Abstract
Ontology design patterns (ODPs) are a proposed solution to facilitate ontology development, and to help users avoid some of the most frequent modeling mistakes. ODPs originate from similar approaches in software engineering, where software design patterns have become a critical aspect of software development. There is little empirical evidence for ODP prevalence or effectiveness thus far. In this work, we determine the use and applicability of ODPs in a case study of biomedical ontologies. We encoded ontology design patterns from two ODP catalogs. We then searched for these patterns in a set of eight ontologies. We found five patterns of the 69 patterns. Two of the eight ontologies contained these patterns. While ontology design patterns provide a vehicle for capturing formally reoccurring models and best practices in ontology design, we show that today their use in a case study of widely used biomedical ontologies is limited.
Biomedical Ontology and Ontology Design Patterns
In this study, we examine the applicability of Ontology Design Patterns (ODPs) to biomedical ontologies. We define ODPs as general modeling solution to solve a recurrent problem in the context of knowledge engineering. ODPs capture common modeling situations, help facilitate ontology development and avoid common mistakes. To ensure a diverse case study set, we select pertinent biomedical ontologies varying in both style (format) and provenance (age). We hypothesize that ODP use differs based on these characteristics. Ontologies created before the introduction of ODPs might not use them. Ontologies created in a format other than OWL might not use the published ODPs, which are OWL centric. Before embarking on an in-depth research program in this area, we carried out a case study.
Ontologies in Biomedicine
Ontologies are machine processable artifacts that capture the relationships between concepts in some domain of interest. In biomedicine, the number and use of ontologies, or more generally, terminologies, has increased dramatically over the last decade.1 As of this publication, the National Center for Biomedical Ontology’s (NCBO) BioPortal, a web-based application for accessing and publishing ontologies, contains over 300 biomedical ontologies.2 BioPortal is quite diverse, with ontologies ranging in size from hundreds of terms to nearly half a million and varying in logical expressivity, age, and development method. Such ontologies capture knowledge about the various subdomains of biomedicine, from medications and diseases to disease processes and genes. The basic science community has used ontologies to assist in understanding the massive amount of data it generates. By annotating experiments and literature with Gene Ontology (GO) terms, researchers are able to integrate results and gain insight about relations that were previously difficult to discover. For example, many software applications can relate differential gene expression to function using GO enrichment analysis.3
In the clinical domain, ontologies have become increasingly important. They are an essential component of Electronic Health Records (EHRs). With the enactment of the HITECH act and its associated meaningful use requirements, EHRs and the use of controlled terminologies in them, are likely to become widespread.4 For example, the National Drug File-Reference Terminology (NDF-RT), produced by the Department of Veteran Affairs, provides clinically relevant information on drugs and pharmacologic classes.5 NDF-RT can serve as a component for a computerized physician order entry and clinical decision support system; implementing either system satisfies meaningful use objectives.4 Additionally, the ONC recommends the use of SNOMED CT, a detailed clinical terminology, by clinical providers to record clinical observations in an EHR (ONC Standards and Certification, Final Rule). The previous examples are but a small sample of the ontologies developed for biomedical applications.
Because of the large and complex nature of biomedical ontologies, they are particularly susceptible to modeling mistakes and errors—much the same way that large complex software projects usually contain bugs. Indeed, recent research focused on analyzing the quality of biomedical ontologies have found numerous modeling problems in ontologies including SNOMED CT and the National Cancer Institute thesaurus.6,7 For example, SNOMED CT contained systematic problems with the use of part-of relationship that caused diabetes to be classified as a disorder of the abdomen—which is incorrect. In summary, the development process for a biomedical ontology is often difficult, especially as the size and complexity of the ontology increases; many developers of ontologies in biomedicine are domain experts rather than logic experts, and focus on the concepts in the domain rather than their ontological representation. Furthermore, an ontology developer must follow best practice and avoid errors while modeling the complexities of the biomedical domain.
