Abstract
Abbreviations are widely used in clinical notes and are often ambiguous. Word sense disambiguation (WSD) for clinical abbreviations therefore is a critical task for many clinical natural language processing (NLP) systems. Supervised machine learning based WSD methods are known for their high performance. However, it is time consuming and costly to construct annotated samples for supervised WSD approaches and sense frequency information is often ignored by these methods. In this study, we proposed a profile-based method that used dictated discharge summaries as an external source to automatically build sense profiles and applied them to disambiguate abbreviations in hospital admission notes via the vector space model. Our evaluation using a test set containing 2,386 annotated instances from 13 ambiguous abbreviations in admission notes showed that the profile-based method performed better than two baseline methods and achieved a best average precision of 0.792. Furthermore, we developed a strategy to combine sense frequency information estimated from a clustering analysis with the profile-based method. Our results showed that the combined approach largely improved the performance and achieved a highest precision of 0.875 on the same test set, indicating that integrating sense frequency information with local context is effective for clinical abbreviation disambiguation.
1. INTRODUCTION
Clinical abbreviations are highly ambiguous. Liu and colleagues1 reported that 33.1% of abbreviations found in the Unified Medical Language System2 (UMLS) 2001 were ambiguous. In a previous study3, we also explored the ambiguity of clinical abbreviations in hospital admission notes using senses from existing knowledge sources (the UMLS and the ADAM4 database), and our results showed that 33.3% – 71.1% abbreviations could be ambiguous, depending on the sources used. It is a challenging task to determine the appropriate meaning of an ambiguous abbreviation in a given context, which is a particular case of the word sense disambiguation (WSD) problem.
WSD has been extensively studied in the field of natural language processing (NLP). Different WSD methods such as knowledge-based and supervised machine learning based methods have been proposed in general English text5–10. A number of studies have focused on WSD in biomedical literature using various types of approaches including supervised, semi-supervised, knowledge-based, and hybrid methods11–20. Similar methods have also been applied to ambiguous terms in clinical text, including abbreviations21–23. Supervised machine learning methods have shown best performance on disambiguation of biomedical terms14. However, it is a costly and time-consuming process to prepare annotated training data for every ambiguous term. In addition, when there exists a majority sense (e.g., relative frequency > 90%) for an ambiguous term, supervised WSD methods do not perform better than a simple strategy that always uses the majority sense, as demonstrated by a simulation study24.
A few studies have investigated methods to automatically generate sense-annotated “pseudo-data” by replacing the long forms (definitions) with the corresponding abbreviations in a corpus, and use the “pseudo-data” to train disambiguation models for abbreviations13,25. The method is very successful in biomedical literature, as definitions are often observed in biomedical papers25. However, this approach may not work very well for many types of clinical notes, especially those directly entered by physicians. Typed-in clinical notes often have a telegraphic style: atypical short phrases, ungrammatical sentences, and pervasive use of abbreviations, which adds additional challenges for clinical NLP systems, when compared with dictated notes26. Our previous study on admission notes directly typed by physicians from New York Presbyterian Hospital (NYPH) showed that about 14.8% of the tokens were abbreviations, and very few definitions of abbreviations (long forms) appeared in the those notes3. Therefore it would be not feasible to create sense-annotated “pseudo data” from those types of clinical notes, using similar approaches.
Pakhomov et al.23 conducted an interesting study to assess the use of external corpora for disambiguating abbreviations in clinical text. They automatically created sense-annotated “pseudo-data” from external corpora such as the web and MEDLINE, and then applied the context of senses to disambiguate abbreviations in the Mayo clinical corpus. They represented training samples and testing samples as context vectors of lexical items and their frequencies. The training vector with the highest cosine similarity to the testing vector was selected and its corresponding sense would be the correct sense for the abbreviation represented by the testing vector. Their evaluation using a set of eight abbreviations showed that the vector similarity based method achieved a best mean accuracy of 67.8% when pseudo-data from both the MEDLINE corpus and Mayo clinical corpus were used. Moreover, their experiments also showed that the vector similarity based method achieved better results than supervised WSD methods, when pseudo-data from a different source was used.
Inspired by Pakhomov et al.23, we proposed to use other types of clinical corpora to help disambiguation of clinical abbreviations in notes physicians directly type. More specifically, we used dictated discharge summaries as an external source to build sense profiles (feature vectors representing different senses) and applied them to disambiguate abbreviations in admission notes via a vector space model. In addition, we integrated the sense frequency information from clustering analysis with the profile-based method to further improve the performance of the clinical abbreviation disambiguation system. To the best of our knowledge, such a method that combines sense profiles with sense frequency information has not been reported for disambiguation of clinical abbreviations.
2. METHODS
This study consisted of two parts: 1) the new profile-based disambiguation method; and 2) the combination approach that integrates the estimated frequency information of senses from clustering analysis with the profile-based disambiguation method. We evaluated both disambiguation methods using a manually annotated independent data set that contained 13 randomly selected abbreviations from admission notes.
