Abstract
We present the construction of three annotated corpora to serve as gold standards for medical natural language processing (NLP) tasks. Clinical notes from the medical record, clinical trial announcements, and FDA drug labels are annotated. We report high inter-annotator agreements (overall F-measures between 0.8467 and 0.9176) for the annotation of Personal Health Information (PHI) elements for a de-identification task and of medications, diseases/disorders, and signs/symptoms for information extraction (IE) task. The annotated corpora of clinical trials and FDA labels will be publicly released and to facilitate translational NLP tasks that require cross-corpora interoperability (e.g. clinical trial eligibility screening) their annotation schemas are aligned with a large scale, NIH-funded clinical text annotation project.
Introduction
Our long-term goal is to develop and publicly release gold standard corpora for medical Natural Language Processing (NLP) tasks and align their annotation schemas with the Strategic Health IT Advanced Research Projects (SHARP) Research Focus Area 4 - Secondary Use of EHR Data’s annotations1. A variety of textual documents written in natural language exist in the medical domain. Accessing and using the information contained in those documents is the goal of medical NLP systems. In order to evaluate the robustness of such systems, high quality gold standards are required. That is, corpora of texts need to be manually annotated with the instances relevant to the specific NLP tasks. In this paper, we present our annotation process for building gold standard corpora from three sources of documents for an NIH funded grant and internally supported patient safety projects2. Clinical notes from the EHR, clinical trial announcements and FDA drug labels were annotated. We describe the annotation of Protected Health Information (PHI)3 elements and the annotation of medications, diseases/disorders, and signs/symptoms. Intended applications of the corpora include automated de-identification of clinical narratives, semi-automated eligibility screening for clinical trial enrollment, and automated detection of adverse drug reactions. We intend to publicly release the annotated gold standard corpora for the clinical trial announcements and the FDA drug labels.
Background
Corpora annotated for PHI elements: Ideally, large-scale corpora including diverse document types of the EHRs should be built for evaluating de-identification systems. However most of the corpora used for measuring the performance of existing systems are composed of only a few document types, such as discharge summaries4, 5, pathology reports6, 7, 8, nursing progress notes5, outpatient follow-up notes9, or medical message boards10. Very few systems have been evaluated on more heterogeneous corpora of documents11, 12. Furthermore, not all of the 18 PHI classes were annotated in previous studies, and important items are often ignored, in particular ages >896, 11, geographical locations6, 11, 13, institution and contact information6, 11, 13, date and IDs6, 13. In numerous cases, because of the sensitivity of PHI data the de-identification performance is measured on synthetically generated PHI (i.e. text manually de-identified and re-identified with fake PHI elements).
Corpora annotated for medical entities: Several studies have built corpora of clinical notes annotated with medical entities to evaluate the performance of NLP systems. Most of them concentrated on a specific entity type such as disorders14 or medications15, on a specific topic such as inflammatory bowel disease16, or on a specific type of notes such as ED reports17 or discharge summaries15. A few reported annotation effort on a larger scale, including multiple entities as well as relations between them18, 19. Due to privacy issues, these resources are not publicly available, with the exception of the corpora from the i2b2 NLP challenges20, which have been manually de-identified and can be accessed with user agreements. Most clinical efforts have been focused on EHR notes, while other types of medical corpora have been much less frequently annotated. Clinical trial announcements have been annotated in a few occasions only: for a preliminary study21 on using Amazon Mechanical Turk (by the senior author of the current work), and for detecting temporal constraints in a sample of 100 eligibility criteria sentences22. Tu et al., annotated 1,000 eligibility criteria sentences but their scope did not include gold standard development for NLP tasks but presenting a practical method of formal eligibility criteria representation23. To our knowledge, no study has yet explored the annotation of FDA drug labels.
Material and Methods
Annotation tasks
The first task consists of annotating PHI elements in clinical notes to build and evaluate a system for the de-identification of clinical narrative text at Cincinnati Children’s Hospital Medical Center (CCHMC).
The second task consists of annotating medical named entities in clinical trial announcements from ClinicalTrials.gov24, clinical notes and FDA drug labels from the DailyMed web site25. Our long term objectives with the corpora are (i) clinical trial eligibility screening, i.e. linking the information in clinical trial announcements with patient data from clinical notes in order to help patient enrollment in clinical trials; and (ii) mining adverse drug reactions based on the information contained in FDA drug labels. Aim (i) requires the annotation of both clinical trial announcements and clinical notes against the same annotation schema. Aim (ii) requires annotating FDA drug labels and notes against the same schema. We focused on a subset of medical entities as a starting set to achieve our objectives and defined two annotation subtasks: (a) Medications (and their attributes): this subtask is relevant to aim (i) and has been accomplished on both clinical notes and clinical trial announcements. (b) Disease/disorders and sign/symptoms: this subtask is relevant to both aims, and has currently been accomplished on clinical trial announcements and FDA drug labels.
