77.143 |
| 0.50< & <0.60 (10 acronyms) | 81.940 | 85.760 | 88.280 | 89.300 | 89.440 | 89.820 | 90.000 | 89.940 | 89.800 | 89.900 | 90.220 | 90.000 | 90.000 | 89.400 | 88.840 |
| 0.60< & <0.70 (8 acronyms) | 85.750 | 88.475 | 90.875 | 91.025 | 90.900 | 91.425 | 91.400 | 91.750 | 91.800 | 91.425 | 91.475 | 91.600 | 91.375 | 90.750 | 90.575 |
| 0.70< & <0.80 (10 acronyms) | 86.060 | 88.280 | 89.540 | 90.320 | 90.480 | 90.480 | 91.140 | 91.320 | 90.900 | 90.820 | 90.860 | 90.780 | 90.900 | 90.500 | 89.040 |
| 0.80< & <0.90 (10 acronyms) | 92.260 | 93.760 | 94.440 | 94.520 | 94.760 | 94.700 | 94.680 | 94.120 | 94.380 | 94.640 | 94.440 | 94.380 | 94.180 | 94.360 | 95.040 |
| 0.90< & <0.95 (5 acronyms) | 93.920 | 94.200 | 95.600 | 95.440 | 95.560 | 95.920 | 96.160 | 96.240 | 96.400 | 96.400 | 96.000 | 96.200 | 96.280 | 96.440 | 97.800 |
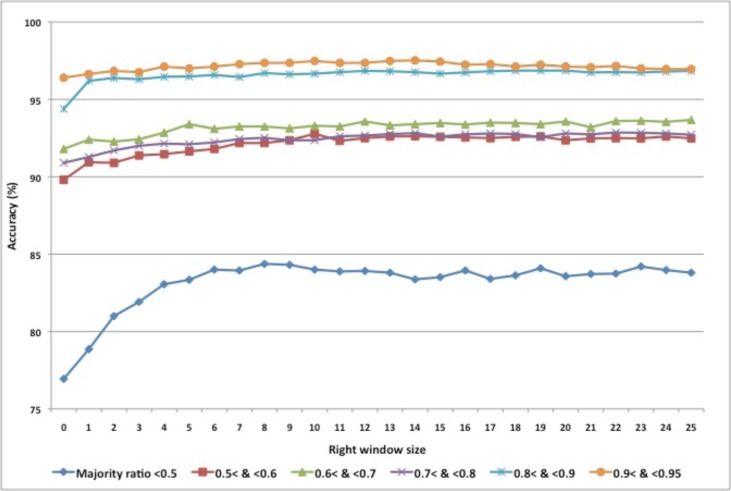
Figure 2 shows accuracy trends for a fixed left window size (40) and an increasing right word window size (X-axis) with different majority sense distributions. In general, good performance is reached with a smaller size right window. The best performance of BoW is 92.88% (over all 50 acronyms and abbreviations) with 40 left side window and 23 right side window.
Figure 2.
Accuracy depending on varying right word window with left 40 word window Majority ratio = majority sense ratio in groups of acronyms and abbreviations
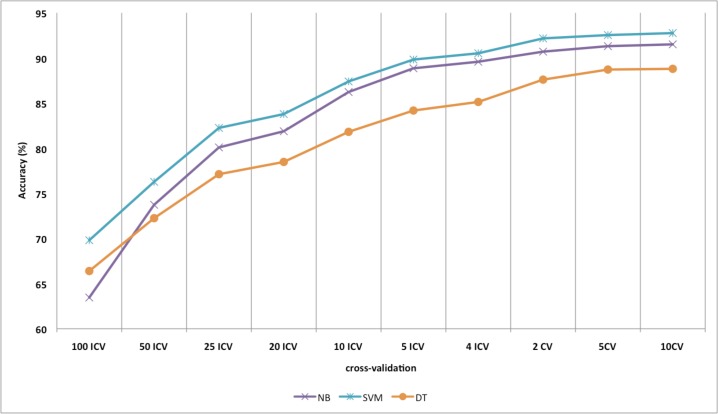
Figure 3 is the aggregated accuracy of 50 abbreviations with both sides of the word windows equal to 40 when using SVM classifiers with BoW with various inverted or standard cross-validation settings. Increasing the training sample size increases the accuracy for disambiguation as expected. Our findings demonstrate that 2, 5 and 10-fold cross-validations show similar performance. Furthermore, increasing the sample size with ICV shows increasing performance when comparing the gradated performance between 100 ICV and 4 ICV. As shown in Figure 3, for a desired accuracy to over 90%, the minimum sample number is 125 (4 ICV) when using SVM classifiers, and approximately 250 (2 CV) when using NB classifiers over the aggregated 50 acronyms and abbreviations. Therefore, when there is little information about majority sense distributions of acronyms and abbreviations, at least 125 training samples is a reasonable baseline required for acronym and abbreviation WSD classification with the SVM classifier.
