Abstract
The enormous amount of data being collected by electronic medical records (EMR) has found additional value when integrated and stored in data warehouses. The enterprise data warehouse (EDW) allows all data from an organization with numerous inpatient and outpatient facilities to be integrated and analyzed. We have found the EDW at Intermountain Healthcare to not only be an essential tool for management and strategic decision making, but also for patient specific clinical decision support. This paper presents the structure and two case studies of a framework that has provided us the ability to create a number of decision support applications that are dependent on the integration of previous enterprise-wide data in addition to a patient’s current information in the EMR.
Introduction
Inpatient and outpatient electronic medical records (EMR) are accumulating enormous amounts of patient, provider, facility, financial and process information. During the early 1990s, this information began to be recognized as an extremely valuable and untapped resource for management and clinical research. However, EMR administrators were concerned about the impact of running large research queries on the clinical database. It was determined that healthcare needed to convert this data into aggregated and separate information systems that could support retrospective and population-based analysis1. Data warehouses had emerged in other industries; however, their adoption by healthcare was slow due to the complexity and heterogeneity of medical, operational, and clinical data2.
As an effort to facilitate access to this wealth of medical information, data warehouses that contained clinical and administrative data from healthcare organization began to be developed3. Using network technologies, interfaces were developed to collect the data from the different databases and stored in a single large database. However, early on, it was recognized that the data from many sources not only needed to be integrated, but also cleansed, and formatted4. Data semantics were then used to regroup and merge patients’ medical data from the autonomous and heterogeneous health information systems5. As expected, this raised concerns of data security and patient privacy. Solutions supporting U.S. and European laws for high level of security, retrieval audit, and user authentication needed to be incorporated to ensure privacy and confidentiality2,5. As further uses for data warehouses were identified, image data using the (digital imaging and communication in medicine) DICOM standard was used to integrate information from picture archiving and communication system (PACS)6–8. The advantage of sharing data owned by different organizations was identified and federated information models were developed9–11. While HL7 is often used as the interface standard for integrating the data from divergent data silos, other data standards including RxNorm12, SNOMED-CT, ICD, CPT, LOINC, UMLS and DRG codes3,13,14 are also often included within the data warehouses. The data stored within these data warehouses can be managed and accessed through direct Structured Query Language (SQL) calls or SQL that is imbedded inside of Application Programming Interfaces (APIs) that are programmed in C++, Java, Perl, etc. User interfaces have also been developed in Visual Basic and distributed as ActiveX objects embedded in an HTML page15 or information retrieval can be performed using metadata-based semantic and full-text search methods10. Web front-ends using i2b2 and caGrid frameworks have facilitated data access from federated information models3. Some have gone one step further by developing a second de-identified data warehouse or framework that is suitable for direct user access through controlled query tools that can support both research and education activities11,17,18.
Most literature describing the use of data warehouses in healthcare didn’t began to appear in the late 1990s. Initially, administrative and business applications showed the benefit of using the data warehouse for managed care contracting, case and provider profiling, utilization management, product management forecasting, capacity planning, medical expense, insurance rejection, hospital volume and patient movement through each hospital19–22. Ad hoc queries could be used to discover hidden knowledge from coded fields, text reports, and laboratory values23. Data mining techniques or Knowledge Discovery in Databases (KDD) borrowed from industry could be used to identify relationships in large databases24. Data mining techniques were able to make business decisions that can influence cost, revenue, and operational efficiency while maintaining a high level of care25. This was followed with examples of using on-line analytical processing (OLAP) from data generated in an on-line transaction processing (OLTP) system26. These new methods have been successfully used for multiple purposes, including patient care, health services research, resource utilization and feasibility studies27. A number of other data query and manipulation methods have successfully been used including support vector machines, exploratory factor analysis, decision tree and Bayesian networks, co-occurrence statistics and Cox analysis24,28–31. In 1998, Kaiser Permanente reported that it not only found ways to save millions of dollars in healthcare costs, but also was able to improve the quality of care with the knowledge gleaned from its data warehouse32.
In addition to the number of administrative uses of data warehouses, a number of clinical uses of data warehouses have been reported over the past 15 years. A search of the literature includes studies on recruitment for clinical trials15,23,33,34, gene-disease associations2,35–38, and family health history data patterns39, public health 40–41, trends in drug use, drug cost and adverse drug interactions42–44, effectiveness of the CPOE system45, diabetes34,46, epilepsy and other neurological disorders9, factors contributing to preterm birth and labor practice patterns19,24, infection surveillance47, medical errors and under documentation22,48. However, most of these studies were used to retrospectively identify medical knowledge that could then be used to change the healthcare process based on population information. For a translational research environment, it is critical to make these data usable at the point-of-need49. An information feedback mechanism is needed that closes the loop of information flow by bringing decision support information from the data warehouse directly to the clinician50. We have found over the past few years, that an enterprise data warehouse (EDW), which provides service for the entire organization, can be a fundamental tool to actively improve the care of specific patients as soon as they are hospitalized, visit clinics or seen by home health nurses. This paper presents the structure and two case studies of a framework that has provided us the ability to create decision support applications that are dependent on the integration of previous enterprise-wide data in addition to current patient information in the EMR.
