Abstract
We present a study that developed and tested three query expansion methods for the retrieval of clinical documents. Finding relevant documents in a large clinical data warehouse is a challenging task. To address this issue, first, we implemented a synonym expansion strategy that used a few selected vocabularies. Second, we trained a topic model on a large set of clinical documents, which was then used to identify related terms for query expansion. Third, we obtained related terms from a large predicate database derived from Medline abstracts for query expansion. The three expansion methods were tested on a set of clinical notes. All three methods successfully achieved higher average recalls and average F-measures when compared with the baseline method. The average precisions and precision at 10, however, decreased with all expansions. Amongst the three expansion methods, the topic model-based method performed the best in terms of recall and F-measure.
Introduction
Free text medical records play a critical role in clinical practice. They also contain a wealth of information far beyond the immediate clinical use. For example, administrators may seek collection of performance measures while researchers may search the data for cohort identification (1, 2). At the same time, finding meaningful information can be a daunting task. The size of clinical data warehouses and repositories are growing exponentially and even a single patient record may contain dozens to hundreds of notes.
Simple text search is often not effective. For instance, a query for patients with “suicide ideation” in clinical notes returns many false positives because this phrase is often negated in the notes. The same query also results in many false negatives because it does not capture related phrases such as “suicidal tendencies” or “plans to commit suicide.”
Query expansion, i.e., adding additional terms to the original query (3), is a common information retrieval (IR) technique to improve the query performance. Typical sources of additional terms are thesauri or the retrieved documents themselves. For instance, retrieval feedback methods analyze the “best” returned documents, as determined by a ranking algorithm or by the user. Query log data, which records the search behavior of previous users, has also become a source for expansion terms especially in Web search engines (4, 5). Some systems automatically expand the original query. In interactive systems, related terms are often presented as suggestions to a user (6). The user may ignore or use the suggested terms to construct new queries. In biomedical informatics there have been a number of applications that have developed query expansion techniques for searching literature (7–12). On clinical notes, several studies have investigated different query expansion methods such as synonym expansion and relevance feedback with mixed results (13–17).
In this paper, we describe our experiments with three different query expansion strategies: synonym-based, topic model-based, and predication-based. The synonym-based expansion used a few selected UMLS source vocabularies and included lexical variants in the expanded queries. In the topic model-based expansion, we added related terms based on a topic-model trained on 100,000 clinical documents. The predication-based expansion made use of a large predication database extracted from medical literature by a natural language processing (NLP) system called SemRep. To the best of our knowledge, the topic model and SemRep predicate-based expansions are approaches that have not been previous explored, especially in the context of clinical text retrieval.
Background
The research on the IR of clinical notes has not been as extensive as the research on the IR of biomedical literature. Past studies in this area mainly clustered around several themes including query log analysis(18–20), temporal relationships (21–24), ontology/dictionary-based query expansion (25, 26), and bundled query sets(27). Particularly worth noting is that the TREC-Medical 2011 led to a set of new studies that tested state-of-the-art IR algorithms on free text notes (13–17). Based on the reported findings, it is clear that user queries require reformulation. However, automated expansions, concept-indexing or relevance feedback has not consistently improved query performance.
Synonym Expansion
In the biomedical domain, multiple studies have described the use of synonyms for query expansion. In 1996, Srinivasan published a study that reported improvements in MEDLINE retrieval performance using synonyms (9). Similarly, Aronson and Rindflesch proposed a UMLS concept-based query expansion method, which performed favorably against relevance feedback (11). On the other hand, a 2000 study by Hersh et al. showed that synonym expansion degraded the query performance rather improving it (10). In 2012, a study by Griffon et al. reported slightly increased recall when using UMLS synonyms and larger performance gains when using a more elaborate strategy focused on MeSH (28).
Several studies from TREC Medical Records 2011 (29) also applied synonym expansion techniques that did not result in consistent performance gain (13–17). UMLS was employed by most of the TREC studies. Given these lessons, our synonym expansion focused on a few UMLS source vocabularies.
Topic Modeling
Topic modeling is a relatively new technique, which analyzes the usage pattern of words or concepts in a corpus. A topic can be defined as a collection of words that occur together frequently and are related to a common subject. Each document is modeled as a mixture of topics. A corpus can be viewed as a matrix M of documents and topics as shown in Table 1.
