Abstract
Complex relationships in biomedical publications are often communicated by diagrams such as bar and line charts, which are a very effective way of summarizing and communicating multi-faceted data sets. Given the ever-increasing amount of published data, we argue that the precise retrieval of such diagrams is of great value for answering specific and otherwise hard-to-meet information needs. To this end, we demonstrate the use of advanced image processing and classification for identifying bar and line charts by the shape and relative location of the different image elements that make up the charts. With recall and precisions of close to 90% for the detection of relevant figures, we discuss the use of this technology in an existing biomedical image search engine, and outline how it enables new forms of literature queries over biomedical relationships that are represented in these charts.
Introduction
Images are an important part of biomedical publications. They are often used to concisely communicate the results of experiments and to visualize models and data sets. Recently, there has been an increasing focus on biomedical images,1 and specialized search engines have been developed that allow users to query the figures of biomedical articles,2,3 which are otherwise not readily accessible. These search engines focus mostly on indexing and making available text within image captions. However, many biomedical images contain much structured information, which is not yet accessible through search. Examples include the fairly regular patterns in diagrams, such as the text of the x- and y-axes, which encode specific relationships of the variables shown in the images. In this work, we are presenting first steps towards breaking down diagrams into components, and extracting and analyzing the components for valuable structured image context. For the purpose of this study, we focus on one particular type of diagram, which we call “axis diagrams”.
We define them as follows:
A diagram is called an axis diagram if it has at least two straight and visible axes that each represent a particular dimension of the underlying data, where the dimension can be nominal, discrete, or continuous in nature and the data itself is represented as dots, lines, bars, etc.
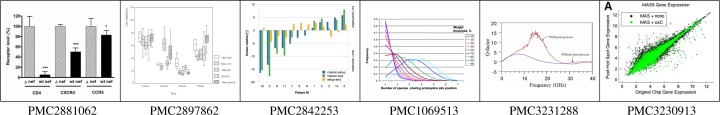
Figure 1 shows six examples of different kinds of axis diagrams. Such diagrams are important for several reasons:
They are abundant in biomedical literature
They are complex in the sense that they combine several dimensions
They follow simple common patterns based on axes
They summarize data for human readers
Figure 1:
Six examples of axis diagrams (with the PMC identifier of the respective article)
The first point illustrates the vast potential of automated diagram identification, retrieval and extraction. The second point highlights the value of a diagram’s internal structure which can be broken down and exploited to relate diagrams to each other, to their context, and to queries. The third point suggests that the analysis and extraction of such diagrams can be done with high quality. The fourth point is perhaps the most important one. It suggests that diagrams are probably the best starting point when navigating the biomedical literature. This makes them prime candidates for the presentation of results by image- and data-focused search engines. Recent query engine approaches like Wolfram Alpha are based upon the idea of presenting results in the form of diagrams instead of links to documents. This requires a formal model of the data, based on which the answers are calculated and diagrams are drawn. This does not seem to be a viable approach for biomedical knowledge within the near future, but we can use the diagrams that are already there, carefully compiled by the authors of scientific articles, retrieve them and present them as results to queries.