Methods
Intermountain Healthcare (Intermountain) is a non-profit healthcare system that operates 22 hospitals and over 179 clinics, instacares, physician offices and home healthcare in Utah and Idaho. Intermountain also has an insurance plan and a medical evacuation unit with five helicopters and three fixed wing aircraft. The mission of Intermountain is to provide comprehensive, clinically effective and cost efficient healthcare throughout Utah and Idaho. As an effort to meet its mission, Intermountain recognized it would need to manage health information across this entire spectrum of care using data from all patient encounters. Intermountain’s EDW began in 1998 under the direction of senior clinical and business leaders. The EDW was formed to provide the infrastructure to measure best practices, costs, and specifically for clinical quality improvement initiatives. The EDW strategy and priorities are governed by a Business Intelligence Council and Executive Committee that is comprised of business and clinical leaders from throughout Intermountain.
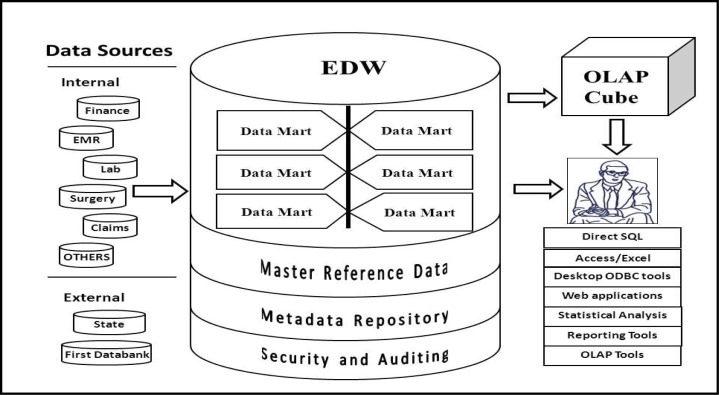
Intermountain’s EDW has grown from a handful of data sources and data sets to having data from nearly all clinical and business areas of the organization, jointly accounting for over 80 billion rows of data in nearly 9,000 ready-to-query tables in a 10 terabyte Oracle 11g database supporting over 100,000 unique queries a month by 1,700 unique logon accounts. The datasets include inpatient and outpatient clinical data from the HELP and HELP2 information systems, financial data from patient accounts to supply chain management and materials management, clinical data from laboratory to imaging, labor and delivery, surgery, numerous data marts supporting Intermountain’s healthcare improvement centers, human resources, payroll, Intermountain’s health plans data and many others (Figure 1).
Figure 1.
Conceptual architecture of the EDW at Intermountain Healthcare.
Each patient in the Intermountain system receives a unique enterprise-wide ID that is used for every inpatient and outpatient encounter. The standard data elements that are used across the many transactional systems within Intermountain such as patient enterprise IDs, provider IDs, facility IDs, etc., are stored in the Master Reference Data and serve as the backbone for data integration across datasets in the EDW. The EDW has a large metadata repository along with a security and auditing infrastructure. The coded data in the EDW is managed by a data dictionary and also contains ICD9, LOINC, and SNOMED coding. The EDW is updated each night from the inpatient and outpatient clinical data and the administrative data. Extraction, transform and loading (ETL) functions are handled by IBM Data Stage. Mortality data from the Utah State Department of Health and data from national databases such as First Data Bank are loaded monthly. The Intermountain EDW also can be linked to a statewide genetic database for studying gene-disease associations. The EDW supports thousands of users ranging from power users writing SQL or PLSQL against the relational tables to business and clinical executives receiving Business Intelligence reports, dashboards and scorecards via an employee portal using reporting tools such as Cognos, Tableau, NetCharts, and SAS and Statit. Intermountain has also invested in the training of data warehouse and analytics experts. Some data is normalized as needed, but in general the data is de-normalized to facilitate user queries and increase response times. The EDW team maintains a web-based metadata tool that provides descriptions for data marts, tables, columns and lists of tables that share common columns.
In 2003, we began to encounter some major patient safety and other clinical issues that required patient data beyond the individual hospital walls. Further root cause analysis led us to determine that enterprise-wide data could provide an integral role in improving individual and population patient care with clinical decision support. However, access to and data from the EDW only would not be able to provide all the required functionality. We have developed a new framework for data collection and decision support applications that demonstrate the benefit of enterprise-wide data access combined with data from the current admission located in the EMR.
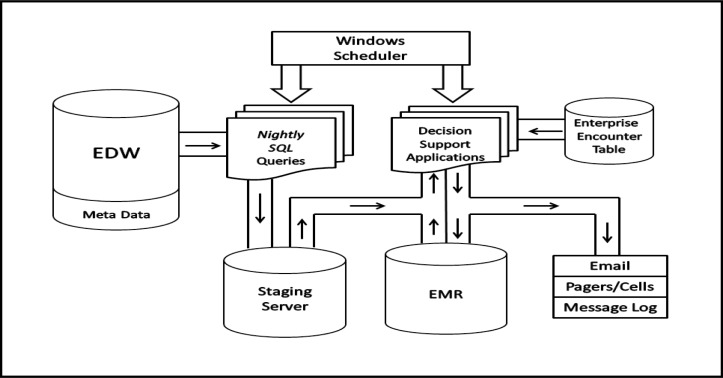
A number of applications using SQL, PLSQL or Java with embedded SQL queries were developed to extract specific data elements from the EDW (Figure 2). Each night after the EDW has been updated with all the clinical and administrative data for the past 24 hours, Windows Scheduler is used to automatically activate the SQL query applications and the extracted data is reformatted and stored in other Oracle tables on staging servers that are maintained 24/7 and backed up each night. The data on the staging servers are stored in data marts oriented to the specific clinical needs of the decision support applications. Windows Scheduler is also used to activate a number of Java applications that run from once a day to every 15 minutes. The first thing each of the applications does is look for patients in the enterprise encounter table that match specific requirements for identifying potential patients of interest. The enterprise encounter table contains the master list of all inpatient and outpatient encounters along with some demographic data. The logic in each of the applications also has a specific list of needed patient data elements with specific time windows. Thus, an application may require a variety of different patient data elements that could have been stored in any inpatient or outpatient facility from a number of years back in time or during the current encounter. Previous data from other facilities is retrieved from the metadata tables on the staging server and current encounter data is retrieved from the EMR. While any data in the EDW is available for use, the main types of data we use are diagnosis and procedure ICD9 codes, laboratory including microbiology and antibiotic susceptibilities, medication fills from community pharmacies in the Health plans data, performed and scheduled surgical procedures, frequency of hospital admissions, demographics, etc. As new decision support applications are requested that require new types of data elements that have not been retrieved from the EDW and stored in the staging files, use of the EDW meta data helps identify where the data is stored and how it is represented, i.e., numeric, character, etc. Alerts generated by the applications can then be sent directly to the healthcare providers through email, pagers, cell phones or the EMR message log depending on the required method and timing requested by the provider. This application framework has enabled us over the past few years to quickly develop and implement a number of new applications that offer decision support directly to healthcare providers.