2.1. Data sets
Two types of clinical corpora were used in this study. One was a collection of dictated discharge summaries from NYPH during the years of 2003 and 2004, which included 38,273 notes in total. The discharge summary corpus was used to automatically generate sense-tagged pseudo data, from which sense profiles were derived using the method described below. The second corpus consisted of physician-typed hospital admission notes from NYPH during 2004–2006, amounting to 16,949 notes. This corpus was used for two purposes: 1) to generate an estimated frequency distribution of senses by performing a clustering analysis; and 2) to construct an annotated data set that would be used as the independent test set for evaluation.
In a previous study3, we identified 977 abbreviations that occurred more than 100 times in the corpus of admission notes. As we are more interested in ambiguous and more clinically relevant (e.g., abbreviations with disease senses) abbreviations, we linked these abbreviations to the UMLS and found 171 abbreviations that had multiple senses and that had at least one disease sense according to the UMLS. We then randomly selected 20 abbreviations from that set. Sentences containing these 20 abbreviations were collected from admission notes and used for clustering analysis in next step. To evaluate the performance of disambiguation methods, we randomly selected up to 200 instances for each abbreviation and a domain expert manually annotated the sense of each instance. After the annotation, we found that 7 abbreviations actually had only one sense in the admission note corpus, though the UMLS linked them to multiple senses. Therefore, the final annotated test set contained 13 ambiguous abbreviations, which were “ad”, “ag”, “bm”, “cm”, “gtt”, “hs”, “ln”, “ls”, “med”, “pt”, “ra”, “si”, and “ss”. There were 2,386 annotated instances in total for all 13 abbreviations.
2.2. The profile-based disambiguation method
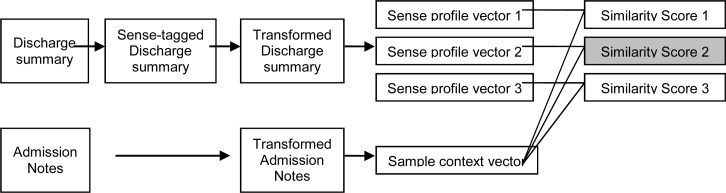
In a previous study27, we used knowledge-based profiles to disambiguate abbreviated gene symbols in the biomedical literature, by using existing knowledge sources of genes in the biology domain. In this study, we modified this method and applied it to clinical abbreviation disambiguation. Figure 1 shows an overview of the profile-based method adapted to disambiguation of clinical abbreviations. For each sense of an abbreviation, a set of instances that contain the fully formed sense string are automatically found in the dictated discharge summaries using an exact string matching method. Then the sense string is replaced with the corresponding abbreviation and the abbreviation is tagged with that sense, which generates the “sense-tagged discharge summaries”. In addition, we applied a transformation step as described below to both discharge summaries and admission notes, to make both corpora look similar to each other. Then sense profiles are built from the sense-tagged and transformed discharge summaries using local contextual words around the target abbreviations. During disambiguation, the cosine-similarities between the context vector of the testing sample and the profile vectors of the possible senses are calculated. The sense corresponding to the highest similarity score will be selected as the correct sense.
Figure 1.
An overview of the profile-based disambiguation method for abbreviations in admission notes.
2.2.1. Build sense profiles from discharge summaries
The dictated discharge summaries are different from the typed-in admission notes in a few aspects. Discharge summaries, which summarize patient information from admission to discharge, contain much broader information than admission notes. As this group were dictated by physicians but then transcribed by transcriptionists, there are less abbreviations and more long forms. The process for generating sense profile vectors from discharge summaries could be described as following:
Tag senses: For each sense of an ambiguous abbreviation (see Table 1), the corpus of discharge summaries was searched for its sense string (also called Long Form – LF) using an exact string matching algorithm. If an instance containing the long form of that sense was found, the long form would be replaced by the corresponding abbreviation (also Short Form - SF). The instance was added to the collection of training data and was tagged with the particular sense. Figure 2 shows an example of the sense tagging process. The instance containing the long form “room air” would have “room air” replaced by its abbreviation “ra”, and the instance labeled as the sense of “room air”. Therefore, we got a set of automatically sense-annotated instances of discharge summaries for each sense of each abbreviation.
Transform discharge summaries: Each sense-tagged instance in discharge summaries was processed by a transformation program, which replaced all the long forms of biomedical terms with their corresponding short forms (abbreviations), based on a predefined SF/LF list. The rationale behind the transformation step was to make the discharge summaries more similar to admission notes. The SF/LF list contains abbreviations from multiple sources: 1) the UMLS abbreviation list (LRABR File); 2) a derived abbreviation list from the UMLS following Liu’s method 1; 3) the ADAM abbreviation database4; and 4) a manually collected abbreviation list from sign-out notes by Stetson et al. 28. Figure 3 shows an example of the transformation on a sense tagged instance. At the end of this step, each abbreviation was associated with a training set that contained instances of transformed discharge summaries, tagged with the different senses for that abbreviation.