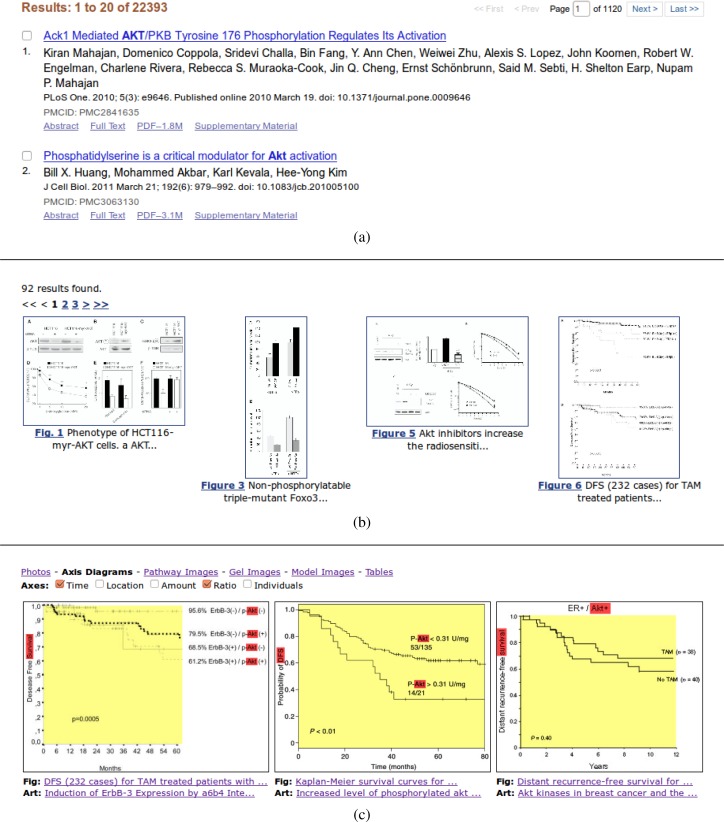
Figure 2 shows what such a query engine could look like, and compares a mockup screen to existing query interfaces. Figure 2a illustrates a classical document-based query interface that is mainly text-based and relies on the users’ ability to parse titles and abstracts to satisfy a particular information need. Figure 2b shows a more specialized biomedical search interface, in this case from the Yale Image Finder, which returns highly specific results from within figures in scientific articles. A large part of such figures, however, consist of multiple parts, only few of which might be relevant for a given query. As a result, there is a considerable burden on the user to parse a figure, and to identify the relevant parts. Our hypothetical search engine and interface, on the other hand, are assisting users in parsing these figures, by extracting the relevant sub-images. This might not look like a radical change, but it could result in considerable time savings, as users may be able to quickly find diagrams of interest and learn more about its context (complete figure, figure caption, article text, authors, etc.). However, this is just one aspect of our approach; analyzing the deeper structure of images, like the axes of diagrams, opens new possibilities for refined queries, and for refined presentations of results. As described in this study, we believe it is feasible to reliably detect the axes of diagrams. Determining the scale type of the axes (nominal, discrete, continuous) and their general content type (time, location, amount, ratio, individuals, etc.) seems feasible too. As a result, we may be able to return very specific diagrams for specific queries. Suppose that a researcher is interested in how the expression of a biomarker, such as AKT, influences disease survival. With our approach, we can retrieve the relevant diagrams and filter them by axis type. Figure 2c illustrates how a query for “AKT” and “survival” could be answered with diagrams depicting time and ratio axes.
Figure 2:
Comparison of three query interface approaches (two real and one fictitious): document-based at the top (PubMed Central), figure-based in the middle (Yale Image Finder), and chart-based at the bottom (fictitious). The query consists of the two keywords “survival” and “AKT”.
In this paper, we present the results of the first step of developing such a diagram-based query interface, that is the detection and localization of axis diagrams within figures. The next steps — axes detection and classification, coverage of other diagram types, and user interface development — are future work.
Background and Related Work
The presented work was performed within the Yale Image Finder (YIF)2 project. Our goal is to use structured information within images to aid in information retrieval and extraction. At the core of the technology, image processing routines are used to parse image content, with a particular focus on detection and extraction of image text, i.e. text embedded in images.4 The YIF search engine, accessible at http://krauthammerlab.med.yale.edu/imagefinder, currently allows for the precise retrieval of images by image text context, and provides navigational support to discover related images, and the associated documents. We are currently developing technologies to allow for more sophisticated image queries, particularly over image text location, i.e. the placement of text within images. This allows for precise queries over specific image elements, such as x-axis labels, or y-axis labels, enabling novel queries.
There exists a fair amount of related work on classifying images from scientific publications. Rafkind et al.5 demonstrated the classification of biomedical images into five classes (Gel, Graph, Image-of-Thing, Mix, and Model). Rodriguez-Esteban and Iossifov6 performed a similar study using four classes (Gel, Pathway, Structure, and Time). Increasingly, such approaches employ figure segmentation and subfigure classification, as presented by Shatkay et al.7 for document classification. Other examples include the work by Murphy et al.8 on the extraction of subcellular location information from images, by Qian and Murphy9 on the detection of panels of fluorescence microscope images, and by Kozhenkov and Baitaluk10 on extracting pathway information from diagrams. To the best of our knowledge, nobody has yet applied such an approach to decompose and analyze biomedical axis diagrams, despite the fact that they constitute a very important and frequent image type.