Figure 2.
Framework for decision support applications dependent on previous enterprise-wide in addition to current patient information.
Case Studies
The two case studies presented in this paper have been published previously51,52. While both applications are clinically used every day at all 22 Intermountain hospitals, some clinics and home health, their impact is not the main focus, but were selected to demonstrate the functionality and capability of the framework. Moreover, neither of the applications described in these case studies would have been possible without this framework.
Multi-drug resistant pathogens
The first case study describes the early and extended functionality of the first application we developed that required enterprise-wide data. Patients with previous multi-drug resistant bacteria are often carriers for years after the initial infection. As these patients enter the hospital or ambulatory facility, the healthcare providers including infection control personnel were usually not aware; even if they were returning to the same hospital within a few days or weeks. Moreover, this situation becomes even more obscure when these patients move from one facility to another. These unknown carriers may then be unknowingly placed in close contact with or even share semi-private rooms with immunocompromised patients. While the spread of multi-resistant pathogens to other patients is an obvious issue, there is also increased risk that the unknown carrier may not be placed on appropriate antibiotics soon enough.
The first data mart we developed on the staging server contained all patients with methicillin-resistant Staphylococcus aureus (MRSA) or vancomycin-resistant Enterococci (VRE). Each night, SQL queries scan all new culture results from all inpatient and outpatients laboratories within the enterprise and look for new patients with MRSA or VRE who are then placed in the multi-drug resistant organism (MDRO) data mart. Although the EDW is updated each night, the SQL queries always look for new cultures during the past three days in case, for any reason, a culture result didn’t make it into the EDW the first night. Often, new MRSA or VRE positive cultures are identified for patients already in the data mart. In that case, the information from the latest results replaces the previous results. Thus, only the newest results are kept in the data mart. We developed a daily report sent to the infection preventionist’s (IPs) printer at LDS Hospital that identified new admissions of patients with previous MRSA and VRE to their hospital and two other hospitals. That report listed the type of MDRO the patient had along with other pertinent information such as the date/time of the latest positive culture, which healthcare facility and the specimen of the latest positive culture (Figure 3). Evaluation of that initial report showed that healthcare providers, including IPs, almost always did not know the new admitted patients had previous MRSA or VRE positive cultures51. Over the past six years, the printout sent to the IPs at one hospital has been replaced by daily email reports sent to the IPs at all 22 Intermountain hospitals, some clinics and home health. Emails sent to laptop computers meant the IPs did not have to be by the printer to receive the report. However, the IPs are not always looking at their laptops or at the hospital 24 hours a day. Thus, for some situations, a daily email was not soon enough to prevent the transmission of MRSA or VRE to another patient or get the infected patient on appropriate antibiotics. In addition to the daily email reports, we have the MDRO application check the enterprise encounter file every 15 minutes. Often the admission information is entered into the encounter file before the patient arrives at the hospital or outpatient clinic. This is certainly the case for scheduled visits. The IPs now receive pages or cell phone text messages for individual patients who are hospitalized or in the emergency department and include the same basic type of information in the daily email report. Since the IPs are not usually at the hospital nights and weekends, nursing supervisors also receive the page or cell phone alerts. The application on the EMR that prints the admission sheet that is placed in the patient’s paper chart also looks at the multi-drug resistant data mart and potential carriers have the appropriate box filled with the needed information. As an additional safety net, another application uses the data from the MDRO data mart and other data pulled each night from the EDW or in the EMR. Other known risk factors for carriers of MRSA include previous hospital visits or the use of certain antibiotics within the past six months, hemodialysis, current hospital stay longer than 30 days, and age > 75 years. Patients who arrive in the ICU with previous MRSA or a combination of the other risk factors have visual alerts sent to the computer screens in their rooms an hour after they arrive to the ICU53. Initially, we sent the ICU alerts as soon as the patient arrived. But, we found that the medical staff had so much to do when the patient first arrived, that they often forgot about the MRSA alerts. Patients not at high risk at admission may be identified afterwards based on the accumulation of new risk factors. Patients with previous MRSA or VRE who have been successfully treated and determined to no longer be carriers based on sequential negative cultures are identified by the IPs and are removed from the MDRO data mart for that specific pathogen type. Some patients may be listed in the data mart with both MRSA and VRE and some get reentered into the data mart based on subsequent positive cultures after they have been removed. There are currently 30,694 unique patients in the data mart with previous MRSA and 2,401 with VRE. In the past 5 years, 194,658 email alerts for MRSA and 22,160 for VRE have been sent to the IPs at the 22 Intermountain hospitals. The log shows that 4,803 pager alerts for MRSA and 838 for VRE have been sent in the past three years. In March of 2011, we were requested to send the MRSA and VRE email alerts to a number of pediatric clinics at 2pm the day before the scheduled visits. During the first year 1,170 MRSA alerts were sent and surprising, no visits by kids with previous VRE were identified. Other MDROs can be easily added to the alerts when requested by infection control.