Corpora
To represent the variety of notes available in the CCHMC EHR, 3,503 clinical notes were selected by stratified random sampling from five million notes composed by CCHMC clinicians during 2010. The notes can be classified into three broad categories: Labeled (created within the EHR system and includes division); Unlabeled (created within the EHR but with no division information); External (written outside of the EHR system (e.g., on a Radiology system and transferred into the EHR)). Table 1 presents the descriptive statistics for each note category in the 3,503 notes sampled for PHI annotation. The study set had the same proportional distribution of the three categories as the five million notes. Table 1 gives details on the different note types within the Labeled category. We included note types in the random sampling only if the number of notes written in that category exceeded the subjective limit of 800 during the previous 12-month period. We oversampled Discharge Summaries because of their richness in de-identification information12 and some of the notes that were less frequent but exceeded the 800-note limit to have at least 20 notes for each type included in the study set. All 3,503 sampled notes were annotated for PHI elements (task 1). Only the 1,655 Labeled notes were used for the annotation of medications (task 2.a).
Table 1.
Statistics of the PHI-annotated clinical corpus
| Documents | Tokens | Non-punctuation tokens | |
|---|---|---|---|
| All Notes | 3,503 | 1,068,901 | 877,665 |
| Unlabeled Notes | 649 | 290,882 | 234,753 |
| External Notes | 1,199 | 88,170 | 75,653 |
| Labeled Notes | 1,655 | 689,849 | 567,259 |
| Asthma Action Plan | 40 | 16,575 | 14,624 |
| Brief OpNote | 40 | 6,504 | 5,019 |
| Communication Body | 40 | 27,809 | 24,057 |
| Consult Note | 40 | 23,241 | 18,699 |
| DC Summaries | 400 | 262,570 | 215,874 |
| ED Medical Student | 40 | 13,456 | 10,957 |
| ED Notes | 218 | 8,202 | 7,118 |
| ED Provider Notes | 111 | 41,058 | 32,693 |
| ED Provider Reassessment | 24 | 2,716 | 2,229 |
| H&P | 20 | 20,148 | 15,771 |
| Med Student | 20 | 26,480 | 20,847 |
| Operative Report | 20 | 11,381 | 9,926 |
| OR Nursing | 20 | 442 | 370 |
| Patient Instructions | 33 | 4,760 | 4,110 |
| Pharmacy Note | 20 | 9,797 | 7,610 |
| Plan of Care Note | 75 | 16,735 | 14,109 |
| Pre-Op Evaluation | 20 | 6,491 | 5,098 |
| Procedure Note | 20 | 9,082 | 7,897 |
| Inpatient Progress Note | 179 | 84,928 | 71,511 |
| Outpatient Progress Note | 128 | 88,678 | 71,036 |
| Referral | 20 | 596 | 494 |
| Telephone Encounter | 127 | 8,200 | 7,210 |
Clinical trial announcements (CTA) were downloaded from the clinicaltrials.gov website24, which resulted in a total 105,598 documents (as of March 2011). We randomly selected a subset of documents for the annotation of medications (task 2.a) and of diseases/disorders and signs/symptoms (task 2.b). We annotated only the eligibility criteria sections of the clinical trial announcements. Table 2 shows the descriptive statistics of the CTA corpora for each annotation task (as task 2.b is still an on-going process, the annotated CTA corpus for this task is only a subset of the CTA corpus for task 2.a). Eventually, the entire CTA corpus will be annotated for medications, diseases/diagnoses, signs/symptoms, procedures, labs, anatomical sites, temporal and negation identifiers.
Table 2.
Statistics for Clinical Trial Announcements (CTA) and FDA Drug Labels (FDA)
| CTA for task 2.a | CTA for task 2.b | FDA | |
|---|---|---|---|
| Documents | 3,000 | 241 | 52 |
| Tokens | 647,246 | 51,793 | 96,675 |
| Non-punctuation tokens | 633,833 | 49,076 | 80,706 |
FDA drug labels were downloaded from the dailymed website25. A sample of labels for both prescriptions drugs and over-the-counter drugs was randomly selected. The prescription labels were selected from the top 200 most frequent drugs26. We annotated only the following sections of the labels (i.e. sections likely to mention medical conditions): Overdosage, Warnings, Warnings and Precautions, Contraindications, Adverse Reactions, Boxed Warning, Indications and Usage, and Precautions. Table 2 shows the size of the FDA corpus. Although the number of annotated documents is small compared to the other corpora, the number of tokens is high (1859 tokens per document on average, which is more than eight times the density of tokens in the CTAs).
CTAs and FDA drug labels are in the public domain, so we will release all our annotated CTA and FDA drug label corpora when the annotation is finalized. We plan to release the CTA corpus in September 2013 and the FDA label corpus in December 2014.
Annotation guidelines
We developed our own guidelines for the annotation of PHI elements. For medications, disease/disorders, and signs/symptoms we followed the annotation guidelines and schema from the SHARPn project27 (those are also consistent with the ShARe (Shared Annotated Resources) project28 guidelines for annotating disorders). New rules and a more detailed list of examples based on the experience of our corpora are being added to the SHARPn guidelines, as necessary. However, our goal is to provide as seamless interoperability with the SHARPn annotations as possible to increase the likelihood that the SHARPn clinical and our medical corpora can be used in cross-domain projects (e.g. computerized clinical trial eligibility screening or health care quality improvement).
Task 1. PHI elements:
We defined 12 classes of PHI, derived from the 18 HIPAA categories (regrouping some of them, refining others):
Name: any first name, middle name, last name or combination of those.
Date: date (e.g. “12/29/2005”, “September 15th”) excluding years occurring on their own (e.g. “in 2005”)
Age: age of the patient (any age, not restricted to ages >98 as specified by HIPAA)
Email
Initials: initials of a person
Institution: hospital names and other organizations
IPAddress: includes IP addresses and URLs
Location: geographical locations such as address, city, state, etc.