Figure 3.
Accuracy depending on CV (size of training sample)
Table 5 summarizes the accuracy of SVM and NB when grouping acronyms and abbreviations according to the majority sense ratios. The highlighted cells are the first points with over 90% aggregated accuracy across inverse cross validation settings. Here, acronyms and abbreviations with high majority sense ratios tend to require fewer samples than acronyms and abbreviations with low majority sense ratios to achieve a threshold of 90% accuracy. In terms of classifiers, SVM and NB classifiers demonstrated better and more stable performance over the DT classifier. SVM had better performance than the NB classifier to classify senses of the acronyms and abbreviations when it has fewer samples.
Table 5.
Comparison among classifiers split by majority sense ratio using NB and SVM
| Majority ratio | SVM classifier | NB classifier | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inverted cross-validation | Cross-validation | Inverted cross-validation | Cross-validation | |||||||||||||||||
| 100 | 50 | 25 | 20 | 10 | 5 | 4 | 2 | 5 | 10 | 100 | 50 | 25 | 20 | 10 | 5 | 4 | 2 | 5 | 10 | |
| <0.50 (7 acronyms) | 42.33 | 53.31 | 64.79 | 67.75 | 73.99 | 77.94 | 79.63 | 82.66 | 82.74 | 83.94 | 39.23 | 52.09 | 62.64 | 65.48 | 72.17 | 76.17 | 78.01 | 79.83 | 80.29 | 81.17 |
| 0.50<& <0.60 (10 acronyms) | 60.07 | 70.76 | 80.27 | 82.83 | 86.76 | 89.36 | 89.98 | 91.12 | 92.84 | 92.80 | 53.52 | 69.17 | 78.17 | 80.94 | 85.56 | 88.62 | 89.24 | 90.14 | 92.12 | 92.20 |
| 0.60< & <0.70(8 acronyms) | 68.34 | 75.77 | 83.24 | 84.66 | 88.83 | 91.20 | 91.58 | 93.03 | 93.15 | 93.28 | 60.35 | 71.83 | 79.55 | 81.53 | 87.15 | 89.68 | 90.28 | 92.35 | 92.83 | 93.15 |
| 0.70< & <0.80 (10 acronyms) | 74.80 | 79.34 | 83.38 | 84.44 | 87.86 | 90.12 | 90.37 | 92.40 | 92.14 | 92.26 | 67.23 | 76.02 | 81.35 | 82.92 | 87.35 | 89.96 | 90.25 | 91.62 | 92.06 | 92.12 |
| 0.80< & <0.90 (10 acronyms) | 84.00 | 87.16 | 89.23 | 89.99 | 92.57 | 94.63 | 95.31 | 96.36 | 96.74 | 96.84 | 79.91 | 85.07 | 87.88 | 88.64 | 91.28 | 93.42 | 93.95 | 94.96 | 95.14 | 95.22 |
| 0.90< & <0.95 (5 acronyms) | 91.72 | 92.44 | 92.90 | 93.02 | 93.88 | 94.99 | 95.83 | 97.36 | 97.04 | 97.00 | 81.83 | 88.99 | 91.12 | 91.72 | 93.45 | 94.43 | 95.27 | 94.04 | 93.52 | 93.32 |
5. DISCUSSION
This study provides important insights into the area of clinical acronym and abbreviation WSD. Our main finding is that the left side of words in a window around the target acronym or abbreviation provides better information for disambiguation than the right side of the window. Therefore, an asymmetrical window larger on the left and smaller on the right maintains performance and allows for a smaller feature space and a more efficient computational process. This phenomenon coincides with the process of sense discrimination by human annotators. When annotators classify senses of acronyms and abbreviations, they mainly focus on the left side of target token. Interestingly, humans require a very small number of tokens for the right window (about 5 words) compared to our automated methods (about 20 word window). One factor that could partially account for this discrepancy is that there may be information lost in the pre-processing steps for features (i.e., lexical normalization and selection of 1,000 frequent words). Another main finding of this study was the observation that a size of around 125 samples with SVM classifiers may be effective as a baseline threshold for training. However, it is important to note that in cases of acronyms and abbreviations with less than 50% majority sense ratios, all accuracies were lower than 90% even in 10-fold cross-validation settings, which warrants future study into the enriching datasets with rarer sense distributions associated with acronyms and abbreviations with these distribution patterns.