Table 1.
Matrix of Documents and Topics
| Doc | C(Topic1) | C(Topic2) | C(Topic3) | … |
|---|---|---|---|---|
| 1 | 0.13 | 0.03 | 0.25 | |
| 2 | 0.02 | 0.86 | 0.01 | |
| 3 | 0.38 | 0.33 | 0.15 | |
| … |
The ith, jth entry of M is the estimate of the contribution of the jth topic to the ith document.
Topic modeling is a method that derives topics from a collection of text documents, which can be implemented as an unsupervised or unsupervised method (30). Steyvers and Griffiths (31), for instance, used this approach to analyze abstracts from the Proceedings of the National Academy of Sciences. Their results show that the extracted topics captured meaningful structure in the data, consistent with the class designations provided by the authors of the articles. In our experiment, we used the efficient, sampling-based implementations of Latent Dirichlet Allocation (LDA) (32, 33).
SemRep
The SemRep (34) is a NLP system, which is built on top of the MetaMap system (35). It parses the MEDLINE citations and extracts semantic relationships (predications) from them. The extraction is accomplished by interpreting linguistic structures. In order to effectively process these constructions, SemRep exploits underspecified syntactic analysis and structured domain knowledge from the UMLS. The predication database we used for this study contains predications extracted from ten years of MEDLINE (1999–2009) and has 25 million predications. Although these predicates were extracted from literature rather than clinical documents, we believe they capture some of the fundamental relations among concepts such as diabetes and insulin, which are relevant to clinical IR as well.
Methods
Synonym Expansion
We identified synonyms and lexical variants using the UMLS. For synonym identification, query terms were first mapped to UMLS concept CUIs, if possible, using MetaMap (35). We also developed a secondary concept matching strategy after some queries did not produce any mappings from MetaMap. MetaMap is effective at using variants to find matches, but can miss some exact matches. This can happen in cases of long phrases, excessive numbers of candidate permutations, and phrase terms that appear in MetaMap’s stop word list. It was for this reason that we made our own term-to-concept table. This table was constructed from the CUI and STR columns of the MRCONSO table, with all records for strings known to be irrelevant to our application removed.
After the matching concept CUI list was obtained, a related term list was built by reverse lookup in the same term-to-concept table. Lexical variants were generated using CUI strings from the MRCONSO table for the mapped CUIs. These CUI strings were used to generate inflections and derivations using LVG (NLM’s Lexical Variant Generation library from the SPECIALIST NLP Tools collection). The related term list and lexical variants were then combined to form the final list of synonyms. In concept identification and synonym lookup, we restricted the use of UMLS to several sources: SNOMED, MeSH, and ICD. This decision was made empirically after we observed many “incorrect” synonyms being added when we used all UMLS source vocabularies. Table 6 includes examples of the synonym expansion.
Table 6.
Examples of synonym-based expansion, topic model-based expansion and predication-based expansion
| Original Query | PTSD | Original Query | Diabetes |
|---|---|---|---|
| Synonym Expansion | ptsd, post traumatic stress syndrome, post traumatic stress syndromes, traumatic neuroses, post-traumatic stress disorder, disorder, post-traumatic stress, post-traumatic neuros, stress disorder, post traumatic, … (102 total) | Synonym Expansion | diabetes, maturity onset diabetes mellitus, non insulin dependent diabetes, disorder diabetes mellitus, diabetes mellitus type ii, non-insulin dependent diabetes, adult onset, diabetes mellitus, maturity-onset, maturity onset diabetes mellitus, … (214 total) |
| Topic model-based Expansion | ptsd, disorder, members, stress, citalopram, sleep, anxiety, trazodone, discussion, session, mood, month | Topic model-based Expansion | diabetes, insulin, glucose, strip, dm, units, diabetic, type, directed, coronary, metformin, ml |
| Predication-based Expansion | ptsd, treatments, cortisol, exposure therapy, cbt, trauma, dissociation, paroxetine, posttraumatic, stress disorder, sertraline | Predication-based Expansion | diabetes, insulin, glucose, streptozotocin, complications, obesity, oxidative stress, hypertension, interventions, hyperglycemia, drugs |
Topic Model Expansion
To identify related terms using topic modeling, we first applied the topic modeling methods to 100,000 clinical documents. 1,000 documents were selected from each of the 100 most frequent document types in VINCI. VINCI is an initiative to improve researchers’ access to Veteran Affairs (VA) data and to facilitate the analysis of those data while ensuring Veterans’ privacy and data security. VINCI hosts the VA’s Corporate Data Warehouse (CDW), which comprises major clinical and administrative domains, including notes, on 25 million patients. The 10 most frequent document types were shown in Table 2:
Table 2.