Phone number: phone and fax numbers
Social security: social security number
IDnum: any identification number such as medical record number, patient ID, etc.
Other: other identifiers not belonging to any specified category (e.g. internal locations inside a hospital).
Task 2.a Medications:
Medication entities were divided in two subtypes:
Medication name corresponds to names of drugs or substances used for treatment.
Medication type is a more general class for entities that do not expressly name a drug but still refer to a medication treatment. This includes drug classes (e.g. “antiobiotics”), types of drug therapy (e.g. “chemotherapy”) and any general references to medications (e.g. “this drug”)
In addition to annotating medication entities, we also annotated attributes linked to those medications (based on the SHARPn guidelines27). Those attributes are divided into 9 classes:
Date: the date associated with the medication. It can be a real date (e.g. “lasix was prescribed on 09/15/2011”) or a relative date (e.g. “day 1 of chemotherapy”)
Strength: the strength number and unit of the prescribed drug (e.g. “aspirin 500 mg”)
Dosage: how many of each drug the patient is taking (e.g. “take 2 pills daily”). We also included references to what type of dose it is (e.g. “high dose of paclitaxel”)
Frequency: how frequently is the drug taken, as well as the time of day (e.g. “1 pill BID”, “2 at bedtime”)
Duration: how long the patient is expected to take the drug or has been taking a drug (e.g. “for 2 weeks”)
Route: Route or method of the medication (e.g. “IV”, “oral”, “by mouth”)
Form: Form of the medication (e.g. “tablet”, “capsule”, “cream”, “liquid”)
Status change: Status refers to whether the medication is currently being taken or not, or is being changed or not (e.g. “started”, “increased”, “stopped”).
Modifier: qualifiers occurring before the drug name (adjectives, quantifiers, pronouns, etc.) that do not belong to any other specified attribute class (e.g. “those drugs”, “concomitant medications”)
All entities were annotated even when they did not refer to a medication taken by a patient (e.g. “penicillin” in “allergy to penicillin”). Discontinuous annotations were allowed (e.g. “Vitamin…D” in “Vitamin C and D”).
Task 2.b Diseases/disorders and signs/symptoms:
Following the SHARPn schema, annotation of disease/disorder and sign/symptom entities is based on the SNOMED CT terminology standard. Annotated entities should be SNOMED CT concepts (or close synonyms) with the following UMLS semantic types:
Diseases/disorders = Congenital Abnormality, Acquired Abnormality, Injury or Poisoning, Pathologic Function, Disease or Syndrome, Mental or Behavioral Dysfunction, Cell or Molecular Dysfunction, Experimental Model of Disease, Anatomical Abnormality, Neoplastic Process
Signs/symptoms = Sign or Symptom
Additionally, (following the SHARPn guidelines) concepts with the UMLS semantic type Finding were also annotated when annotators judged that the concepts corresponded to signs/symptoms or diseases/disorders.
Annotators were instructed to annotate only the most specific mentions of SNOMED CT concepts. That is, in the sentence “the patient has chronic pain”, “chronic pain” is to be annotated because it corresponds to a SNOMED CT concept ([82423001] Chronic pain, UMLS CUI [C0150055]). “Pain” on its own is less specific and should not be annotated alone in that sentence. The goal is to annotate only those entities that directly correspond to SNOMED CT concepts. However, acknowledging the shortcomings of existing terminologies, annotators were allowed to make exceptions and annotate entities that could not be found in SNOMED CT when they clearly belonged to a disease/disorder or sign/symptom category. The benefit of using an existing terminology is that it provides a standardized way of annotating and that entities are normalized to a knowledge source which is necessary to remove ambiguities and use the information in subsequent computational tasks such as decision support.
All entities were annotated even when they did not refer to the condition of a patient in the specific sentence but in other context they would be interpreted as a medical condition (e.g. “influenza” in “vaccination against influenza”). An attribute is added later to indicate if the condition belongs to a patient. Discontinuous annotations were allowed (e.g. in “muscle…weakness” in “muscle tenderness and weakness”).
Annotators
All annotation tasks were performed by two annotators. Annotation of PHI elements, which did not require any medical knowledge, was performed by two non-clinicians (with Bachelor degrees). Medications, which are often easy to identify for non-clinicians, were also annotated by non-clinicians. Diseases/disorders and signs/symptoms are more difficult to identify and distinguish. Only one of our annotators for this task had a clinical background (BSN, RN). We hypothesized that with sufficient training and the help of existing medical terminologies, we could reach a good level of agreement between the two annotators. Chapman et al.29 also used both clinician and non-clinician annotators and found that the annotation was high quality but non-clinicians needed longer training time.
Software
The Protégé plug-in Knowtator30 was used for annotating our corpora. Knowtator was installed on a Linux server for server side annotation of the corpora. We did not store any document on laptops or desktop computer because of HIPAA and IRB requirements. We also tested a remote access configuration to allow annotators to work from home. Knowator was remotely accessed through the CITRIX31 client software (from home) and NoMachine’s NX32 client (from the office), which both provided secure connections and good Graphical User Interface performance for Knowtator. For task 2.b (annotation of diseases/disorders and signs/symptoms), annotators also used a SNOMED CT browser (the UMLS Terminology Services SNOMED CT browser33 to look up terms found in documents.