Figure 3.
Example of the daily secure MRSA/VRE email alerts sent to the infection preventionists at all 22 Intermountain hospitals. Pager alerts do not include the patient’s name.
High risk of venous thromboembolism
Venous thromboembolism (VTE) includes deep vein thrombosis (DVT) and pulmonary embolism (PE); both of which are frequent complications associated with hospitalization and affect the morbidity, mortality, length of stay and costs of millions of patients each year throughout the world. Medical as well as surgical patients are at risk during their hospital stay. There are a number of known and suspected combinations of risk factors for VTE and manual surveillance for the high-risk patients was too time-consuming to be done reliably every day. We selected a previously published high-risk score that included individual risk factors that were found in our EDW54. A VTE data mart and new SQL queries were developed that collected and stored the presence of each risk factor identified for any patient found in the encounter table during the nightly search. Patients with a score of 4 points or higher are identified as high risk for VTE. Cancer, previous VTE and hypercoagulability is scored as 3 points, surgery duration greater than one hour as 2 points and age > 70 years, bed rest, body mass index (BMI) > 29 kg/m2 and use of hormone-replacement therapy or oral contraceptives as 1 point each. Thus, patients could be identified as high-risk at admission or later as they accumulate more risk factors such as surgery duration or bed rest. The cancer risk, previous DVT or PE are identified from ICD9 codes or natural language programs that screen admission diagnoses, vascular studies for DVTs and CT-angiograms and VQ scans for PEs. Hypercoagulability is identified from laboratory results and surgery duration greater than one hour within the past 30 days from the surgical data, and the use of hormone-replacement therapy or oral contraceptives from the health plans data in the EDW that is collected from community pharmacies. Current age, bed rest and BMI based on the patient’s height and weight need to be retrieved from the current encounter data in the EMR. The EMR is also checked for the diagnosis of cancer and new hypercoagulability laboratory results. Anticoagulation coordinators and pharmacists at each hospital receive an email every morning that identifies each new high-risk patient and lists their individual risk factors (Figure 4). The VTE decision support application also checks the high-risk patients EMR for the use of appropriate anticoagulation and leg compression therapy. Evaluation of the VTE high risk alerts found a sensitivity of 98% and a positive predictive value of 99%52. A new program is being tested that looks for new high risk patients every hour and sends the VTE high risk alerts to the pharmacist’s task list in the pharmacy menu in the EMR. Likewise, the VTE decision support application now has patient specific logic that checks for the appropriate use of anticoagulation based on the drug and the current dosage. When discrepancies are found, pager text messages are sent directly to hospitalists and the message log.
Figure 4.
Example of the daily secure VTE high risk email alerts sent to pharmacists at all 22 Intermountain hospitals. Pager alerts do not contain patient names.
The case studies presented in this paper were included to demonstrate the functionality of the framework. We have a number of other decision support applications that use this same framework and are developing new ones. Some only need to look for one or two data elements in the EDW while some perform natural language processing (NLP) on dictated emergency department notes, history and physical notes, radiographic findings, etc. in the EDW.
Discussion
More and more health care institutions are developing data warehouses and decision support tools for management, strategic and clinical decision making. Enterprise-wide data extends the data warehouse beyond the single facility’s walls. We have been able to develop a number of decision support applications that are dependent on the integration of previous enterprise-wide data in addition to current patient information through use of our EDW. The framework presented in this paper has allowed us to reuse the same methods and data marts to reduce development time for new applications.
We have emphasized the importance of using an enterprise-wide data warehouse for patient specific decision support in this paper. Other organizations have also reported on the value of enterprise-wide data for clinical decision support at the point of care55. Likewise, the development of an EDW by just a few affiliated hospitals has been extremely valuable. In fact, the need to monitor and prevent infection transmission provides an ideal case for sharing data between multiple facilities. An early paper reporting the advantage of sharing information among different hospitals used the data warehouse to monitor antimicrobial resistance, measure antimicrobial use, detect hospital-acquired bloodstream infections, measure the cost of infections, and detect antimicrobial prescribing errors56. They were also able to estimate the amount of time and money saved through use of the data warehouse. However, clinical data warehouses from single facilities also provide a valuable resource to improve patient care. One hospital has used its data warehouse to provide lists of high-risk patients linked to the patient’s next scheduled visit. This enabled the targeted delivery of H1N1 vaccine to high volume clinics44. Moreover, in some situations, organizations have decided that it would not be desirable to create a combined central data warehouse and use a federated model where each facility manages and maintains control over their local databases that can be queried by the other facilities through a standard web service API57.