The 10 most frequent document types in VINCI
| ADDENDUM | NURSING INPATIENT NOTE |
| NURSING NOTE | PRIMARY CARE OUTPATIENT NOTE |
| UNTITLED | MENTAL HEALTH NOTE |
| PRIMARY CARE NOTE | PREVENTIVE MEDICINE NOTE |
| TELEPHONE ENCOUNTER NOTE | SOCIAL WORK NOTE |
We then employed the MALLET program to identify 1,000 topics (co-occurring word clusters). MALLET is a topic modeling software that includes an extremely fast and highly scalable implementation of Gibbs sampling, efficient methods for document-topic hyperparameter optimization, and tools for inferring topics for new documents given trained models (36). Table 3 shows 10 highly prevalent topics.
Table 3.
Example showing the 10 topics with the highest total weights.
| active, mg, mouth, tab, tablet, day, hcl, cap, medications, bedtime, needed, outpatient, capsule, tablets, times, status, supplies, hours, half |
| active, mouth, mg, tab, day, tablet, cap, medications, capsule, outpatient, hcl, tablets, bedtime, times, status, supplies, vitamin, needed, acid |
| ou, od, os, clear, flat, add, lens, dfe, iris, full, apd, macula, vision, cc, rx, mild, exam, cornea, ocular |
| active, tab, mg, mouth, tablet, day, medications, outpatient, half, status, supplies, bedtime, tablets, hcl, including, na, metoprolol, simvastatin, sa |
| active, po, tab, mg, prn, bid, inpatient, oral, cap, medications, qhs, status, hold, acetaminophen, supplies, qam, sa, tid, ec |
| active, mg, mouth, va, tab, medications, day, tablet, status, outpatient, cap, supplies, total, aspirin, ec, hcl, bedtime, including, half |
| mouth, mg, tablet, tab, day, needed, hcl, cap, tablets, bedtime, times, capsule, half, hours, morning, capsules, acetaminophen, ec, food |
| continue, cont, abd, cv, bp, bs, stable, po, ext, soft, rrr, heparin, code, nt, gen, labs, pulm, pending, nad |
| mouth, mg, tablet, tab, day, blood, pressure, half, heart, bedtime, morning, cholesterol, sa, tablets, capsule, lisinopril, simvastatin, hcl, cap |
| skin, risk, moisture, pressure, color, turgor, normal, assessment, problems, interventions, ulcer, temperature, braden, dry, protocol, limits, current, warm, score |
The following are a few examples showing the most salient topic for several document types (Table 4):
Table 4.
The most salient topic for several document types
| Note Type | Top Topic |
|---|---|
| Anesthesiology note | surgery op post pre operative surgical anesthesia mar date surgeon |
| Cardiology note | icd jul sn guidant rate lead interrogation shocks vf episodes |
| Dentistry note | dental dx teeth care visit completed status treatment primary dob |
| Emergency dept note | dose iv time med route medication rate date effective po |
| Gastroenterology attending note | hepatitis factor antibody risk ab hcv year pcr rna negativ |
| Home health note | days day remission affected area dry topical dt skin eye |
| Mental health attending note | wnl veteran zoloft time note good reports labs shirt wife |
Clearly not all topic words are clinically related to each other, but the majority of them have some semantic connections. To improve performance, we filtered out the top 200 words (e.g. test and medication) on the VINCI corpus from the related terms. To identify the most frequent words we passed the entire corpus through a tokenizer. We filtered out all non-word tokens, tallied the number of occurrences of each word token, and identified the most frequent word tokens. We empirically chose 200 as the cutoff after examining the word frequencies.