Annotation process
We followed the same annotation process for all annotations tasks. All documents were double-annotated by two annotators, and disagreements were resolved during “consensus sessions”. Annotation started with an initial training period (of variable length, depending on the task and its level of difficulty) during which annotators familiarized themselves with the annotation guidelines, the software and the type of documents to be annotated. During that time, inter-annotator agreement was computed at the end of each day and a consensus session to resolve disagreements was held, supervised by an NLP researcher. Annotation guidelines were also regularly updated according to issues raised during the consensus sessions: annotation rules were clarified and additional examples were included in the guide. After the training period, i.e. when a high level of inter-annotator agreement was reached, guidelines were stabilized for the most part, and consensus sessions were held on a less frequent basis, every 3–4 days or so (depending on the task), and with less supervision (e.g. supervision for cases where agreement was difficult to reach). An exception was made in the case of the annotation of diseases/disorders and sign/symptoms, for which consensus sessions were held frequently even after the training period, because of the difficulty of the task. For task 2.a, which included not only the annotation of entities but also of their attributes, we divided the annotation process into two steps: first the annotation of medication entities (medication names and medication types) and second the annotation of attributes based on the consolidated annotated set from the first step.
Measures of inter-annotator agreement
We consider and discuss two commonly used measures of inter-annotator agreement (IAA).
Cohen’s Kappa. Cohen’s kappa coefficient34 (κ) is defined using observed agreement (Ao) and agreement expected by chance (Ae): . The observed agreement Ao (or percentage of agreement) is the number of instances the annotators agree on divided by the total number of instances. The observed agreement Ae is based on the probability of the two annotators agreeing on any given category, which is the product of the chance of each annotator assigning an instance to that category. Ae is then the sum of this product over all categories. The chance of each annotator assigning a given category is estimated by looking at the observed distribution.
F-measure. Agreement between annotators can also be measured using standard performance measures in information retrieval and NLP35, 36: precision, recall, and more particularly the F-measure. Precision (or positive predictive value) is the number of correct answers divided by the total number of answers a system has predicted. Recall (or sensitivity) is the number of correct answers divided by the total number of answers in the gold standard. F-measure is the harmonic mean of precision and recall and is written as (with P=precision, R=recall, and β usually equal to 1): F=((1+β2)P*R)/β2*(P+R). In the case of IAA, we treat the annotations of one annotator as the reference and the annotations of the other annotator as the answers of a system. Switching the roles of the two annotators does not change the value of the F-measure so it does not matter which one plays the role of the gold standard35, 36.
Kappa vs. F-measure. Cohen’s kappa is often the standard measure of inter-annotator agreement used for classification tasks. However, as pointed out by Hripcsak36, kappa is not the most appropriate measure for named entity annotation in textual documents. Indeed kappa requires the number of negative cases to be computed, which is unknown in the case of named entities. Named entities are sequences of words, and there is no pre-existing fixed number of items to consider when annotating a text. A simple solution is to consider individual tokens as the items to be marked, and to compute a “token-level” kappa37, 38. However this has two major drawbacks. First evaluating IAA like this does not properly reflect the annotation task, because annotators do not label tokens individually, but look at sequences of one or more tokens. So the information of whether the annotators annotated the same sequence of tokens as one named entity will be lost when remaining at the level of individual tokens. Second, the number of negative cases (all tokens that have not been annotated) will be much larger than the number of positive cases, and kappa will be calculated on a very imbalanced data. It has been observed that in this case the value of kappa is close to the value of positive specific agreement39 (which is equivalent to the F-measure). Other proposed solutions40 include considering only noun phrases, considering all possible n-grams in a text (sequence of n tokens) or considering only items by one or two of the annotators, although none of those are fully accurate40. For this reason, the F-measure, which does not require the number of negative cases, is usually recognized as a better way to measure inter-annotator agreement for named entity annotation tasks. In this paper, we compute the F-measure as the main measure of inter-annotator agreement, and provide in addition the “token-level” kappa. We present IAA results computed during the training period, as well the IAA computed after this initial training period.
Results
Descriptive statistics
Table 3 shows the number of annotations in each corpus, for each annotation task. The most frequent PHI categories are Date and Name. Email, IPAddress, Initials and Social Security are very rare. Medication names are more frequent in clinical notes than in clinical trial announcements, which contain much more medication types. Consequently, attributes are more numerous in clinical notes, except for modifier, a general attribute that most often applies to medication types. Annotation of PHI elements took 40 days (∼ 10.9 documents per hour). Annotation of medication entities took 21 days for CTAs (∼ 17.9 documents per hour) and 10 days for clinical notes (∼ 20.7 documents per hour). Annotation of medication attributes took 19 days for CTAs (∼ 19.7 documents per hour) and 11 days for clinical notes (∼ 18.8 documents). Annotation of disease/disorder and sign/symptoms was the most time-consuming, especially on FDA drug labels which are very long documents with a high density of entities. It took 10 days for FDA drug labels (∼ of 0.65 document per hour) and 5 days for CTAs (∼ 6 documents per hour).
Table 3.