We extracted the most frequently used 440 acronyms and abbreviations with a cut-off frequency of 500 occurrences from a large corpus consisting of various types of clinical notes and annotated these with experts. To examine questions about training sample size, we carefully selected the acronyms and abbreviations according to the majority sense ratio. While it is possible that these findings are specific to the corpus of text that we used, these results are still helpful to identify representative trends in acronym and abbreviation sense disambiguation in the clinical domain. The large size of the dataset (50 acronyms) is also helpful in elucidating the amount of variability that exists in WSD of acronyms in clinical texts. Some of the parameters are slightly different in these experiments compared to previous studies, several findings from this study on acronym and abbreviation WSD in clinical notes are consistent with several other previous studies3, 10, 25, 27 of word, acronyms, and abbreviation sense disambiguation in biomedical literature and clinical notes. (i.e., the BoW feature is a powerful feature and SVM algorithm has good performance for WSD). The defining contribution of this work was its use of a large set of clinical acronyms and abbreviations and the examination of both window orientation and size as well as looking at the question about minimum training sample numbers with a systematic approach.
The combination of using all features dropped performance in our results. A possible explanation is the presence of duplicative or conflicting information between different features (especially POS tag feature) with larger window sizes (up to document level). Another possible reason is that CUIs and semantic features may contain noise from inaccuracies in MetaMap, which was used for CUI mapping. There is also a tendency for clinical texts to contain incomplete sentences and other poorly-formed text. Furthermore, windows for WSD tasks are typically based on centering acronyms and abbreviations in our experience and also sometimes do not maintain full sentences for the Stanford POS tagger or using by MetaMap. As such, the Stanford POS tagger or MetaMap may generate incorrect POS tags or concepts from any partial sentence phrase, which may deteriorate the overall ML performance. Lastly, the Stanford POS tagger may not be optimized for dealing with clinical notes because it is trained and designed for general English.
It is important to note that this is another example where MetaMap may need future optimization as a core of the UMLS. Because the tool was not developed for the clinical domain, it may suffer in performance for certain clinical tasks. According to Savova et al.3, 20% of pertinent ambiguous terms overlapping between biomedical and clinical domains possess more senses in the clinical domain than the biomedical domain. Xu et al.2 also found that terms in clinical corpora have low coverage in UMLS. Therefore, we may miss CUI and semantic information in the clinical domains. We also attempted to enhance semantic information by adding semantic grouping information of McCray et al. but found that this did not significantly improve the performance because one of the semantic groups dominates (48.8%): “Chemicals & Drugs”. Furthermore, some groups such as “Genes & Molecular Sequences”, “Geographic Areas”, “Occupations”, etc, are proportionally too small (only 0.1%).
Certain limitations are important to note with this study in its interpretation. The main limitation is that the features utilized here are based on words and are mostly dependent upon one another. In other words, CUI or semantic information from MetaMap contains overlapped information with BoW. Therefore, performance using BoW features shows similar performance using the combination of knowledge features (BoW+CUI+Semantic information). Another issue is that there is no systematic management implemented for the number of features in this study. The average number of features per instance was 849 for BoW, 2,427 for CUI, and 134 for semantic information when we fix the word window size to 40 symmetrically. In other words, MetaMap features may offer insufficient information for the machine to learn compared to BoW features. There is also the important issue of dealing with rare senses, which drop the system performance significantly and require specific methodologies to address adequately. We did not eliminate these rare senses in this experiment in order to reflect the difficulty of this task with clinical notes, and all rare senses, as well as typographical and other errors in the samples were included in this experiment.