We determined which topics contained query terms and referred to them as matching topics. We then identified co-occurring terms within those matching topics. Within each topic, a term has a weight assigned by the topic modeling algorithm. This weight represents the term’s overall contribution to the topic. A query term may map to multiple topics and a co-occurring term may also appear in multiple matched topics. The terms from matching topics were ranked by summing their weights in all matching topics. The 10 highest-ranking were then used in the query expansion. Examples of the topic model-based expansion are shown in Table 6.
Predication-based Expansion
A database of predications was used to identify subjects where the object of the predication was one of the query terms. To create the predication database, the SemRep program was applied to 25 million MEDLINE abstracts. The following are a few rows from the predication database:
Table 5.
| Subject | Predicate | Object |
|---|---|---|
| Suicide | ISA | Psychiatric disorder |
| Human cardiac tissue | PART_OF | Cardiac |
| Beta-sitosterol | INHIBITS | Cholesterol |
| PF | LOCATION_OF | IL-1 receptor antagonist |
| Dexamethasone | AUGMENTS | GILZ |
| Calcineurin inhibitors | TREATS | Ulcerative colitis |
| Tumor necrosis factor-alpha | ASSOCIATED_WITH | Inflammatory diseases |
To improve performance, we also filtered out the top 200 words on the VINCI corpus from the subject terms.
Subjects of matching predications were ranked by frequency, and the 10 highest-ranking subjects were used as terms in the query expansion. Examples of the predication-based expansion are shown in Table 6.
Document Ranking
Retrieved documents were ranked by the sum of individual TF-IDF scores for the original query and expansion terms contained in each document. There was no difference in the calculations for the original query and the expanded queries. The term frequency TF for term t in document d is multiplied by inverse document frequency IDF and then used as the weight. For a document set D, IDF is calculated as:
TF-IDF scores were pre-calculated for each term in the document sets.
Evaluation
Two sample data sets were obtained from the VINCI database. We randomly selected two samples of 300 notes each. One sample was selected from patients with two or more encounters with ICD9 code (309.81) for post-traumatic stress disorder (PTSD). The other sample was selected from patients with two or more encounters with ICD9 codes (250.*) for diabetes mellitus (DM). No documents were shared between the two sample data sets and the 100,000 document set used to construct the topic model.
A clinician reviewed six PTSD (e.g. “suicide ideation”) and six DM topics (e.g. “change in DM symptoms”) on the 2 data sets respectively. These topics were chosen because of our current research projects on VINCI. The reviewer ranked the relevance of the documents to each topic using a 3-point Likert scale: 0: Irrelevant, 1: Possibly relevant, and 2:Defintiely relevant. The “Possibly relevant” category is needed because individual notes without context sometimes do not provide enough information to determine its relevance. For instance, a mere mention of changing the patient’s reactions to the stressful memories suggests that the note might be relevant to the topic of cognitive therapy, but does not offer sufficient evidence.
We applied all 3 query-expansion methods to the 2 data sets along with a baseline method without query expansion. The baseline queries were “PTSD” and “diabetes” (for the PTSD and DM data sets, respectively). A total of 19 queries were used in the testing. The document set TF-IDF scores were calculated separately for 300 PTSD documents and 300 Diabetes documents.
We calculated the typical IR performance measures. These were precision (positive predictive value), recall (sensitivity), and F-measure (the harmonic mean of precision and recall). We also calculated the precision for the top 10 ranked documents (P@10). The reason for choosing these performance measures is that there are two typical types of clinical text retrieval use cases: one is cohort-corpus identification, for which recall is more important, and the other is locating the most relevant notes on a topic in clinical care, for which the query results should ranked and precision is more important.