Number of annotations for each task
| Task 1 (PHI elements) | Task 2.a (medications) | Task 2.b (diseases/disorders and signs/symptoms) | |||||
|---|---|---|---|---|---|---|---|
| Entity Type | Entity Type | CTA | Clinical notes | Entity Type | FDA | CTA | |
| Age | 2,109 | Medication name | 9,968 | 12,517 | Disease_Disorder | 5,842 | 3,601 |
| Date | 13,060 | Medication_Type | 11,789 | 4,275 | Sign_symptom | 2,782 | 163 |
| 14 | Date | 16 | 121 | ||||
| IDnum | 1,117 | Dosage | 645 | 1,884 | |||
| Institution | 1,994 | Duration | 644 | 619 | |||
| IPAddress | 16 | Form | 482 | 4,413 | |||
| Initials | 10 | Frequency | 381 | 4,553 | |||
| Location | 396 | Route | 894 | 3,235 | |||
| Name | 7,776 | Status change | 598 | 2,983 | |||
| Other | 3,446 | Strength | 409 | 6,484 | |||
| Phone Number | 876 | Modifier | 5,827 | 1,770 | |||
| Social_Security | 1 | ||||||
| All classes | 30,815 | All classes | 31,653 | 42,854 | All classes | 8,624 | 3,764 |
Inter-Annotator Agreement (IAA)
Table 4 show IAA results during the training period for each task (training for task 2.a and task 2.b was performed on clinical trial announcements). Agreement is fair for PHI categories (0.7694 overall IAA): it is already high for Date and Phone, but especially low for IDnum and Location. It is lower for medications (0.7383 overall IAA): medication names have a good agreement, but most attributes have low or medium agreement. Agreement is the lowest for task 2.b (0.7079 overall IAA), particularly for signs/symptoms.
Table 4.
Inter-Annotator Agreement (IAA) during training period for each annotation task
| Task 1 (PHI elements) | Task 2.a (medications) | Task 2.b (disease/disorder and sign/symptom) | ||||||
|---|---|---|---|---|---|---|---|---|
| Entity Type | IAA | Kappa | Entity Type | IAA | Kappa | Entity Type | IAA | Kappa |
| Age | 0.6408 | 0.7871 | Medication name | 0.8459 | 0.8751 | Disease_Disorder | 0.7365 | 0.8389 |
| Date | 0.8799 | 0.9621 | Medication_Type | 0.6927 | 0.8152 | Sign_symptom | 0.4417 | 0.4736 |
| 0 | 0 | Date | 0.0000 | −0.0001 | ||||
| IDnum | 0.2762 | 0.3092 | Dosage | 0.2569 | 0.2973 | |||
| Institution | 0.7834 | 0.9424 | Duration | 0.3913 | 0.4961 | |||
| IPAddress | _ | _ | Form | 0.4634 | 0.3198 | |||
| Initials | _ | _ | Frequency | 0.3103 | 0.5845 | |||
| Location | 0.3520 | 0.8462 | Route | 0.7467 | 0.7091 | |||
| Name | 0.7768 | 0.9684 | Status change | 0.0952 | −9.27e−05 | |||
| Other | 0.6635 | 0.6963 | Strength | 0.5000 | 0.7161 | |||
| Phone Number | 0.9036 | 0.9825 | Modifier | 0.7383 | 0.6936 | |||
| Social_Security | _ | _ | ||||||
| All classes | 0.7694 | 0.9054 | All classes | 0.7383 | 0.8298 | All classes | 0.7079 | 0.8188 |
IAA for PHI elements after the training period is high (0.9176 overall IAA, see Table 5), for all PHI types except for Other, Initials and Social Security Numbers (SSN). Other is the most ambiguous category, and Initials and SSN are very rare in the corpus so even missing one will bring down the agreement. IAA for medications (Table 5) is high for both corpora (overall IAA of 0.8999 for CTAs and of 0.8965 for clinical notes) with only a couple attributes having low or medium agreement (Date and Duration). IAA for task 2.b (Table 6) is lower than for the two previous tasks, but is still at a good level for both corpora (overall agreement of 0.8875 for CTAs and of 0.8467 for FDA drug labels). In all cases, we can observe that the Kappa value is close to and sometimes even higher than the F-measure (i.e. when the number of tokens that constitute the named entity is high for many named entities then agreeing on the majority of the entities will inflate the Kappa value). Thus we agree with previous studies that there is no advantage in using Kappa when annotating entities in texts. However, using Kappa in addition to the F-measure adds useful token-level performance evaluation although token-level F-measure can serve the same purpose.
Table 5.
Inter-annotator agreement after the training period for task 1 and task 2.a
| Task 1 (PHI elements) | Task 2.a (medications) | ||||||
|---|---|---|---|---|---|---|---|
| CTA | Clinical notes | ||||||
| Entity type | F-measure | Kappa | Entity type | F-measure | Kappa | F-measure | Kappa |
| Age | 0.9151 | 0.8618 | Medication name | 0.9415 | 0.9325 | 0.9001 | 0.9519 |
| Date | 0.9595 | 0.9679 | Medication_Type | 0.8822 | 0.8938 | 0.8865 | 0.8987 |
| 0.8571 | 1 | Date | 0.5185 | 0.6769 | 0.2252 | 0.2421 | |
| IDnum | 0.9389 | 0.9499 | Dosage | 0.7972 | 0.8098 | 0.8386 | 0.8124 |
| Institution | 0.9474 | 0.9474 | Duration | 0.6303 | 0.6732 | 0.6833 | 0.7126 |
| IPAddress | 0.1739 | 0.0145 | Form | 0.8624 | 0.8585 | 0.8810 | 0.8726 |
| Initials | 0.8696 | 0.9249 | Frequency | 0.8889 | 0.8865 | 0.9468 | 0.9003 |
| Location | 0.8183 | 0.9583 | Route | 0.8980 | 0.9126 | 0.8815 | 0.8835 |
| Name | 0.9534 | 0.9596 | Status change | 0.7053 | 0.6993 | 0.7636 | 0.7760 |
| Other | 0.6860 | 0.7327 | Strength | 0.8460 | 0.8901 | 0.9250 | 0.9220 |
| Phone Number | 0.9546 | 0.9718 | Modifier | 0.9268 | 0.9295 | 0.8677 | 0.8605 |
| Social_Security | 0 | 0 | |||||
| All classes | 0.9176 | 0.9263 | All classes | 0.8999 | 0.8986 | 0.8965 | 0.8993 |
Table 6.