Future work is needed to determine if our methods and findings are scalable for other clinical note corpora. We used a heuristic approach to detect section information which may require modification for other documents, as over 25,000 lexically unique section headers were found in this document repository. Finally, although we assumed that there was “one sense per-discourse”, this may not apply throughout an entire clinical document8 when considering section information, which is an issue that we plan to explore further.
6. CONCLUSION
In this study we investigated a large group of clinical acronyms and abbreviations from our clinical notes corpus to understand issues related to practical clinical acronym and abbreviation WSD. Using 50 clinical acronyms and abbreviations with a majority sense < 95%, we found BoW to be an efficient feature set. When looking at window orientation and size, a symmetric window of ∼40 words was found to have good performance with the left side of the window providing more valuable information compared to the right side. Our experiments also demonstrate that an SVM classifier with at minimum 125 training samples was needed to achieve at least 90% accuracy for clinical WSD tasks. These findings provide important insight into the application of clinical acronym and abbreviation WSD in clinical NLP system modules.
7. ACKNOWLEDGMENTS
The authors would like to thank Fairview Health Services for ongoing support of this research. This work was supported by the University of Minnesota Institute for Health Informatics Research Support Grant (SM, GM, SP), the American Surgical Association Foundation Grant (GM) and the National Library of Medicine (#R01 LM009623-01) (SP).
8. REFERENCES
- 1.Stetson PD, Johnson SB, Scotch M, Hripcsak G. The sublanguage of cross-coverage. Proc AMIA Symp; 2002. pp. 742–746. [PMC free article] [PubMed] [Google Scholar]
- 2.Xu H, Stetson PD, Friedman C. A study of abbreviations in clinical notes. AMIA Annu Symp Proc; 2007. pp. 821–825. [PMC free article] [PubMed] [Google Scholar]
- 3.Savova GK, Coden AR, Sominsky IL, et al. Word sense disambiguation across two domains: biomedical literature and clinical notes. J Biomed Inform. 2008 Dec;41(6):1088–1100. doi: 10.1016/j.jbi.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 4.Friedman C, Liu H, Shagina L, Johnson S, Hripcsak G. Evaluating the UMLS as a source of lexical knowledge for medical language processing. Proc AMIA Symp; 2001. pp. 189–193. [PMC free article] [PubMed] [Google Scholar]
- 5.Pakhomov S, Pedersen T, Chute CG. Abbreviation and acronym disambiguation in clinical discourse. AMIA Annu Symp Proc; 2005. pp. 589–593. [PMC free article] [PubMed] [Google Scholar]
- 6.Schuemie MJ, Kors JA, Mons B. Word sense disambiguation in the biomedical domain: an overview. J Comput Biol. 2005 Jun;12(5):554–565. doi: 10.1089/cmb.2005.12.554. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan A. An experimental study of ambiguity and context. Mechanical Translation. 1950;2(2):39–46. [Google Scholar]
- 8.Xu H, Stetson PD, Friedman C. Methods for building sense inventories of abbreviations in clinical notes. J Am Med Inform Assoc. 2009 Jan-Feb;16(1):103–108. doi: 10.1197/jamia.M2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz AS, Hearst MA. A simple algorithm for identifying abbreviation definitions in biomedical text. Pac Symp Biocomput. 2003:451–462. [PubMed] [Google Scholar]
- 10.Joshi M, Pakhomov S, Pedersen T, Chute CG. A comparative study of supervised learning as applied to acronym expansion in clinical reports. AMIA Annu Symp Proc; 2006. pp. 399–403. [PMC free article] [PubMed] [Google Scholar]
- 11.NIH Unified Medical Language System (UMLS) 2011. [ http://www.nlm.nih.gov/research/umls/]