Results
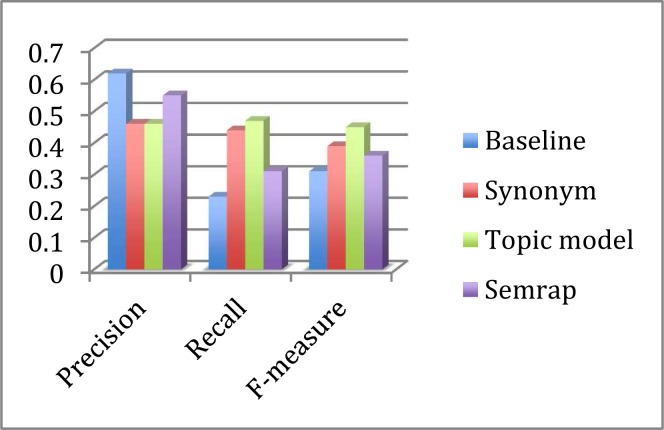
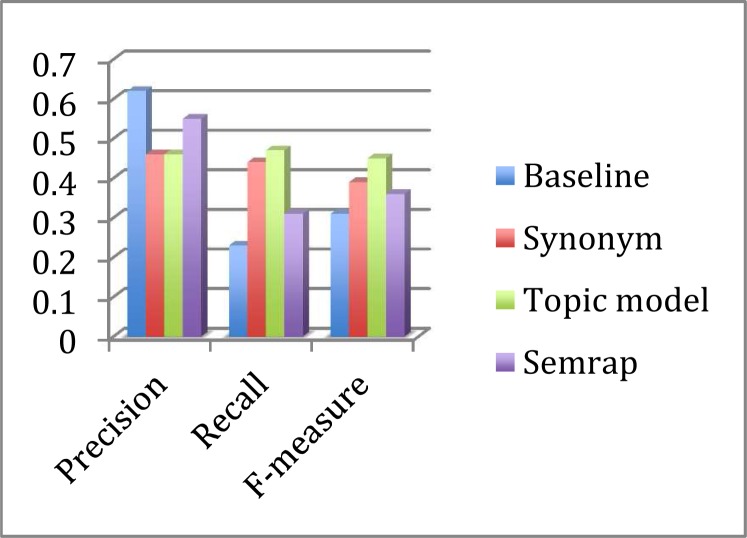
Compared to queries without expansion, all three query expansion methods resulted in clear improvements in the average F-measure (Table 7 and 8, Figure 1 and 2). (Please note that the average F-measure was not calculated using the average precision and recall, but calculated as the mean of the F-measure of each of the queries.) As all methods resulted in decreased precision, this overall performance improvement was driven by markedly increased recall. The topic-model expansion resulted in the best recall and F-measure, followed by synonym expansion and SemRep. SemRep demonstrated the least drop in precision.
Table 7.
Average performance measures for PTSD and MD treating possibly relevant as relevant
| Precision | Recall | F-measure | |
|---|---|---|---|
| Baseline | 0.62 | 0.23 | 0.31 |
| Synonym | 0.46 | 0.44 | 0.39 |
| Topic model | 0.46 | 0.47 | 0.45 |
| SemRep | 0.55 | 0.31 | 0.36 |
Table 8.
Average performance measures for PTSD and MD treating possibly relevant as irrelevant
| Precision | Recall | F-measure | |
|---|---|---|---|
| Baseline | 0.48 | 0.25 | 0.29 |
| Synonym | 0.34 | 0.47 | 0.35 |
| Topic model | 0.35 | 0.48 | 0.38 |
| SemRep | 0.43 | 0.33 | 0.34 |
Figure 1.
Treating Possibly Relevant as Relevant
Figure 2.
Treating Possibly Relevant as Irrelevant
Treating possibly relevant as relevant or irrelevant did not change the overall trend. The more lenient criteria (i.e. consider possibly relevant as relevant) did produce higher precision, lower recall, and higher F-measure (Table 7 and 8, Figure 1 and 2). Treating possibly relevant as relevant or irrelevant, the baseline recall measures were 23% and 25%. The synonym recall measures were 44% and 47%. The topic-model-expansion recall measures were from 47% and 48%.
Another observation we made was that the number of expansion terms does not correlate with the precision or recall. Synonym expansion added the most number of terms to the queries, sometimes many times more than the topic model and predicate expansion (Table 4). Although one might assume more terms leads to better recall, more synonyms are not sufficient. On the other hand, we had not expected synonyms to reduce precision in a significant way.
Since precision, recall and F-measures are set-based measures, they do not inform the rankings of documents. In the cohort or corpus identification context, document ranking is often not important because a user typically wants to obtain all relevant cases or a random subset of all relevant cases. Indeed, presenting top ranking results for review in the context of cohort or corpus identification may mislead users in terms of the quality of the documents being returned. Ranking, however, would be important to a doctor who wants to find the most relevant note relating to PTSD or a researcher who wants to annotate only the most relevant documents on PTSD.