Over the past 14 years, the EDW department at Intermountain has not only increased the amount and types of stored data, but has also made a number of changes to improve and extend the value of the data to help us meet numerous internal and external reporting requirements. Maintaining an EDW the size of Intermountain’s requires a large commitment by the organization and Intermountain was fortunate to get early support from upper management. In addition to the obvious hardware expense, a new department must be organized for maintenance and customer support. There are a number of different methods and ways to develop and maintain a clinical data warehouse and some work better for different organizations. Integrating the data is a required initial step but the method depends on the different structures and location of the various data sources. An essential part of the integration is the record linkage58. Intermountain is fortunate to assign each patient a unique enterprise number during the first encounter with the system. This greatly facilitates the data integration process but duplicate records still need to be identified. This is a common issue faced by all clinical data warehouses and some novel methods have been developed59. Many clinical research data integration platforms rely on the relational Entity-Attribute-Value model for data storage because of its flexibility60. In most cases we have preferred to de-normalize the tables in the EDW at Intermountain to facilitate query formulation and reduce execution time.
Although the EDW is updated from most of the different data sources each night, we always run the nightly SQL queries used for our decision support applications for three days back in time in case there are network problems or scheduled EDW maintenance downtimes. Since the data from the queries is almost always collected before it is needed for the patient’s next encounter, this is rarely an issue. Keep in mind, data warehouses were not originally developed to be up 24/7. While the EDW metadata has been an extremely valuable resource for locating needed data, we always verify and validate the required data elements for the decision support logic by using the data from multiple known patients for comparison. Numerous different data elements can be stored in similar sounding tables with similar column names. Also, since the exact date and time of the data element is almost always critical in the decision support logic, we need to make sure we know when a specimen for a laboratory result was collected, when a problem was recorded or a procedure occurred. How these times are stored can vary in our EDW. Some date/times are stored as numeric values and some as “dates” or characters. Moreover, some date/times are stored as Greenwich Mean times and thus require a conversion to Mountain Time. The EDW team converts some of the nonstandard date/times during the ETL process. Also, there are some laboratory values that are stored in the EMR and sent to the EDW as integers, floating point, real numbers, etc. Thus, there are a number of special cases that need to be learned and accounted for when querying and using data from the EDW. This exemplifies the need to test and retest the decision support logic before it goes into production. As an effort to reduce these types of problems and facilitate data access, the EDW department is developing an Analytic Health Repository which will be another layer over the EDW that will provide permanent data integration and data quality processes and structures, and services that will allow users to retrieve and use the data in a standard method. In addition, the EDW team is looking at an Operational Data Store (ODS) that will receive more real-time feeds from the EMR or other systems.
There are some obvious limitations to the current EDW at Intermountain and the decision support framework we present here. While we have an extensive EDW, it still only contains the data from the Intermountain system. Although that accounts for around half of the healthcare in Utah, we still encounter numerous patients with previous critical information from outside our system. That supports the case for Intermountain to also be able to share their data within a federated model with other healthcare organizations within and outside of Utah. The federated model is successfully being used today by other organizations for the sharing of genomic data2. Many of our decision support applications rely on the use of ICD9 codes from previous or outside facilities. Cancer is a risk factor for the VTE alerts and a previous study has shown that not all patients with ICD9 codes for cancer have corresponding documentation in pathology reports61. Another limitation is that we do not have an enterprise database that contains daily updated patient lists. While some physicians maintain their daily patient lists, we sometimes need to rely on the attending physician listed in the patients EMR in order to send patient specific alerts to email, pagers, cell phones or the message log. In some cases this lack of precision prevents us from sending some potential time critical alerts to the correct healthcare providers. The other method for alert review we do not use is through CPOE when any designated provider with the appropriate status based on their logon could be sent the alerts. While many healthcare facilities have this capability today, Intermountain is still in the process of making that a reality.
While schema integration may be more difficult for medical databases because of the complexity of the concepts and the associated relationships, the data warehouse can provide a unique advantage to the healthcare organization2. Incentives for healthcare organizations have shifted from volume-based to results-based payment. Thus, patient outcomes and safety, and cost effectiveness will continue to drive reimbursements. Health care management must plan and implement a strategy using a best practice approach. By using data in the warehouse, healthcare organizations can not only provide accurate data for management and resource utilization, but use the information in the data warehouse to improve patient care. Management can also save millions of dollars through the implementation of clinical pathways in better resource utilization and changing physician behavior to best practices based on evidence-based medicine62. Likewise, a challenge for next-generation health care providers is to improve on how they consolidate and manage information across the continuum of care. This involves building a warehouse of clinical and financial information that can be shared and leveraged by health care professionals, regardless of the location or type of care setting63. As integrated delivery systems are formed and patient care delivery is restructured, health care professionals must also be able to distribute, access, and evaluate information across care settings.
Conclusion
We have found the EDW to be a vital tool for management and strategic decision making. Moreover, we have developed a framework that provides us the ability to create decision support applications for specific patients that are dependent on the integration of previous enterprise-wide data in addition to their current encounter information.
Acknowledgments
We thank the following people for their input and direction on the previous projects used for the case studies in this paper. Rouett H. Abouzelof MSN, CIC, Caroline W. Taylor MSN, CIC, Vickie R. Anderson RN, Valerie T. Aston, RRT, BS, Scott C. Woller, MD, C. Greg Elliott, MD, Scott M. Stevens, MD, Jacob S. Tripp, PhD.