Inter-annotator agreement after the training period for task 2.b (disease/disorders and sign/symptoms)
| Entity Type | FDA | CTA | ||
|---|---|---|---|---|
| F-measure | Kappa | F-measure | Kappa | |
| Disease_Disorder | 0.8552 | 0.8930 | 0.8935 | 0.9388 |
| Sign_symptom | 0.8285 | 0.8182 | 0.7594 | 0.8039 |
| All classes | 0.8467 | 0.8855 | 0.8875 | 0.9356 |
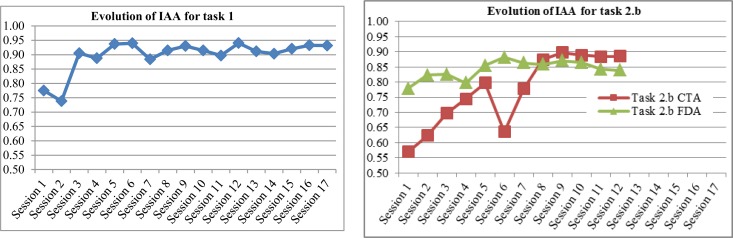
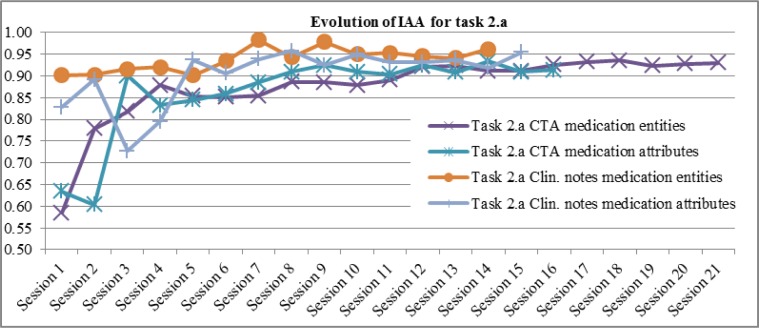
Figure 1 and Figure 2 show IAA computed at each consensus session (including training periods), for each annotation task and corpus. For task 2.a (Figure 2) we show the evolution of IAA for medication entities (medication names and types) and for their attributes separately, since the annotation was done in two steps.
Figure 1.
Evolution of Inter-Annotator Agreement at each consensus session for task 1 and task 2.b
Figure 2.
Evolution of Inter-Annotator Agreement (F-measure) at each consensus session for task 2.a
Discussion
Inter-annotator agreement was the highest for the annotation of PHI elements. They are well-defined entities, less ambiguous than most medical entities. Because most of the annotators did not have clinical backgrounds, their lower agreement in identifying medical entities could be due to their lack of expertise, and it took them longer to adjust to the task. However, they were still able to reach a good agreement with sufficient training for all annotation tasks, and the supervisor did not observe any significant difference in annotation quality between the non-expert annotator and the annotator who had a clinical background in task 2.b. Chapman et al. found that non-clinicians can annotate clinical text at high quality after a more extensive training period29. In a future step, we intend to have a physician validate the quality of medical annotations on a sample of our corpora.
IAA increased after a period of training. We believe a combination of factors can be involved. First of all, annotators familiarize themselves with the guidelines, resolving potential difficulty in understanding them. Second, they also get more accustomed to the medical texts they are working on, and become more consistent when annotating recurring cases. A potential problem that can bias IAA is that annotators could influence each other when resolving their disagreements, especially if one annotator advocates more strongly her opinions. However, we prevented this situation by having a third-party to supervise the consensus sessions between the two annotators. Supervision was only relaxed when high inter-annotator agreement had already been reached and consistently maintained.
We can observe (Figure 1 and 2) that IAA increases more significantly at the beginning, and then reaches a high level with slight fluctuations, i.e. small increases or decreases. Task 2.a on clinical notes (Figure 2) and task 2.b on FDA labels (Figure 1 left side) start at a higher level of IAA than for the other corpus on the same task, because annotators were already trained for the task. The fact that IAA fluctuates even after the training period is an argument in favor of double-annotation, to reduce cases of mislabeling and build stronger gold standards.