Build sense profile vectors: A feature vector was created for each instance in the training set of an abbreviation. Similar to a previous study29, we used three types of features to form the feature vector of an instance: 1) stemmed words within a window size of 5 of the target abbreviation; 2) positional information + stemmed words within a window size of 5 of the target abbreviation (e.g., “L2_acute” represents a feature of the second left word “acute”); 3) section header of the admission note where the abbreviation occurs, by using a list of frequent section headers. For each abbreviation, features of its instances were weighted using the TF-IDF weighting schema 30, which is widely used in the vector space model for information retrieval. Given a document d, the Term Frequency (TF) of term t is defined as the frequency of t occurring in d. The Inverse Document Frequency (IDF) of term t is defined as the logarithm of the number of all documents in the collection divided by the number of documents containing the term t. Then term t in document d is weighted as TF*IDF. In this case, we treated each instance of an abbreviation as a document and all three types of features from that instance would be terms in that document. At this stage, each instance in the training set of an abbreviation was represented by a weighted feature vector, in which each feature was coupled with an associated TF-IDF weight. To create a sense profile vector, all the weighted feature vectors of the instances tagged with that sense were combined. The final weight of a feature in the sense profile vector would be the averaged weight of the feature across all the instances with that sense.
Table 1.
Senses and their frequency distributions based on the manually annotated test set. Majority senses based on the annotated test set from admission notes, automatically generated sense-tagged pseudo-data from discharge summaries, and clustering analysis based on admission notes were also marked respectively in the columns 4–6.
| Abbreviation (# of senses) | Sense | Relative Frequency annotations | Majority Sense Admission notes annotations | Majority Sense Discharge Summaries | Majority Sense Admission notes Clustering Analysis |
|---|---|---|---|---|---|
|
| |||||
| ad (4) | Advertisement | 0.021 | |||
| Add | 0.007 | ||||
| Alzheimer’s disease | 0.790 | X | X | ||
| Adenosine | 0.182 | X | |||
|
| |||||
| ag (3) | anion gap | 0.520 | X | X | |
| Antigen | 0.160 | ||||
| Adrian Gonzalez (Name Initials) | 0.320 | X | |||
|
| |||||
| bm (2) | bowel movement | 0.883 | X | X | X |
| bone marrow | 0.117 | ||||
|
| |||||
| cm (5) | Cardiomyopathy | 0.071 | X | ||
| costal margin | 0.015 | ||||
| cardiac monitoring | 0.005 | ||||
| Centimeter | 0.828 | X | X | ||
| Cardiomegaly | 0.081 | ||||
|
| |||||
| gtt (2) | Drop | 0.152 | |||
| Drip | 0.848 | X | X | X | |
|
| |||||
| hs (8) | Hepatosplenomegaly | 0.005 | |||
| History | 0.020 | X | |||
| Hepatospleno | 0.005 | ||||
| Hours | 0.010 | ||||
| Hudson South | 0.005 | ||||
| high school | 0.010 | ||||
| at bedtime | 0.551 | X | X | ||
| heart sounds | 0.393 | ||||
|
| |||||
| ln (2) | lymph node | 0.995 | X | X | X |
| natural logarithm | 0.005 | ||||
|
| |||||
| ls (3) | lung sounds | 0.190 | |||
| Lumbosacral | 0.805 | X | X | ||
| Lymphocytes | 0.005 | X | |||
|
| |||||
| med (2) | Medication | 0.615 | X | X | X |
| Medicine/Medical | 0.385 | ||||
|
| |||||
| pt (4) | posterior tibial | 0.005 | |||
| Patient | 0.905 | X | X | X | |
| physical therapy | 0.035 | ||||
| prothrombin time assay | 0.055 | ||||
|
| |||||
| ra (3) | right atrium | 0.030 | |||
| room air | 0.900 | X | X | X | |
| rheumatoid arthritis | 0.070 | ||||
|
| |||||
| si (5) | small intestine | 0.010 | |||
| Staten Island | 0.010 | ||||
| suicidal ideation | 0.934 | X | X | X | |
| Sacroiliac | 0.020 | ||||
| Sign | 0.025 | ||||
|
| |||||
| ss (7) | Steve Shea (Name Initials) | 0.289 | |||
| sliding scale | 0.086 | ||||
| Hemoglobin SS | 0.437 | X | X | X | |
| Serosanguinous | 0.005 | ||||
| social security | 0.010 | ||||
| Substernal | 0.127 | ||||
| single strength | 0.046 | ||||
Figure 2.
An example of the sense tagging step for the profile-based disambiguation method.
Figure 3.
An example of the transformation step for the profile-based disambiguation method.