References
- 1.Ledbetter CS, Morgan MW. Toward best practice: leveraging the electronic patient record as a clinical data warehouse. J Healthc Inf Manag. 2001 Summer;15(2):119–31. [PubMed] [Google Scholar]
- 2.Chute CG, Beck SA, Fisk TB, Mohr DN. The Enterprise Data Trust at Mayo Clinic: a semantically integrated warehouse of biomedical data. J Am Med Inform Assoc. 2010 Mar-Apr;17(2):131–5. doi: 10.1136/jamia.2009.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thadani SR, Weng C, Bigger JT, Ennever JF, Wajngurt D. Electronic Screening Improves Efficiency in Clinical Trial Recruitment. JAMIA. 2009 Nov;16(6):869–873. doi: 10.1197/jamia.M3119. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verma R, Harper J. Life cycle of a data warehousing project in healthcare. J Healthc Inf Manag. 2001 Summer;15(2):107–17. [PubMed] [Google Scholar]
- 5.Kerkri EM, Quantin C, Allaert FA, et al. An approach for integrating heterogeneous information sources in a medical data warehouse. J Med Syst. 2001 Jun;25(3):167–76. doi: 10.1023/a:1010728915998. [DOI] [PubMed] [Google Scholar]
- 6.Zhang M, Zhang H, Tjandra D, Wong ST. DBMap: a space-conscious data visualization and knowledge discovery framework for biomedical data warehouse. IEEE Trans Inf Technol Biomed. 2004 Sep;8(3):343–53. doi: 10.1109/titb.2004.832550. [DOI] [PubMed] [Google Scholar]
- 7.Wong ST, Hoo KS, Jr, Knowlton RC, et al. Design and applications of a multimodality image data warehouse framework. J Am Med Inform Assoc. 2002 May-Jun;9(3):239–54. doi: 10.1197/jamia.M0988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubin DL, Desser TS. A data warehouse for integrating radiologic and pathologic data. J Am Coll Radiol. 2008 Mar;5(3):210–7. doi: 10.1016/j.jacr.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 9.Cao X, Wong ST, Hoo KS, Jr, Tjandra D, Fu JC, Lowenstein DH. A web-based federated neuroinformatics model for surgical planning and clinical research applications in epilepsy. Neuroinformatics. 2004;2(1):101–18. doi: 10.1385/NI:2:1:101. [DOI] [PubMed] [Google Scholar]
- 10.Livne OE, Schultz ND, Narus SP. Federated Querying Architecture with Clinical & Translational Health IT Application. J Med Syst. 2011 Oct;35(5):1211–24. doi: 10.1007/s10916-011-9720-3. Epub 2011 May 3. [DOI] [PubMed] [Google Scholar]
- 11.McDonald CJ, Dexter P, Schadow G, Chueh HC, Abernathy G, Hook J, Blevins L, Overhage JM, Berman JJ. SPIN query tools for de-identified research on a humongous database. AMIA Annu Symp Proc. 2005:515–9. [PMC free article] [PubMed] [Google Scholar]
- 12.Hernandez P, Podchiyska T, Weber S, Ferris T, Lowe H. Automated mapping of pharmacy orders from two electronic health record systems to RxNorm within the STRIDE clinical data warehouse. AMIA Annu Symp Proc. 2009 Nov 14;:244–8. 2009. [PMC free article] [PubMed] [Google Scholar]
- 13.Kamal J, Liu J, Ostrander M, et al. Information warehouse - a comprehensive informatics platform for business, clinical, and research applications. AMIA Annu Symp Proc. 2010 Nov 13;:452–6. 2010. [PMC free article] [PubMed] [Google Scholar]
- 14.Grant A, Moshyk A, Diab H, et al. Integrating feedback from a clinical data warehouse into practice organisation. Int J Med Inform. 2006 Mar-Apr;75(3–4):232–9. doi: 10.1016/j.ijmedinf.2005.07.037. Epub 2005 Sep 8. [DOI] [PubMed] [Google Scholar]
- 15.Murphy SN, Barnett GO, Chueh HC. Visual query tool for finding patient cohorts from a clinical data warehouse of the partners HealthCare system. Proc AMIA Symp. 2000:1174. [PMC free article] [PubMed] [Google Scholar]
- 16.Cuggia M, Garcelon N, Campillo-Gimenez B, et al. Roogle: an information retrieval engine for clinical data warehouse. Stud Health Technol Inform. 2011;169:584–8. [PubMed] [Google Scholar]
- 17.Liu J, Erdal S, Silvey SA, et al. Toward a fully de-identified biomedical information warehouse. AMIA Annu Symp Proc. 2009 Nov 14;:370–4. 2009. [PMC free article] [PubMed] [Google Scholar]
- 18.Erdal BS, Liu J, Ding J, et al. A Database De-identification Framework to Enable Direct Queries on Medical Data for Secondary Use. Methods Inf Med. 2012 Feb 7;51(2) doi: 10.3414/ME11-01-0048. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 19.Hall ES, Thornton SN. Extracting nursing practice patterns from structured labor and delivery data sets. AMIA Annu Symp Proc. 2007 Oct 11;:309–13. [PMC free article] [PubMed] [Google Scholar]
- 20.Dewitt JG, Hampton PM. Development of a data warehouse at an academic health system: knowing a place for the first time. Acad Med. 2005 Nov;80(11):1019–25. doi: 10.1097/00001888-200511000-00009. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, Matsumura Y, Nakagawa K, Ji S, Nakano H, Teratani T, Zhang Q, Mineno T, Takeda H. Analysis of yearly variations in drug expenditure for one patient using data warehouse in a hospital. J Med Syst. 2007 Feb;31(1):17–24. doi: 10.1007/s10916-006-9039-7. [DOI] [PubMed] [Google Scholar]