Annotation of diseases/disorders and signs/symptoms (task 2.b) was perceived as the most difficult and time-consuming task. Since annotation was based on SNOMED CT concepts and UMLS semantic types, it required looking up terms in a SNOMED CT browser, which slowed down the process. Using the browser was both helpful and confusing. It helped the annotators when they encountered terms they did not know and were unsure whether to annotate or not. But they also had difficulty mapping phrases from the text to SNOMED CT concepts. Since the browser mostly does an exact match without deep linguistic processing, annotators had sometimes to think of alternative ways of wording a term in order to find a match. In some cases they found only a very general concept or a really specific one, or no match at all (e.g. no SNOMED CT concept was found for “dysproteinemia”). Using the UMLS semantic types to classify entities as diseases/disorders or signs/symptoms was also problematic in some cases. Annotators found inconsistencies in the types provided by the UMLS for some concepts, or semantic types that contradicted their intuition, which confused them. For instance “chronic back pain” has semantic type Sign or Symptom, but “chronic low back pain” has semantic type Disease or Syndrome, this difference did not make sense to either annotator. Finally, the semantic type that gave most difficulty was Finding, a very broad type which was found to correspond sometimes to signs/symptoms (e.g. “fever”), sometimes to disease/disorders (e.g. “severe asthma”), sometimes to laboratory or test results (e.g. “abnormal ECG”), sometimes to a general observation (e.g. “in good health”). Annotators found it helpful to have a large set of examples to rely on. In addition to examples provided in the guidelines, they also took notes and built “cheat sheets” listing terms with their appropriate category. The annotation of diseases/disorders and signs/symptoms is still an on-going process. We will annotate the complete set of clinical trial announcements, and the contextual features of the entities (e.g. severity, body side, negation, etc.). We will experiment with annotating the attributes as a second step to entity annotation (as for medications) as well as at the same time as the entities, and get feedback from the annotators. We are also exploring ways of helping annotators, mostly by pre-annotation. This has been tried for annotating disorders using the MetaMap tool but was found to slow down the annotation process by generating too much noise14. Since we already have a fair amount of annotated documents, we will focus on pre-annotating based on the list of already annotated terms.
Future work will include annotating additional medical entities, such as labs, procedures and anatomical sites.
Conclusion
We described the construction of annotated corpora with good inter-annotator agreement for multiple types of documents and several NLP tasks. Most important contributions are that we built a large-scale corpus of clinical notes including the variety of notes available in the EHR, and we explored new types of corpora (clinical trial announcements and FDA drug labels). Aside from providing gold standards for NLP tasks, annotating corpora of different types with the same entities (e.g. medications for CTAs and clinical notes) against the same or very similar annotation schema and guideline allows inter-corpus comparisons, both for descriptive statistics (e.g. number of entities, IAA) and for domain adaption experiments. Most importantly, annotating synergistic corpora against the same guideline allows interoperability for translational research tasks (e.g. patient safety, quality improvement projects, and cohort discovery for trial eligibility screening) which require multi-domain approach to solve.
Acknowledgments
This work was partially supported by NIH grant 5R00LM010227-04. We would like to thank the SHARPn PIs and Dr. Guergana Savova for providing advice and sharing the annotation guideline and Knowtator schema.
REFERENCES
- [1].http://informatics.mayo.edu/sharp/index.php/Main_Page
- [2].http://projectreporter.nih.gov/project_info_description.cfm?aid=8215715&icde=11732766
- [3].http://www.hhs.gov/ocr/privacy/hipaa/understanding/summary/index.html
- [4].Uzuner Özlem, Luo Y, Szolovits P. Evaluating the State-of-the-Art in Automatic De-identification. J Am Med Inform Assoc. 2007;14(5):550–563. doi: 10.1197/jamia.M2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Neamatullah I, Douglass MM, Lehman LH, Reisner A, Villarroel M, Long WJ, et al. Automated de-identification of free-text medical records. BMC Med Inform DecisMak. 2008;8(1):32. doi: 10.1186/1472-6947-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Thomas SM, Mamlin B, Schadow G, McDonald C. A successful technique for removing names in pathology reports using an augmented search and replace method. Proceedings of the AMIA Annual Symposium. 2002:777–781. [PMC free article] [PubMed] [Google Scholar]
- [7].Gupta D, Saul M, Gilbertson J. Evaluation of a deidentification (De-Id) software engine to share pathology reports and clinical documents for research. Am J Clin Pathol. 2004;121(2):176–86. doi: 10.1309/E6K3-3GBP-E5C2-7FYU. [DOI] [PubMed] [Google Scholar]