2.2.2. Disambiguation using sense profiles
Before disambiguation, the admission notes containing testing samples were processed by the same transformation program. Similar to sense profile vectors, a feature vector of a testing sample from a transformed admission note was formed by the same three types of features. Features of a testing sample were weighted using the TF-IDF weights of the corresponding features in the sense profile vectors when calculating vector similarity. Cosine similarities between a testing sample feature vector and its possible sense profile vectors were calculated. The sense whose profile vector had the highest similarity with the testing sample feature vector was selected as the correct sense.
2.3. Integrating frequency information with the profile-based disambiguation method
The profile-based disambiguation method, as well as other supervised machine learning methods for WSD, uses the local contextual information, such as words around the target term, for disambiguation. Other information, such as the sense frequency distribution of an ambiguous term, also affects the performance of disambiguation, as demonstrated in 24. Therefore we proposed to combine both types of information for word sense disambiguation in this study.
2.3.1. Estimated sense distribution from clustering analysis
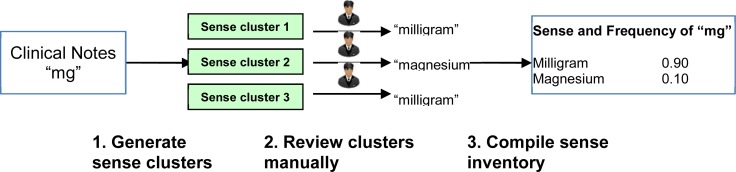
We have developed a clustering-based method to build sense inventories of clinical abbreviations in a semiautomated fashion29. Figure 1 shows an overview of this method, which consists of three steps: 1) collect instances of an abbreviations and cluster them into different sense clusters; 2) select one instance that is closest to the centroid from each sense cluster and manually determine its sense; and 3) combine senses from all clusters to form the sense inventory of an abbreviation. We have demonstrated that the sense detection method could group instances into different clusters and determine the sense of a cluster by affordable annotation29. Such sense clustering analysis could provide estimation about frequency distribution of abbreviations.
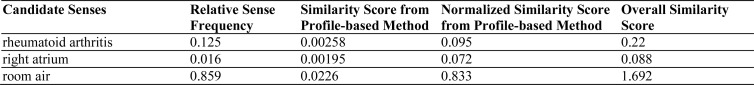
For each abbreviation in this study, up to 1000 instances were randomly selected and clustered using a sense clustering algorithm called Tight Clustering for Rare Senses (TCRS) 31. One instance from each cluster was reviewed and its annotated sense was assigned to the cluster. Estimated sense frequency distributions of an ambiguous abbreviation in admission notes were obtained in the following way. For example, the clustering analysis of 1000 instances of abbreviation “ra” showed that 125 instances mapped to the sense “rheumatoid arthritis”, 16 mapped to “right atrium”, and 859 mapped to “room air”. The relative frequency of each sense in the corpus was therefore 0.125(125/1000), 0.016(16/1000), and 0.859 (859/1000) respectively. Therefore, the estimated sense frequency distribution for (“rheumatoid arthritis”, “right atrium”, “room air”) would be (0.125, 0.016, 0.859). Based on the estimated frequency distribution from the clustering analysis, a classifier could be built by selecting the estimated majority sense from clustering analysis, which is the sense with the highest estimated relative sense frequency.
2.3.2. The combination model
In this study, we used a simple stragety to combine the sense frequency information with the profile-based classifier. For a testing sample, its individual similarity score with a possible sense profile vector was normalized by dividing the sum of its similarity scores with all candidate senses. The overall similarity score of a candidate sense was considered to be the sum of the normalized similarity score from the profile-based classifier and the relative sense frequency from the estimated sense distribution. The sense with the highest overall similarity was selected as the correct sense. Figure 4 shows an example of how to calculate the overall similarity score for the abbreviation “ra”. The relative sense frequency from clustering analysis is listed in the second column. Similarity scores from the profile-based method, as well as the normalized similarity scores, are listed in the third and fourth columns respectively. The overall similarity scores in the fifth column were calculated by adding values from the second and fourth columns. In this example, the sense “room air” was selected as the correct sense because it had the highest overall similarity score.
Figure 4.
The clustering-based method for building abbreviations sense inventory and estimating sense frequency from clincial corpus.
Figure 4.
An example of calculating overall similarity scores using the combination model.
2.4. Evaluation
For each ambiguous abbreviation, the randomly selected and manually sense-annotated test set from admission notes served as gold standard for evaluation. Results from different disambiguation methods were compared with the gold standard, and two measurements were reported. One was Precision, which was defined as the ratio between the number of correctly disambiguated testing samples by the profile-based method and the number of testing samples where the profile-based method could make a decision. Sometimes, the profile-based disambiguation method may not be able to make a decision on a testing sample, e.g. when two sense profiles have the same similarity scores with regard to the testing sample. Therefore we also defined Recall, which was the ratio between the number of testing samples for which the disambiguation method could make a decision and the total number of testing samples. When we built sense profiles by replacing long forms with corresponding abbreviations, some long forms were never found in discharge summaries and no sense profile could be built. Therefore we also measured the coverage for building sense profiles, which was defined as the ratio between the number of senses whose long forms could be found in the corpus and the total number of senses of all abbreviations in the testing set.