Methods
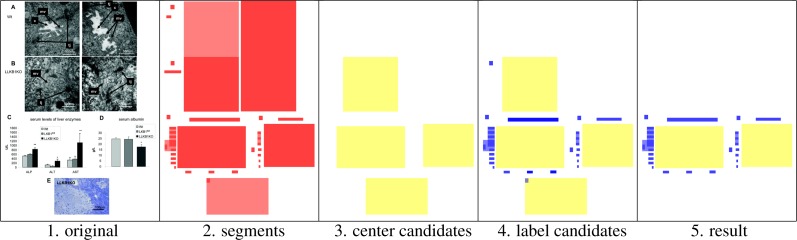
We present an algorithm for detecting axis diagrams within figures and to extract their exact location. The algorithm exploits the fact that axis diagrams commonly consist of one large center segment depicting image data, and several smaller text label segments arranged around the center segment. After experimentation on a small sample of images, we came up with a specific five step algorithm and we gradually adjusted the involved parameters. The different steps of the algorithm are illustrated in Figure 3; the steps are:
Retrieve the original figure as a bitmap image
Preprocess the image with an image text detection algorithm that we developed in previous work,4 which returns a set of (possibly overlapping) rectangle-shaped text segments for each figure; in addition, OCR is run on the individual segments and the recognized text is returned
Find potential center segments: take all segments of a certain size (at least 1% of the area of the entire image) and of a certain shape (width/height ratio of at most 4 to 1); if two of the potential center segments overlap: remove both of them and add their intersection instead
Search in a rectangle-shaped band along the border of the center segment for potential label segments: the band is partly outside the center segment (this part of the band is of width 0.25s with where w and h are width and height, respectively, of the center segment) and partly inside it (of width 0.13s); keep as potential labels all segments that are fully contained in this band and are not too large (less than 10% of the area of the center segment)
Remove center segments that have too few labels (less than 5 labels, possibly overlapping), do not have enough total label area (less than 2% of the area of the center segment), or do not have much text in their labels (less than 4 recognized characters); the remaining center segments and their label segments are recognized as axis diagrams
Figure 3:
The different steps of the algorithm demonstrated on an exemplary figure (from PMC3262187).
For evaluation purposes, we created a gold standard corpus of images with and without axis diagrams. We took a random sample of 100 open-access articles from PubMed Central that have at least one figure in the form of a bitmap image and a figure caption. Altogether, these 100 articles contain 404 figures, an average of about 4 figures per article.
For these figures, the number of axis diagrams was subsequently annotated by hand, resulting in 508 axis diagrams, i.e. more than the number of figures (because many figures contain more than one diagram), demonstrating that axis diagrams are indeed a very important image type in the biomedical literature. 177 figures (44%) contain at least one axis diagram; 24 thereof (6%) contain more than 5 axis digrams. This corpus was collected and annotated after we finished implementing the algorithm presented here. During the development of the algorithm we used another small corpus, which was not part of the evaluation corpus.
We first evaluated the algorithm for its ability to distinguish two classes: figures with at least one axis diagram (class 1), and figures without (class 0). As a baseline, we used a Support Vector Machine (SVM)11 to perform the classification task using features extracted from the image itself and from the caption text. For the image properties, we used 64 grayscale histogram features of the type presented by Shatkay et al.7 and 13 texture features based on Haralick et al.12 For the caption features, we tokenized and stemmed the caption text and transformed it into a word vector. The output of the segment-based algorithm was used as an additional feature to investigate whether combining the two approaches leads to improved results. The SVM evaluation was done via 10-fold cross-validation on our gold standard corpus. Concretely, we used the Weka environment13 with the Lovins stemmer14 and the Sequential Minimal Optimization (SMO)15 implementation of SVM using the polynomial kernel.
The evaluation procedure described above only classifies the presence or absence of an axis diagram; it does not evaluate whether we correctly locate the axis diagrams within the figures. For that reason, we performed a second, more fine-grained and more specific evaluation round, investigating how many diagrams are correctly recognized at the correct location. In this case, we can not easily compare it to a baseline method, as the SVM classifiers cannot predict the location of diagrams. Still, we can use the SVM classifiers to improve precision by disregarding the figures that do not contain any diagrams according to SVM and by applying the segment-based algorithm on the remaining figures.
Results
Table 1 shows the results. The segment-based algorithm is able to detect figures that contain axis diagrams with a high precision of 86.6% and with a recall of 65.5%. The SVM classifiers based on image features and caption text alone have lower precision than the segment-based algorithm, but higher recall. Overall, the SVM classifier using image and caption features performs better than the segment-based classifier with F-measures of 84.3% and 74.6%, respectively. Combining the two approaches leads to higher values with an F-measure of almost 90%.