- 22.Studnicki J, Berndt DJ, Luther SL, Fisher JW. The application of volume-outcome contouring in data warehousing. J Healthc Inf Manag. 2004 Fall;18(4):49–55. [PubMed] [Google Scholar]
- 23.Murphy SN, Morgan MM, Barnett GO, Chueh HC. Optimizing healthcare research data warehouse design through past COSTAR query analysis. Proc AMIA Symp. 1999:892–6. [PMC free article] [PubMed] [Google Scholar]
- 24.Prather JC, Lobach DF, Goodwin LK, Hales JW, Hage ML, Hammond WE. Medical data mining: knowledge discovery in a clinical data warehouse. Proc AMIA Annu Fall Symp. 1997:101–5. [PMC free article] [PubMed] [Google Scholar]
- 25.Silver M, Sakata T, Su HC, Herman C, Dolins SB, O’Shea MJ. Case study: how to apply data mining techniques in a healthcare data warehouse. J Healthc Inf Manag. 2001 Summer;15(2):155–64. [PubMed] [Google Scholar]
- 26.Ebidia A, Mulder C, Tripp B, Morgan MW. Getting data out of the electronic patient record: critical steps in building a data warehouse for decision support. Proc AMIA Symp. 1999:745–9. [PMC free article] [PubMed] [Google Scholar]
- 27.Myers DL, Burke KC, Burke JD, Jr, Culp KS. An integrated data warehouse system: development, implementation, and early outcomes. Manag Care Interface. 2000 Mar;13(3):68–72. [PubMed] [Google Scholar]
- 28.Zhou X, Chen S, Liu B, et al. Development of traditional Chinese medicine clinical data warehouse for medical knowledge discovery and decision support. Artif Intell Med. 2010 Feb-Mar;(2–3):48. 139–52. doi: 10.1016/j.artmed.2009.07.012. Epub 2010 Feb 1. [DOI] [PubMed] [Google Scholar]
- 29.Cho IS, Haug PJ. The contribution of nursing data to the development of a predictive model for the detection of acute pancreatitis. Stud Health Technol Inform. 2006;122:139–42. [PubMed] [Google Scholar]
- 30.Cao H, Markatou M, Melton GB, Chiang MF, Hripcsak G. Mining a clinical data warehouse to discover disease-finding associations using co-occurrence statistics. AMIA Annu Symp Proc. 2005:106–10. [PMC free article] [PubMed] [Google Scholar]
- 31.Botsis T, Anagnostou VK, Hartvigsen G, Hripcsak G, Weng C. Developing a multivariable prognostic model for pancreatic endocrine tumors using the clinical data warehouse resources of a single institution. Appl Clin Inform. 2010;1(1):38–49. doi: 10.4338/ACI-2009-12-RA-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hollis J. Deploying an HMO’s data warehouse. Health Manag Technol. 1998 Jul;19(8):46–8. [PubMed] [Google Scholar]
- 33.Brooks CJ, Stephens JW, Price DE, et al. Use of a patient linked data warehouse to facilitate diabetes trial recruitment from primary care. Prim Care Diabetes. 2009 Nov;3(4):245–8. doi: 10.1016/j.pcd.2009.06.004. Epub 2009 Jul 14. [DOI] [PubMed] [Google Scholar]
- 34.Weng C, Bigger JT, Busacca L, Wilcox A, Getaneh A. Comparing the effectiveness of a clinical registry and a clinical data warehouse for supporting clinical trial recruitment: a case study. AMIA Annu Symp Proc. 2010 Nov 13;:867–71. 2010. [PMC free article] [PubMed] [Google Scholar]
- 35.Shah SP, Huang Y, Xu T, Yuen MM, Ling J, Ouellette BF. Atlas - a data warehouse for integrative bioinformatics. BMC Bioinformatics. 2005 Feb 21;6:34. doi: 10.1186/1471-2105-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen J, Zhao P, Massaro D, et al. The PEPR GeneChip data warehouse, and implementation of a dynamic time series query tool (SGQT) with graphical interface. The PEPR GeneChip data warehouse, and implementation of a dynamic time series query tool (SGQT) with graphical interface. [DOI] [PMC free article] [PubMed]
- 37.Sagynaliev E, Steinert R, Nestler G, Lippert H, Knoch M, Reymond MA. Web-based data warehouse on gene expression in human colorectal cancer. Proteomics. 2005 Aug;5(12):3066–78. doi: 10.1002/pmic.200402107. [DOI] [PubMed] [Google Scholar]
- 38.Györffy B, Lage H. A Web-based data warehouse on gene expression in human malignant melanoma. J Invest Dermatol. 2007 Feb;127(2):394–9. doi: 10.1038/sj.jid.5700543. Epub 2006 Aug 31. [DOI] [PubMed] [Google Scholar]
- 39.Hulse NC, Wood GM, Haug PJ, Williams MS. Deriving consumer-facing disease concepts for family health histories using multi-source sampling. J Biomed Inform. 2010 Oct;43(5):716–24. doi: 10.1016/j.jbi.2010.04.003. Epub 2010 Apr 9. [DOI] [PubMed] [Google Scholar]
- 40.Hristovski D, Rogac M, Markota M. Using data warehousing and OLAP in public health care. Proc AMIA Symp. 2000:369–73. [PMC free article] [PubMed] [Google Scholar]
- 41.Berndt DJ, Hevner AR, Studnicki J. CATCH/IT: a data warehouse to support comprehensive assessment for tracking community health. Proc AMIA Symp. 1998:250–4. [PMC free article] [PubMed] [Google Scholar]