2.4.1. Evaluation of the profile-based disambiguation method
Two baseline methods were also implemented and compared with the profile-based disambiguation method. One was a random sense selection method, which randomly selected a possible sense as the correct sense. The second one was a majority-sense based method that always used the majority sense as the correct sense. In this method, the majority sense was determined by the automatically sense tagged data sets from discharge summaries.
2.4.2. Evaluation of the combined disambiguation method
Two individual disambiguation methods: the profile-based method and the majority-sense based method that always selects the sense with the highest estimated frequency from admission notes were reported together with the combined method. The same test set from admission notes were used for this evaluation as well.
3. RESULTS
Table 1 shows the 13 ambiguous abbreviations used in this study. The senses and their frequency distribution information were based on the manually annotated test set from admission notes. We also reported majority senses determined by different data sources or methods, including 1) majority senses based on the annotated test set from admission notes; 2) majority senses from automatically generated sense-tagged pseudo-data from discharge summaries; and 3) majority senses based on estimated frequency information from clustering analysis on admission notes. The majority senses generated from the clustering analysis were exactly same as those from manual annotation. However, five out of thirteen abbreviations would have different majority senses if we used pseudo data from discharge summaries.
Table 2 shows the precision and recall of the profile-based disambiguation method, as well as two baseline methods, on the test set. The first column shows the 13 abbreviations with their numbers of senses in the parenthesis. As all disambiguation methods made decisions regarding most testing samples, the recall remained as 1.00. The profile-based disambiguation method with transformation reached the highest average precision of 0.792. The average precision was much higher for the profile-based methods than for the baseline methods (the Random and the Majority-sense based methods). The improvement of the transformation step for the profile-based method was very small (from 0.784 to 0.792). We had a total of 50 senses for 13 ambiguous abbreviations. When building sense profiles using the long form replacement method based on exact string matching algorithm, we did not find any matches for the long forms of two senses (“cm” – “costal margin” and “ln” – “natural logarithm”). So the coverage for building sense profiles was 48/50 = 96%.
Table 2.
Results of different profile-based method, when compared with two baseline methods.
| Abbr | Random | Majority (from discharge summaries) | Profile Un-transformed | Profile Transformed | ||||
|---|---|---|---|---|---|---|---|---|
| Pre | Rec | Pre | Rec | Pre | Rec | Pre | Rec | |
| ad(4) | 0.217 | 1.00 | 0.182 | 1.00 | 0.643 | 1.00 | 0.734 | 1.00 |
| ag(3) | 0.228 | 1.00 | 0.320 | 1.00 | 0.890 | 1.00 | 0.846 | 1.00 |
| bm(2) | 0.529 | 1.00 | 0.884 | 1.00 | 0.910 | 1.00 | 0.931 | 1.00 |
| cm(5) | 0.227 | 1.00 | 0.071 | 1.00 | 0.591 | 1.00 | 0.597 | 1.00 |
| gtt(2) | 0.523 | 1.00 | 0.848 | 1.00 | 0.934 | 1.00 | 0.929 | 1.00 |
| hs(8) | 0.092 | 1.00 | 0.021 | 1.00 | 0.744 | 1.00 | 0.759 | 1.00 |
| ln(2) | 0.427 | 1.00 | 0.995 | 1.00 | 0.995 | 1.00 | 0.995 | 1.00 |
| ls(3) | 0.271 | 1.00 | 0.005 | 1.00 | 0.975 | 1.00 | 0.975 | 0.995 |
| med(2) | 0.538 | 1.00 | 0.613 | 1.00 | 0.774 | 1.00 | 0.804 | 1.00 |
| pt(4) | 0.209 | 1.00 | 0.903 | 1.00 | 0.536 | 1.00 | 0.490 | 1.00 |
| ra(3) | 0.300 | 1.00 | 0.900 | 1.00 | 0.945 | 1.00 | 0.955 | 1.00 |
| si(5) | 0.178 | 1.00 | 0.934 | 1.00 | 0.614 | 1.00 | 0.574 | 1.00 |
| ss(7) | 0.100 | 1.00 | 0.437 | 1.00 | 0.643 | 1.00 | 0.707 | 1.00 |
| AVG | 0.295 | 1.00 | 0.547 | 1.00 | 0.784 | 1.00 | 0.792 | 1.00 |
Table 3 shows the results of the combined method, as well as individual methods. As the results show, the individual methods (the profile-based and the estimated majority sense-based method) had similar average precision values (0.792/0.770). But the combined method had a much higher average precision value (0.875) than any individual method did.
Table 3.