- 12.IHTSDO Systematized Nomenclature of Medicine-Clinical Terms (SNOMED CT) 2011. [ http://www.ihtsdo.org/snomed-ct/] [DOI] [PubMed]
- 13.McInnes BT, Pedersen T, Pakhomov SV. UMLS-Interface and UMLS-Similarity : open source software for measuring paths and semantic similarity. AMIA Annu Symp Proc. 2009;2009:431–435. [PMC free article] [PubMed] [Google Scholar]
- 14.Fung KW, Hole WT, Nelson SJ, Srinivasan S, Powell T, Roth L. Integrating SNOMED CT into the UMLS: an exploration of different views of synonymy and quality of editing. Journal of the American Medical Informatics Association : JAMIA. 2005;12(4):486–494. doi: 10.1197/jamia.M1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 16.McInnes BT, Pedersen T, Carlis J. Using UMLS Concept Unique Identifiers (CUIs) for word sense disambiguation in the biomedical domain. AMIA Annu Symp Proc; 2007. pp. 533–537. [PMC free article] [PubMed] [Google Scholar]
- 17.Leroy G, Rindflesch TC. Effects of information and machine learning algorithms on word sense disambiguation with small datasets. Int J Med Inform. 2005 Aug;74(7–8):573–585. doi: 10.1016/j.ijmedinf.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 18.Leroy G, Rindflesch TC. Using symbolic knowledge in the UMLS to disambiguate words in small datasets with a naive Bayes classifier. Stud Health Technol Inform. 2004;107(Pt 1):381–385. [PubMed] [Google Scholar]
- 19.Rindflesch TC, Aronson AR. Ambiguity resolution while mapping free text to the UMLS Metathesaurus. Proc Annu Symp Comput Appl Med Care. 1994:240–244. [PMC free article] [PubMed] [Google Scholar]
- 20.Toutanova K, Klein D, Manning CD, Singer Y. Feature-rich part-of-speech tagging with a cyclic dependency network. Proceedings of the 2003 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology - Volume 1; Edmonton, Canada. Association for Computational Linguistics; 2003. pp. 173–180. [Google Scholar]
- 21.Smith L, Rindflesch T, Wilbur WJ. MedPost: a part-of-speech tagger for bioMedical text. Bioinformatics. 2004 Sep 22;20(14):2320–2321. doi: 10.1093/bioinformatics/bth227. 2004. [DOI] [PubMed] [Google Scholar]
- 22.Long WJ. Parsing free text nursing notes. AMIA Annu Symp Proc; 2003. p. 917. [PMC free article] [PubMed] [Google Scholar]
- 23.Mohammad S, Pedersen T. Combining lexical and syntactic features for supervised word sense disambiguation. Paper presented at: Proc of the CoNLL; 2004. [Google Scholar]
- 24.Gale WA, Church KW, Yarowsky D. A Method for Disambiguating Word Senses in a Large Corpus. Comput Humanities. 1992 Dec;26(5–6):415–439. [Google Scholar]
- 25.Liu H, Teller V, Friedman C. A multi-aspect comparison study of supervised word sense disambiguation. J Am Med Inform Assoc. 2004 Jul-Aug;11(4):320–331. doi: 10.1197/jamia.M1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denny JC, Spickard A, 3rd, Johnson KB, Peterson NB, Peterson JF, Miller RA. Evaluation of a method to identify and categorize section headers in clinical documents. J Am Med Inform Assoc. 2009 Nov-Dec;16(6):806–815. doi: 10.1197/jamia.M3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H, Markatou M, Dimova R, Liu H, Friedman C. Machine learning and word sense disambiguation in the biomedical domain: design and evaluation issues. BMC Bioinformatics. 2006;7:334. doi: 10.1186/1471-2105-7-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc Annu Symp Comput Appl Med Care; 1994. pp. 235–239. [PMC free article] [PubMed] [Google Scholar]
- 29.Manning CD, Schütze H. Foundations of statistical natural language processing. Cambridge, Mass: MIT Press; 1999. [Google Scholar]
- 30.McCray AT, Burgun A, Bodenreider O. Aggregating UMLS semantic types for reducing conceptual complexity. Stud Health Technol Inform. 2001;84(Pt 1):216–220. [PMC free article] [PubMed] [Google Scholar]