- 42.Bahl V, McCreadie SR, Stevenson JG. Developing dashboards to measure and manage inpatient pharmacy costs. Am J Health Syst Pharm. 2007 Sep 1;64(17):1859–66. doi: 10.2146/ajhp060596. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Q, Matsumura Y, Teratani T, et al. The application of an institutional clinical data warehouse to the assessment of adverse drug reactions (ADRs). Evaluation of aminoglycoside and cephalosporin associated nephrotoxicity. Methods Inf Med. 2007;46(5):516–22. [PubMed] [Google Scholar]
- 44.Ferranti JM, Langman MK, Tanaka D, McCall J, Ahmad A. Bridging the gap: leveraging business intelligence tools in support of patient safety and financial effectiveness. J Am Med Inform Assoc. 2010;17:136e143. doi: 10.1136/jamia.2009.002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kamal J, Rogers P, Saltz J, Mekhjian Hx. Information warehouse as a tool to analyze Computerized Physician Order Entry order set utilization: opportunities for improvement. AMIA Annu Symp Proc. 2003:336–40. [PMC free article] [PubMed] [Google Scholar]
- 46.Breault JL, Goodall CR, Fos PJ. Data mining a diabetic data warehouse. Artif Intell Med. 2002 Sep-Oct;(1–2):26. 37–54. doi: 10.1016/s0933-3657(02)00051-9. [DOI] [PubMed] [Google Scholar]
- 47.Hota B, Lin M, Doherty JA, et al. Formulation of a model for automating infection surveillance: algorithmic detection of central-line associated bloodstream infection. J Am Med Inform Assoc. 2010;17:42–48. doi: 10.1197/jamia.M3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chase HS, Radhakrishnan J, Shirazian S, Rao MK, Vawdrey DK. Under-documentation of chronic kidney disease in the electronic health record in outpatients. J Am Med Inform Assoc. 2010;17:588e594. doi: 10.1136/jamia.2009.001396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hu H, Correll M, Kvecher L, et al. DW4TR: A Data Warehouse for Translational Research. J Biomed Inform. 2011 Dec;44(6):1004–19. doi: 10.1016/j.jbi.2011.08.003. Epub 2011 Aug 22. [DOI] [PubMed] [Google Scholar]
- 50.Bréant C, Borst F, Nkoulou R, Irion O, Geissbuhler A. Closing the loop: bringing decision support clinical data at the clinician desktop. Stud Health Technol Inform. 2007;129(Pt 2):890–4. [PubMed] [Google Scholar]
- 51.Evans RS, Lloyd JF, Abouzelof RH, Taylor CW. System-wide surveillance for clinical encounters by patients previously identified with MRSA and VRE. pp. 212–216. MEDINFO 2004. [PubMed]
- 52.Evans RS, Lloyd JF, Aston VT, et al. Computer surveillance of patients at high risk for and with venous thromboembolism. AMIA Annu Symp Proc. 2010 Nov 13;:217–21. [PMC free article] [PubMed] [Google Scholar]
- 53.Evans RS, Wallace CJ, Lloyd JF, et al. Rapid identification of hospitalized patients at high risk for MRSA carriage. J Am Med Inform Assoc. 2008;15:506–12. doi: 10.1197/jamia.M2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kutcher N, Koo S, Quiroz R, et al. Electronic alerts to prevent venous thromboembolism among hospitalized patients. N Engl J Med. 2005;352:969–77. doi: 10.1056/NEJMoa041533. [DOI] [PubMed] [Google Scholar]
- 55.Gilbreath RE. Provider-sponsored coordinated care organizations: designing systems for patient-centered care. Healthc Inf Manage. 1996 Fall;10(3):43–66. [PubMed] [Google Scholar]
- 56.Wisniewski MF, Kieszkowski P, Zagorski BM, Trick WE, Sommers M, Weinstein RA. Development of a clinical data warehouse for hospital infection control. J Am Med Inform Assoc. 2003 Sep-Oct;10(5):454–62. doi: 10.1197/jamia.M1299. Epub 2003 Jun 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weber GM, Murphy SN, McMurry AJ, et al. The Shared Health Research Information Network (SHRINE): A Prototype Federated Query Tool for Clinical Data Repositories. J Am Med Inform Assoc. 2009;16:624–630. doi: 10.1197/jamia.M3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horvath MM, Winfield S, Evans S, Slopek S, Shang H, Ferranti J. The DEDUCE Guided Query tool: providing simplified access to clinical data for research and quality improvement. J Biomed Inform. 2011 Apr;44(2):266–76. doi: 10.1016/j.jbi.2010.11.008. Epub 2010 Dec 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Duvall SL, Fraser AM, Kerber RA, Mineau GP, Thomas A. The impact of a growing minority population on identification of duplicate records in an enterprise data warehouse. Stud Health Technol Inform. 2010;160(Pt 2):1122–6. [PubMed] [Google Scholar]
- 60.Wade TD, Hum RC, Murphy JR. A Dimensional Bus model for integrating clinical and research data. J Am Med Inform Assoc. 2011 Dec;18(Suppl 1):i96, i102. doi: 10.1136/amiajnl-2011-000339. Epub 2011 Aug 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Botsis T, Hartvigsen G, Chen F, Weng C. Secondary Use of EHR: Data Quality Issues and Informatics Opportunities. AMIA Summits Transl Sci Proc. 2010 2010 Mar 1;:1–5. [PMC free article] [PubMed] [Google Scholar]
- 62.Shams K, Farishta M. Data warehousing: toward knowledge management. Top Health Inf Manage. 2001 Feb;21(3):24–32. [PubMed] [Google Scholar]
- 63.Waldo BH. Decision support and data warehousing tools boost competitive advantage. Nurs Econ. 1998 Mar-Apr;16(2):91–3. [PubMed] [Google Scholar]