Results of the combined method for disambiguation of clinical abbreviations.
| Abbr | Profile Only | Sense Frequency Only | Combined | |||
|---|---|---|---|---|---|---|
| Precision | Recall | Precision | Recall | Precision | Recall | |
| ad(4) | 0.734 | 1.00 | 0.790 | 1.00 | 0.867 | 1.00 |
| ag(3) | 0.846 | 1.00 | 0.520 | 1.00 | 0.875 | 1.00 |
| bm(2) | 0.931 | 1.00 | 0.883 | 1.00 | 0.894 | 1.00 |
| cm(5) | 0.597 | 1.00 | 0.827 | 1.00 | 0.837 | 1.00 |
| gtt(2) | 0.929 | 1.00 | 0.848 | 1.00 | 0.853 | 1.00 |
| hs(8) | 0.759 | 1.00 | 0.549 | 1.00 | 0.933 | 1.00 |
| ln(2) | 0.995 | 1.00 | 0.995 | 1.00 | 0.995 | 1.00 |
| ls(3) | 0.975 | 0.995 | 0.805 | 1.00 | 0.860 | 1.00 |
| med(2) | 0.804 | 1.00 | 0.613 | 1.00 | 0.789 | 1.00 |
| pt(4) | 0.490 | 1.00 | 0.903 | 1.00 | 0.908 | 1.00 |
| ra(3) | 0.955 | 1.00 | 0.900 | 1.00 | 0.915 | 1.00 |
| si(5) | 0.574 | 1.00 | 0.934 | 1.00 | 0.934 | 1.00 |
| ss(7) | 0.707 | 1.00 | 0.437 | 1.00 | 0.721 | 1.00 |
| AVG | 0.792 | 1.00 | 0.770 | 1.00 | 0.875 | 1.00 |
4. DISCUSSION
In this study, we proposed a profile-based method that automatically generates abbreviations’ sense profiles from dictated discharge summaries and uses them to disambiguate clinical abbreviations in typed-in admission notes. Our evaluation showed this method was very effective and it performed better than other baseline methods. We further developed a combined approach that integrates estimated sense frequency information derived form clustering analysis with the profile-based method. The evaluation showed that the combined approach achieved a much higher precision (0.875) than any single classifier (0.792/0.770), indicating an effective yet feasible (e.g., only minimum annotation is required) approach to clinical abbreviation disambiguation. We expect this method can be easily integrated with exiting NLP systems such as MedLEE32 (e.g., as a post-processing program) to further improve their capability of handling clinical abbreviations.
The profile-based method was originally developed to disambiguate gene symbols in biomedical literature27; but this study demonstrated that it performs well for disambiguation in clinical text as well. The method is similar to Pakhomov et al. 23; but there are a few differences: 1) we build a profile vector for each sense of an ambiguous abbreviation by merging training sample vectors with the same sense, and we compute similarity between a testing sample vector and its possible sense profile vectors, not individual training sample vectors; 2) the weighting schema used in our method is the TF-IDF weighting, not just term frequency; and 3) we use the corpus of discharge summaries for training and we add an additional transformation step for both the discharge summaries and the admission notes to make them more similar, when building sense profiles and performing disambiguation. The profile-based method seemed to be more tolerant of the dissimilarity between the training and testing sets, as the transformation step for building sense profiles did not improve the performance very much. Therefore, it is more useful when the training data is from a different corpus than the corpus containing the testing data. In this study, we used a corpus of dictated discharge summaries, which are widely available in many hospitals. When dictated discharge summaries are not available, other clinical text containing full forms of abbreviations could be used, but the quality of sense profiles from those corpora need to be investigated further.
Based on results in Table 3, we also noticed that the simple majority-sense based approach actually achieved a reasonable performance (average precision 0.770). Our observation showed that a large portion of clinical abbreviations probably had a dominant sense. Among the 13 abbreviations in this study, 9 of them had a majority sense with a relative frequency > 70%. We also investigated the sense frequency distribution of 16 abbreviations from the study by Joshi et al. 22 and found 10 out of 16 had a majority sense with a relative frequency > 70%. These statistics indicated that the majority-sense based method could be effective for disambiguating clinical abbreviations. However, it is not straightforward to determine the majority sense of an abbreviation. Manual annotation of randomly selected samples (such as the test set in this study) is one way to obtain a sense frequency distribution, but it is very time consuming and costly. Automatically sense-tagged pseudo data (such as the sense-tagged discharge summaries) can provide estimated sense frequency information as well; but it may not reflect the real distribution of senses in the original corpus. As shown in Table 1, majority senses obtained from discharge summaries were not good enough (5 out of 13 were wrong) and could be detrimental to performance (see Table 2). For example, for the abbreviation “cm”, the true majority sense with the long form “centimeter” occurred infrequently in discharge summaries and therefore the majority sense incorrectly became “cardiomyopathy”. By selecting the “wrong” majority sense of “cardiomyopathy”, the precision of the Majority-Sense based method was very low for “cm” (0.071) when applied to the testing samples from admission notes. On the other hand, the clustering-based sense detection method obtained the correct majority sense for every abbreviation in our study, indicating its usefulness in this context.