Table 1:
Evaluation of axis diagram detection. The first task (top) is to classify figures with respect to whether they contain axis diagrams or not. The second task (bottom) is to find the location of the diagrams within the figures. The used methods are the segments-based algorithm and SVM-based classifiers using image data and caption text.
| task | method | precision | recall | F-measure |
|---|---|---|---|---|
|
| ||||
| detection of figures with axis diagrams | segments | 0.866 | 0.655 | 0.746 |
| SVM (image only) | 0.663 | 0.898 | 0.763 | |
| segments + SVM (image only) | 0.801 | 0.729 | 0.763 | |
| SVM (caption only) | 0.835 | 0.774 | 0.804 | |
| segments + SVM (caption only) | 0.899 | 0.853 | 0.875 | |
| SVM (image + caption) | 0.851 | 0.836 | 0.843 | |
| segments + SVM (image + caption) | 0.897 | 0.887 | 0.892 | |
|
| ||||
| extraction of axis diagram locations | segments | 0.845 | 0.396 | 0.539 |
| segments + SVM (image only) | 0.844 | 0.394 | 0.537 | |
| segments + SVM (caption only) | 0.876 | 0.390 | 0.539 | |
| segments + SVM (image + caption) | 0.893 | 0.394 | 0.546 | |
Looking at the specific cases where the combined algorithm gave an incorrect result reveals that 11% of the errors concern borderline cases, i.e. diagrams for which it is not perfectly clear whether they should be considered axis diagrams according to our definition. These errors are certainly excusable. 50% of the false positive cases concern general diagrams that are not axis diagrams but do not fit into a typical category. Because of this, they probably lack features that would allow the SVM classifier to recognize that they are not axis diagrams. In the case of the false negatives, 15% are probably due to the bad quality of the respective images, but apart from that, no obvious patterns can be identified.
When it comes to the actual extraction of the diagrams, the segment-based algorithm on its own predicts the location of the diagrams with a high precision of 84.5%, but a rather low recall of 39.6%. Combining it with the SVM classifiers can only improve precision but not recall (because no new diagrams can be detected), and it indeed improves precision to 89.3% with only a marginal decrease on the side of the recall (39.4%).
Discussion
Our results are on the upper-end or above of what has been achieved with other types of images in biomedical publications. Rodriguez-Esteban and Iossifov6 ran classifiers for four classes of images: gel images, pathway diagrams, protein structure depictions, and diagrams with a time axis. Their resulting F-measures range from 63% to 84%, depending on the class. It is interesting that, out of their classes, the one that is most similar to axis diagrams — that is time diagrams — got the poorest classification results: their time diagrams got F-measures of 63%, whereas we achieved 89% for axis diagrams. Rafkind et al.5 performed a similar study using five classes: gel images, graphs (bar/line charts, plots, etc.), image-of-thing (photographs), model images (of a biological process, experimental design, etc.), and mixed images. Their “graphs” class seems to be very close to what we defined as “axis diagrams”. For this class, they achieved an F-measure of 78.7%, which is considerably lower than what we achieved. In both cases, we should be careful when comparing these values, because different methods are applied and different images are used for evaluation. Still, it shows that our approach is promising.
Since axis diagrams are so frequent, the low recall of the location extraction is not a severe problem at this point. One factor that makes the recall so low is the fact that some figures contain arrays of large numbers of very small axis diagrams that are hard to detect. Our algorithm fails mostly on these figures, with a considerable negative impact on the recall.
Conclusion
We think that many queries that researchers post to scientific search engines are best answered by diagrams. These diagrams can be found in biomedical articles, but they are not accessible to common document-based search engines. In this article, we show that a special and very frequent type of diagram — axis diagrams — can be automatically extracted in a reliable way. We plan to exploit this in the future by implementing a diagram-based search engine that allows us to answer specific questions by precisely retrieving the relevant diagrams. We believe that such a tool would be of great value to researchers working in the biomedical domain.
Acknowledgments
We thank Songhua Xu for his feedback on questions regarding image segmentation and OCR. This research has been funded by NLM Grant No. 5R01LM009956.