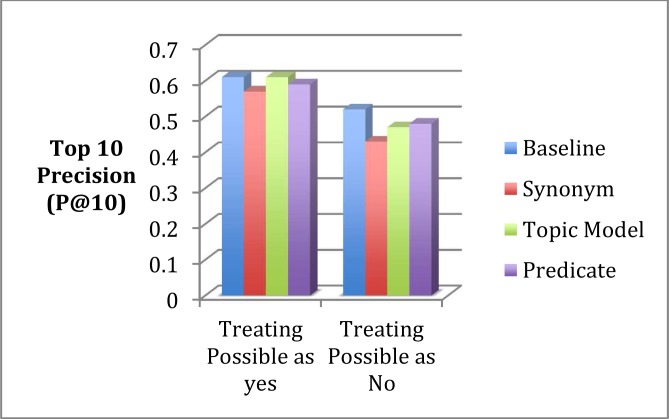
We expect users who care about precision to be particularly concerned about the precision of top n returns since human reviewers can only review a limited number of documents that are ranked near the top. We measured p@10 (i.e. precision at 10) and found that the decrease in P@10 with query expansion was much smaller than the overall precision (Table 9 and Figure 3). In the case of topic model-based expansion, its P@10 was the same as the baseline when we treated “possibly relevant” as “relevant”.
Table 9.
P@10 measures for Baseline vs. 3 query expansion methods
| Baseline | Synonym | Topic Model | Predicate | |
|---|---|---|---|---|
| Treating Possible as yes | 0.61 | 0.57 | 0.61 | 0.59 |
| Treating Possible as No | 0.52 | 0.43 | 0.47 | 0.48 |
Figure 3.
P@10 measures for Baseline vs. 3 query expansion methods
Discussion
The Query expansion methods were successful in improving the F-measure and recall, but reduced precision. While the improvements of 5 to 14 percentage points in F-measure were not trivial, the increases of 8 to 24 percentage points in recall are more significant. Amongst the three expansion methods, the topic model-based expansion performed the best in terms of recall and F-measure. The baseline method presented the highest precision and lowest recall.
Previous IR studies have not experimented with the topic model or SemRep predicate-based expansion. The findings of this study suggest that they are promising approaches that deserve further exploration. The synonym expansion has been implemented and assessed by a number of studies with mixed results. We attribute its effectiveness in our experiment to the selective use of source vocabularies from the UMLS. Indeed, we have observed much worse performance when the whole UMLS was used in synonym expansion.
The three expansion methods draw on very different knowledge and data sources: controlled vocabulary, clinical notes, and biomedical literature. As a result, the three sets of expanded terms had very small overlap and complement each other. They could also potentially be combined to achieve better performance.
Since our methods decreased false negatives at the cost of false positives, a logical follow-up question is how to improve the precision. For certain queries, using NLP techniques to filter out the negative and hypothetical assertions may be extremely helpful. The identification and interpretation of templated text as well as disambiguation may also improve the precision.
There are many clinical and research contexts where IR is needed. Some may require high precision and some may require high recall. In other cases, a balance between precision and recall is desired. For instance, a surveillance study to detect rare adverse events of drugs may want to maximize recall. A decision support application that aims at presenting physicians with the most relevant note on a topic (e.g. suicide attempt) would want to maximize precision. There is no one best method for all search goals. Method selection needs to be matched to the specific goal.
One limitation of the study is that we did not evaluate the expansion methods using the TREC Medical Records 2011 data. This is partially due to the timing of our study, which was carried out between the 2011 and 2012 TRECs. We are currently participating in the 2012 TREC. We also have a strong reason to test the methods on the VA data since our IR research is funded by the VA. The query topics in this study were real search tasks presented to us by VA researchers. Our document set is also relatively small in this study. In future studies, we plan to increase both the number of documents as well as the search topics.
Since this is our first attempt to use topic model and SemRep predicates in query expansion, improvements can be made to both methods. For instance, our topic model was built using words/tokens as features. We hypothesize conducting on the basis of term or concept features will improve the quality of the topic model. Most of the detailed information provided by SemRep (e.g. predicate type) was not used in the study. A more selective use of the SemRep could lead to better performance.