In this study, we used a very simple addition method to combine the sense frequency information and the context similarity information. The results showed that this simple combination method was very effective. The average precision of the combined method increased 8.3% when compared with the profile-based method alone. Most of the 13 abbreviations showed improved performance when the simple combination model was applied. But there were four abbreviations (bm, gtt, ls, and ra), which showed decreased performance when the combined method was used. We noticed that all those four abbreviations had very high performance (precision over 90%) when the profile-based method was used alone. We looked into the instances where the combined method made an error, while the profile-based method alone made a correct decision. For example, for the abbreviation “ls”, the profile-based method generated a very high normalized similarity score (0.751) for the correct sense “lung sounds”. But after the relative sense frequency was added, the overall similarity score for the correct sense “lung sounds” was lower than the sense “lumbosacral”. Therefore, the incorrect sense “lumbosacral” was selected as the final sense. A possible solution to this type of error is to ignore the sense distribution information when the profile-based method generates a very high similarity score for a certain sense. This will involve work to develop more sophisticated combination models and to determine the thresholds. In the future, we will build different combination models, such as a logistic regression model, to combine the sense distribution information and the context similarity information by assigning different weights for each type of information.
5. CONCLUSION
The profile-based method could disambiguate abbreviations in typed-in admission notes in an unsupervised fashion and our evaluation demonstrated its robustness. When sense frequency information estimated from a clustering analysis was combined with the profile-based method, the performance of our WSD system was largely improved (precision from 0.792 to 0.875), indicating that integrating sense frequency information with local context is effective for clinical abbreviation disambiguation.
Acknowledgments
This study was supported by grants from the US NIH: NLM R01LM010681 (HX), R01LM8635 (CF), and R01LM010016 (CF).
Reference
- 1.Liu H, Lussier YA, Friedman C. A study of abbreviations in the UMLS. Proc AMIA Symp; 2001. pp. 393–397. [PMC free article] [PubMed] [Google Scholar]
- 2.UMLS US Dept of Health and Human Services, NIH, NLM.
- 3.Xu H, Stetson PD, Friedman C. A study of abbreviations in clinical notes. AMIA … Annual Symposium proceedings / AMIA Symposium. AMIA Symposium; 2007. pp. 821–825. [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou W, Torvik VI, Smalheiser NR. ADAM: another database of abbreviations in MEDLINE. Bioinformatics. 2006 Nov 15;22(22):2813–2818. doi: 10.1093/bioinformatics/btl480. [DOI] [PubMed] [Google Scholar]
- 5.Bruce R, Wiebe J. Word-sense disambiguation using decomposable models. Proceedings of the 32nd annual meeting on Association for Computational Linguistics; Morristown, NJ, USA: Association for Computational Linguistics; 1994. pp. 139–146. [Google Scholar]
- 6.Lee YK, Ng HT. An empirical evaluation of knowledge sources and learning algorithms for word sense disambiguation. Proceedings of the ACL-02 conference on Empirical methods in natural language processing - Volume 10; Morristown, NJ, USA: Association for Computational Linguistics; 2002. pp. 41–48. [Google Scholar]
- 7.Li L, Roth B, Sporleder C. Topic models for word sense disambiguation and token-based idiom detection. Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics; Uppsala, Sweden: Association for Computational Linguistics; 2010. pp. 1138–1147. [Google Scholar]
- 8.Magnus M, Mikael A. Combination of contextual features for word sense disambiguation: LIU-WSD. SENSEVAL-2 Workshop. 2001:123–127. [Google Scholar]
- 9.Mohammad S. Combining Lexical and Syntactic Features for Supervised Word Sense Disambiguation; Proceedings of the Conference on Computational Natural Language Learning; 2004. p. 8. [Google Scholar]
- 10.Navigli R. Word sense disambiguation: A survey. ACM Comput. Surv. 2009;41(2):1–69. [Google Scholar]
- 11.Humphrey SM, Rogers WJ, Kilicoglu H, Demner-Fushman D, Rindflesch TC. Word Sense Disambiguation by Selecting the Best Semantic Type Based on Journal Descriptor Indexing: Preliminary Experiment. J Am Soc Inf Sci Technol. 2006 Jan 1;57(1):96–113. doi: 10.1002/asi.20257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leroy G, Rindflesch TC. Effects of information and machine learning algorithms on word sense disambiguation with small datasets. Int J Med Inform. 2005 Aug;74(7–8):573–585. doi: 10.1016/j.ijmedinf.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Liu H, Lussier YA, Friedman C. Disambiguating ambiguous biomedical terms in biomedical narrative text: an unsupervised method. Journal of biomedical informatics. 2001 Aug;34(4):249–261. doi: 10.1006/jbin.2001.1023. [DOI] [PubMed] [Google Scholar]