References
- 1.Peng Hanchuan. Bioimage informatics: a new area of engineering biology. Bioinformatics. 2008 Sep;24(17):1827–1836. doi: 10.1093/bioinformatics/btn346. URL http://dx.doi.org/10.1093/bioinformatics/btn346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu Songhua, McCusker James, Krauthammer Michael. Yale Image Finder (YIF): a new search engine for retrieving biomedical images. Bioinformatics. 2008 Sep;24(17):1968–1970. doi: 10.1093/bioinformatics/btn340. URL http://dx.doi.org/10.1093/bioinformatics/btn340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hearst Marti A, Divoli Anna, Guturu Harendra, Ksikes Alex, Nakov Preslav, Wooldridge Michael A, et al. Biotext search engine. Bioinformatics. 2007 Aug;23(16):2196–2197. doi: 10.1093/bioinformatics/btm301. URL http://dx.doi.org/10.1093/bioinformatics/btm301. [DOI] [PubMed] [Google Scholar]
- 4.Songhua Xu, Krauthammer Michael. A new pivoting and iterative text detection algorithm for biomedical images. J. of Biomedical Informatics. 2010 Dec;43(6):924–931. doi: 10.1016/j.jbi.2010.09.006. URL http://dx.doi.org/10.1016/j.jbi.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rafkind Barry, Lee Minsuk, Chang Shih-Fu, Yu Hong. Proceedings of the Workshop on Linking Natural Language Processing and Biology: Towards Deeper Biological Literature Analysis. Stroudsburg, PA, USA: Association for Computational Linguistics; 2006. Exploring text and image features to classify images in bioscience literature; pp. 73–80. BioNLP ’06. URL http://dl.acm.org/citation.cfm?id=1567619.1567632. [Google Scholar]
- 6.Rodriguez-Esteban Raul, Iossifov Ivan. Figure mining for biomedical research. Bioinformatics. 2009 Aug;25(16):2082–2084. doi: 10.1093/bioinformatics/btp318. URL http://dx.doi.org/10.1093/bioinformatics/btp318. [DOI] [PubMed] [Google Scholar]
- 7.Shatkay Hagit, Chen Nawei, Dorothea Blostein. Integrating image data into biomedical text categorization. Bioinformatics. 2006 Jul;22(14):e446–e453. doi: 10.1093/bioinformatics/btl235. URL http://dx.doi.org/10.1093/bioinformatics/btl235. [DOI] [PubMed] [Google Scholar]
- 8.Murphy Robert F, Kou Zhenzhen, Hua Juchang, Joffe Matthew, Cohen William W. Extracting and structuring subcellular location information from on-line journal articles: The subcellular location image finder. Proceedings of the IASTED International Conference on Knowledge Sharing and Collaborative Engineering (KSCE-2004); ACTA Press; 2004. pp. 109–114. URL http://www.actapress.com/Abstract.aspx?paperId=17244. [Google Scholar]
- 9.Qian Yuntao, Murphy Robert F. Improved recognition of figures containing fluorescence microscope images in online journal articles using graphical models. Bioinformatics. 2008 Feb;24(4):569–576. doi: 10.1093/bioinformatics/btm561. URL http://dx.doi.org/10.1093/bioinformatics/btm561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozhenkov Sergey, Baitaluk Michael. Mining and integration of pathway diagrams from imaging data. Bioinformatics. 2012 Mar;28(5):739–742. doi: 10.1093/bioinformatics/bts018. URL http://dx.doi.org/10.1093/bioinformatics/bts018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortes Corinna, Vapnik Vladimir. Support-vector networks. Machine learning. 1995;20(3):273–297. [Google Scholar]
- 12.Haralick Robert M, Shanmugam K, Dinstein Its’Hak. Textural Features for Image Classification. IEEE Transactions on Systems, Man, and Cybernetics. 1973 Nov;3(6):610–621. URL http://dx.doi.org/10.1109/TSMC.1973.4309314. [Google Scholar]
- 13.Hall Mark, Frank Eibe, Holmes Geoffrey, Pfahringer Bernhard, Reutemann Peter, Witten Ian H. The WEKA data mining software: An update. ACM SIGKDD Explorations Newsletter. 2009;11(1):10–18. [Google Scholar]
- 14.Lovins Julie Beth. Development of a stemming algorithm. Mechanical Translation and Computational Linguistics. 1968;1(1,2):22–31. [Google Scholar]
- 15.Platt John C. Advances in kernel methods. chapter Fast training of support vector machines using sequential minimal optimization. MIT Press; Cambridge, MA, USA: 1999. pp. 185–208. URL http://dl.acm.org/citation.cfm?id=299094.299105. [Google Scholar]