Another future step is to add these query expansion functions to the Voogo search engine we have developed for the VINCI data. This search engine already has a synonym expansion function which is slightly different from the method we describe here. Topic model and predicate-based modeling have also been added recently.
Acknowledgments
This work is funded by VA grants CHIR HIR 08-374 and VINCI HIR-08-204.
References
- 1.Hu H, Correll M, Kvecher L, Osmond M, Clark J, Bekhash A, et al. DW4TR: A Data Warehouse for Translational Research. J Biomed Inform. 2011;44(6):1004–19. doi: 10.1016/j.jbi.2011.08.003. Epub 2011/08/30. [DOI] [PubMed] [Google Scholar]
- 2.Horvath MM, Winfield S, Evans S, Slopek S, Shang H, Ferranti J. The DEDUCE Guided Query tool: providing simplified access to clinical data for research and quality improvement. J Biomed Inform. 2011;44(2):266–76. doi: 10.1016/j.jbi.2010.11.008. Epub 2010/12/07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frakes WB, Baeza-Yates R. Modern Information Retrieval. Addison Wesley; 1999. [Google Scholar]
- 4.Cui H, Wen J-R, Nie J-Y, Ma W-Y, editors. Query Expansion by Mining User Logs. IEEE Transaction on Knowledge and Data Engineering; 2003. [Google Scholar]
- 5.Stenmark D, editor. Query Expansion on a Corporate Intranet: Using LSI to Increase Precision in Explorative Search. Proceedings of the 38th Hawaii International Conference on System Sciences; 2005 03–06 Jan. [Google Scholar]
- 6.Harman D, editor. Towards interactive query expansion. 11th annual international ACM SIGIR conference on Research and development in information retrieval; May 1988; Grenoble, France: ACM Press; [Google Scholar]
- 7.Doszkocs TE. AID, an Associative Interactive Dictionary for Online Searching. Online Review. 1978;2(2):163–72. [Google Scholar]
- 8.Pollitt S. CANSEARCH: An expert systems approach to document retrieval. Information Processing & Management. 1987;23(2):119–38. [Google Scholar]
- 9.Srinivasan P. Retrieval feedback in MEDLINE. J Am Med Inform Assoc. 1996;3(2):157–67. doi: 10.1136/jamia.1996.96236284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hersh W, Price S, Donohoe L. Assessing thesaurus-based query expansion using the UMLS Metathesaurus. Proc AMIA Symp; 2000. pp. 344–8. [PMC free article] [PubMed] [Google Scholar]
- 11.Aronson AR, Rindflesch TC. Query expansion using the UMLS Metathesaurus. Proc AMIA Annu Fall Symp; 1997. pp. 485–9. [PMC free article] [PubMed] [Google Scholar]
- 12.Nenadic G, Mima H, Spasic I, Ananiadou S, Tsujii J. Terminology-driven literature mining and knowledge acquisition in biomedicine. Int J Med Inform. 2002;67(1–3):33–48. doi: 10.1016/s1386-5056(02)00055-2. [DOI] [PubMed] [Google Scholar]
- 13.Ozturkmenoglu O, Alpkocak A. DEMIR at TREC-Medical 2011: Power of Term Phrases in Medical Text Retrieval. 20th Anniversary of Text Retrieval Conference; 2011. [Google Scholar]
- 14.Mariam Daoud DK, Miao Jun, Huang Jimmy. York University at TREC 2011: Medical Records Track. Gaithersburg, Maryland, USA: 2011. TREC 2011. 2011. [Google Scholar]
- 15.Martijn Schuemie DT, Meij Edgar. DutchHatTrick: Semantic query modeling, ConText, section detection, and match score maximization. TREC 20112011.
- 16.Karimi S, Martinez D, Ghodke S, Zhang L, Suominen H, Cavedon L. Search for Medical Records:NICTA at TREC 2011 Medical Track. TREC 20112011.