- [8].Beckwith BA, Mahaadevan R, Balis UJ, Kuo F. Development and evaluation of an open source software tool for deidentification of pathology reports. BMC Med Inform DecisMak. 2006;6:12. doi: 10.1186/1472-6947-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Morrison FP, Li L, Lai AM, Hripcsak G. Repurposing the Clinical Record: Can an Existing Natural Language Processing System De-identify Clinical Notes? J Am Med Inform Assoc. 2008;16(1):37–39. doi: 10.1197/jamia.M2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Benton A, Hill S, Ungar L, Chung A, Leonard C, Freeman C, et al. A system for de-identifying medical message board text. BMC Bioinformatics. 2011;12(3):S2. doi: 10.1186/1471-2105-12-S3-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Taira RK, Bui AAT, Kangarloo H. Identification of patient name references within medical documents using semantic selectional restrictions. Proceedings of the AMIA Annual Symposium. 2002:757–761. [PMC free article] [PubMed] [Google Scholar]
- [12].Aberdeen J, Bayer S, Yeniterzi R, Wellner B, Clark C, Hanauer D, et al. The MITRE Identification Scrubber Toolkit: Design, training, and assessment. Int J Med Inform. 2010;79(12):849–859. doi: 10.1016/j.ijmedinf.2010.09.007. [DOI] [PubMed] [Google Scholar]
- [13].Gardner J, Xiong L. HIDE: An Integrated System for Health Information De-identification. Proceedings of the 21st IEEE International Symposium on Computer-Based Medical Systems. 2008:254–259. [Google Scholar]
- [14].Ogren P, Savova G, Chute C. Constructing Evaluation Corpora for Automated Clinical Named Entity Recognition. Proceedings of the Sixth International Conference on Language Resources and Evaluation; 2008. [Google Scholar]
- [15].Uzuner Özlem, Solti I, Xia F, Cadag E. Community annotation experiment for ground truth generation for the i2b2 medication challenge. J Am Med Inform Assoc. 2010;17(5):519–523. doi: 10.1136/jamia.2010.004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].South BR, Shen S, Jones M, Garvin J, Samore MH, Chapman WW, et al. Developing a manually annotated clinical document corpus to identify phenotypic information for inflammatory bowel disease. BMC Bioinformatics. 2010;10(Suppl 9):S12. doi: 10.1186/1471-2105-10-S9-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chapman WW, Dowling JN. Inductive creation of an annotation schema for manually indexing clinical conditions from emergency department reports. J Biomed Inform. 2006;39:196–208. doi: 10.1016/j.jbi.2005.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Roberts A, Gaizauskas R, Hepple M, Demetriou G, Guo Y, Roberts I, et al. Building a semantically annotated corpus of clinical texts. J Biomed Inform. 2009;42:950–966. doi: 10.1016/j.jbi.2008.12.013. [DOI] [PubMed] [Google Scholar]
- [19].Uzuner Özlem, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18:552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].https://www.i2b2.org/NLP
- [21].Yetisgen-Yildiz M, Solti I, Xia F. Using Amazon’s Mechanical Turk for Annotating Medical Named Entities. Proceedings of the AMIA Annual Fall Symposium; 2010. [PMC free article] [PubMed] [Google Scholar]
- [22].Luo Z, Johnson SB, Lai AM, Weng C. Extracting Temporal Constraints from Clinical Research Eligibility Criteria Using Conditional Random Fields. Proceedings of the AMIA Annual Symposium; 2011. [PMC free article] [PubMed] [Google Scholar]
- [23].Tu SW, Pelega M, Carini S, Bobak M, Ross J, Rubin D, et al. A practical method for transforming free-text eligibility criteria into computable criteria. J Biomed Inform. 2011;44(2):239–250. doi: 10.1016/j.jbi.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].http://www.clinicaltrials.gov/ct2/home
- [25].http://dailymed.nlm.nih.gov/dailymed/about.cfm
- [26].http://www.drugs.com/top200.html
- [27].SHARPn annotation guidelines. www.sharpn.org. Expected release, September, 2012. (Oral communication from SHARPn’s NLP PI)
- [28].Shared Annotated Resources. https://www.clinicalnlpannotation.org/index.php/Main_Page
- [29].Chapman WW, Dowling JN, Hripcsak G. Evaluation of training with an annotation schema for manual annotation of clinical conditions from emergency department reports. Int J Med Inform. 2008;77(2):107–113. doi: 10.1016/j.ijmedinf.2007.01.002. [DOI] [PubMed] [Google Scholar]
- [30].Ogren PV. Knowtator: a protégé plug-in for annotated corpus construction. Proceedings of the 2006 Conference of the North American Chapter of the Association for Computational Linguistics on Human Language Technology; 2006. pp. 273–275. Available from: http://knowtator.sourceforge.net/ [Google Scholar]
- [31].http://www.citrix.com/lang/English/home.asp
- [32].http://www.nomachine.com/download-client-linux.php
- [33].UMLS Terminology Services https://uts.nlm.nih.gov/home.html
- [34].Cohen J. A coefficient of agreement for nominal scales. Educ Psychol Meas. 1960;20:37–46. [Google Scholar]
- [35].Brants T. Inter-annotator agreement for a German newspaper corpus. Proceedings of the Second International Conference on Language Resources and Evaluation (LREC 2000); Athens, Greece. 2000. [Google Scholar]
- [36].Hripcsak G, Rothschild AS. Agreement, the F-Measure, and Reliability in Information Retrieval. J Am Med Inform Assoc. 2005;12:296–298. doi: 10.1197/jamia.M1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tomanek K, Hahn U. Timed Annotations - Enhancing MUC7 Metadata by the Time It Takes to Annotate Named Entities. Proceedings of the Linguistic Annotation Workshop; 2009. pp. 112–115. [Google Scholar]
- [38].Becker M, Hachey B, Alex B, Grover C. Optimising selective sampling for bootstrapping named entity recognition. Proceedings of the ICML Workshop on Learning with Multiple Views; 2005. pp. 5–11. [Google Scholar]
- [39].Hripcsak G, Heitjan DF. Measuring agreement in medical informatics reliability studies. J Biomed Inform. 2002;35:99–110. doi: 10.1016/s1532-0464(02)00500-2. [DOI] [PubMed] [Google Scholar]
- [40].Grouin C, Rosset S, Zweigenbaum P, Fort K, Galibert O, Quintard L. Proposal for an extension of traditional named entities: from guidelines to evaluation, an overview. Proceedings of the 5th Linguistic Annotation Workshop (LAW V ’11); Portland, Oregon. 2011. pp. 92–100. [Google Scholar]