- 14.Schuemie MJ, Kors JA, Mons B. Word sense disambiguation in the biomedical domain: an overview. J Comput Biol. 2005 Jun;12(5):554–565. doi: 10.1089/cmb.2005.12.554. [DOI] [PubMed] [Google Scholar]
- 15.Jimeno-Yepes A, McInnes BT, Aronson AR. Collocation analysis for UMLS knowledge-based word sense disambiguation. BMC bioinformatics. 2011;12(Suppl 3):S4. doi: 10.1186/1471-2105-12-S3-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jimeno-Yepes AJ, Aronson AR. Knowledge-based biomedical word sense disambiguation: comparison of approaches. BMC bioinformatics. 2010;11:569. doi: 10.1186/1471-2105-11-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McInnes BT, Pedersen T, Liu Y, Melton GB, Pakhomov SV. Knowledge-based method for determining the meaning of ambiguous biomedical terms using information content measures of similarity. AMIA ... Annual Symposium proceedings / AMIA Symposium. AMIA Symposium; 2011. pp. 895–904. [PMC free article] [PubMed] [Google Scholar]
- 18.Stevenson M, Agirre E, Soroa A. Exploiting domain information for Word Sense Disambiguation of medical documents. Journal of the American Medical Informatics Association : JAMIA. 2012 Mar 1;19(2):235–240. doi: 10.1136/amiajnl-2011-000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevenson M, Guo Y. Disambiguation of ambiguous biomedical terms using examples generated from the UMLS Metathesaurus. Journal of biomedical informatics. 2010 Oct;43(5):762–773. doi: 10.1016/j.jbi.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 20.Yepes AJ, Aronson AR. Knowledge-based and knowledge-lean methods combined in unsupervised word sense disambiguation. Proceedings of the 2nd ACM SIGHIT International Health Informatics Symposium; 2012. pp. 733–736. [Google Scholar]
- 21.Liu H, Teller V, Friedman C. A multi-aspect comparison study of supervised word sense disambiguation. Journal of the American Medical Informatics Association : JAMIA. 2004 Jul-Aug;11(4):320–331. doi: 10.1197/jamia.M1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joshi M, Pakhomov S, Pedersen T, Chute CG. A comparative study of supervised learning as applied to acronym expansion in clinical reports. AMIA … Annual Symposium proceedings / AMIA Symposium. AMIA Symposium; 2006. pp. 399–403. [PMC free article] [PubMed] [Google Scholar]
- 23.Pakhomov S, Pedersen T, Chute CG. Abbreviation and acronym disambiguation in clinical discourse. AMIA … Annual Symposium proceedings / AMIA Symposium. AMIA Symposium; 2005. pp. 589–593. [PMC free article] [PubMed] [Google Scholar]
- 24.Xu H, Markatou M, Dimova R, Liu H, Friedman C. Machine learning and word sense disambiguation in the biomedical domain: design and evaluation issues. BMC bioinformatics. 2006;7:334. doi: 10.1186/1471-2105-7-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu H, Kim W, Hatzivassiloglou V, Wilbur WJ. Using MEDLINE as a knowledge source for disambiguating abbreviations and acronyms in full-text biomedical journal articles. Journal of biomedical informatics. 2007 Apr;40(2):150–159. doi: 10.1016/j.jbi.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 26.Zheng K, Mei Q, Yang L, Manion FJ, Balis UJ, Hanauer DA. Voice-dictated versus typed-in clinician notes: linguistic properties and the potential implications on natural language processing. AMIA ... Annual Symposium proceedings / AMIA Symposium. AMIA Symposium; 2011. pp. 1630–1638. [PMC free article] [PubMed] [Google Scholar]
- 27.Xu H, Fan JW, Hripcsak G, Mendonca EA, Markatou M, Friedman C. Gene symbol disambiguation using knowledge-based profiles. Bioinformatics. 2007 Apr 15;23(8):1015–1022. doi: 10.1093/bioinformatics/btm056. [DOI] [PubMed] [Google Scholar]
- 28.Stetson PD, Johnson SB, Scotch M, Hripcsak G. The sublanguage of cross-coverage. Proc AMIA Symp; 2002. pp. 742–746. [PMC free article] [PubMed] [Google Scholar]
- 29.Xu H, Stetson PD, Friedman C. Methods for building sense inventories of abbreviations in clinical notes. Journal of the American Medical Informatics Association : JAMIA. 2009 Jan-Feb;16(1):103–108. doi: 10.1197/jamia.M2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salton G, Buckley C. Term-weighting approaches in automatic text retrieval. Information Processing & Management. 1988;24(5):513–523. [Google Scholar]
- 31.Xu H, Wu Y, Elhadad N, Stetson PD, Friedman C. A new clustering method for detecting rare senses of abbreviations in clinical notes. Submitted to Journal of Biomedical Informatics. 2012 doi: 10.1016/j.jbi.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. Journal of the American Medical Informatics Association : JAMIA. 1994 Mar-Apr;1(2):161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]