- 17.Dinh D, Tamine L. IRIT at TREC 2011: evaluation of query expansion techniques for medical record retrieval. Text Retrieval Conference; Gaithersburg, Maryland, USA. 2011. TREC 2011. [Google Scholar]
- 18.Yang L, Mei Q, Zheng K, Hanauer DA. Query log analysis of an electronic health record search engine. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium; 2011. pp. 915–24. Epub 2011/12/24. [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy SN, Morgan MM, Barnett GO, Chueh HC. Optimizing healthcare research data warehouse design through past COSTAR query analysis. Proc AMIA Symp; 1999. pp. 892–6. [PMC free article] [PubMed] [Google Scholar]
- 20.Natarajan K, Stein D, Jain S, Elhadad N. An analysis of clinical queries in an electronic health record search utility. Int J Med Inform. 2010;79(7):515–22. doi: 10.1016/j.ijmedinf.2010.03.004. Epub 2010/04/27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klimov D, Shahar Y, Taieb-Maimon M. Intelligent interactive visual exploration of temporal associations among multiple time-oriented patient records. Methods Inf Med. 2009;48(3):254–62. doi: 10.3414/ME9227. Epub 2009/04/24. [DOI] [PubMed] [Google Scholar]
- 22.Bellika JG, Sue H, Bird L, Goodchild A, Hasvold T, Hartvigsen G. Properties of a federated epidemiology query system. Int J Med Inform. 2007;76(9):664–76. doi: 10.1016/j.ijmedinf.2006.05.040. Epub 2006/09/05. [DOI] [PubMed] [Google Scholar]
- 23.Deshpande AM, Brandt C, Nadkarni PM. Temporal query of attribute-value patient data: utilizing the constraints of clinical studies. Int J Med Inform. 2003;70(1):59–77. doi: 10.1016/s1386-5056(02)00183-1. Epub 2003/04/23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorda W, Gall W, Duftschmid G. Clinical data retrieval: 25 years of temporal query management at the University of Vienna Medical School. Methods Inf Med. 2002;41(2):89–97. Epub 2002/06/14. [PubMed] [Google Scholar]
- 25.Mabotuwana T, Warren J. An ontology-based approach to enhance querying capabilities of general practice medicine for better management of hypertension. Artif Intell Med. 2009;47(2):87–103. doi: 10.1016/j.artmed.2009.07.001. Epub 2009/08/28. [DOI] [PubMed] [Google Scholar]
- 26.Schulz S, Daumke P, Fischer P, Muller M. Evaluation of a document search engine in a clinical department system. AMIA Annu Symp Proc; 2008. pp. 647–51. Epub 2008/11/13. [PMC free article] [PubMed] [Google Scholar]
- 27.Hanauer DA. EMERSE: The Electronic Medical Record Search Engine. AMIA Annu Symp Proc; 2006. p. 941. Epub 2007/01/24. [PMC free article] [PubMed] [Google Scholar]
- 28.Griffon N, Chebil W, Rollin L, Kerdelhue G, Thirion B, Gehanno JF, et al. Performance evaluation of Unified Medical Language System(R)’s synonyms expansion to query PubMed. BMC medical informatics and decision making. 2012;12(1):12. doi: 10.1186/1472-6947-12-12. Epub 2012/03/02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.NIST Guidelines for the 2011 TREC Medical Records Track. 2011. http://wwwnlpir.nist.gov/projects/trecmed/2011/
- 30.Steyvers M, Griffiths T. Probabilistic topic models. Lawrence Erlbaum; 2007. [Google Scholar]
- 31.Griffiths TL, Steyvers M. Finding scientific topics. National Acad Sciences. 2004:5228–35. doi: 10.1073/pnas.0307752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blei DM, Ng AY, Jordan MI. Latent Dirichlet Allocation. Journal of Machine Learning Research. 2003;3:993–1022. [Google Scholar]
- 33.Smith KB, Ellis SA. Standardisation of a procedure for quantifying surface antigens by indirect immunofluorescence. Journal of immunological methods. 1999;228(1–2):29–36. doi: 10.1016/s0022-1759(99)00087-3. [DOI] [PubMed] [Google Scholar]
- 34.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J Biomed Inform. 2003;36(6):462–77. doi: 10.1016/j.jbi.2003.11.003. Epub 2004/02/05. [DOI] [PubMed] [Google Scholar]
- 35.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 36.McCallum AK. MALLET: A Machine Learning for Language Toolkit. http://malletcsumassedu2002.