One possible way of tackling the complexities of ontology development is to use an ontology development methodology that incorporates best practice into the design and construction of an ontology. In the world of ontologies, this best practice comes in the form of style guidelines accompanied with the application and use of ontology design patterns (ODPs). A design pattern is a general modeling solution to solve a recurrent problem. Thus, an ontology design pattern is simply general modeling solution to solve a recurrent problem in the context of knowledge engineering. The basic ideas behind ontology design patterns are very similar to the ideas behind software design patterns. 9 Software design patterns are standard solutions to common problems in the context of software engineering. The proponents of Ontology Design Patterns claim that they help to improve ontology quality, improve ontology reuse, provide documentation of design rationale, make ontologies more maintainable, and make ontologies easier to understand.10 If these claims are true, the authors, maintainers, and users of biomedical ontologies could clearly benefit from the incorporation of patterns into the design of such ontologies. ODPs have become popular over the last few years, with workshops held at the International Semantic Web Conferences and with the creation of online ODP libraries.
Given the recent research results about biomedical ontology quality, and the popularity of design patterns, presented to ameliorate problems with ontology quality, the aim of this paper is to present a case study that looks at different biomedical ontologies and the application of ontology design patterns to these ontologies. Specifically, this case study is part of an analysis examining if biomedical ontologies contain instantiations of cataloged ontology design patterns (deliberate or otherwise), and whether design patterns might be beneficial to typical biomedical ontologies. This work is part of a much larger research program that ultimately aims to look at the prevalence of patterns in biomedical ontologies and how best to use them.
Background
Ontology vs. Terminology
ODPs, as defined, are applied to ontologies. They are used to assist in correctly representing knowledge about a domain. Many biomedical terminologies are represented via description logics, and capture knowledge about more than just a set of terms, including relationships among concepts. Therefore, in this study we also consider biomedical terminologies, which their creators may label as a terminology, but are in fact, much closer to an ontology, where ODPs are indeed useful.
OWL
The latest standard in ontology languages is OWL, which became a World Wide Web Consortium (W3C) Recommendation in October 2009. An OWL ontology is a set of axioms, or statements, that are used to describe concepts and the relationships between them in the domain of interest.11 Because OWL is based on an expressive Description Logic (i.e., decidable fragments of First Order Logic), statements made in the language have precisely defined semantics.12 Being a W3C Recommendation, there is widespread tool support, including APIs, editors, and reasoners for working with OWL ontologies. Protégé, one of the most commonly used ontology development environments in biomedicine, has native support for browsing and editing OWL ontologies.
OBO
In the world of biomedical ontologies, there is another widely used ontology language called OBO. There is a close relationship between OBO and OWL, and it is possible to translate faithfully an OBO ontology into an OWL ontology. The relationship between OBO and OWL has grown closer over the last few years, so much so, that latest OBO specification defines the semantics of the OBO language by mapping OBO syntax into OWL syntax.13 This close relationship between OBO and OWL means that all of the tools available for working with OWL ontologies are also available for working with OBO ontologies.14
Ontylog
The Ontylog language is a Description Logic based language that is used by various consortia to manage and edit their ontologies. In particular, SNOMED CT is written in Ontylog. For the sake of brevity, we do not present details of this language, however it should be noted that a description logic underpins the Ontylog language. Given these description logic underpinnings of Ontylog, it is straightforward to translate Ontylog ontologies into OWL ontologies. The SNOMED CT distribution provides a method to do just that.
Protégé-Frames
Many large biomedical ontologies have been constructed with Protégé-Frames, including the Foundational Model of Anatomy (FMA). The Protégé system now supports OWL, but has its roots as a frame based system. Frames are record like structures with slots that capture information about concepts in a domain. In contrast to Ontylog and OBO, which can essentially be considered as simple syntactic variants of OWL, the knowledge representation formalism that underpins Protégé-Frames is not logic based, and there is no (trivially) simple translation from frames into OWL. In particular, there are several non-trivial translations of the FMA into the OWL language. Therefore we only considered a particular translation of this ontology in our analysis.15
Ontology Design Patterns
Ultimately, a design pattern provides a solution to a common modeling problem. It does this by specifying what the problem is and supplies a description of how an application or domain ontology should be modified to solve the problem. Catalogs, or resources that list ontology design patterns for reference, have been created. The Manchester catalogue of ontology design patterns for bio-ontologies (MBOP) is one of two main pattern catalogues, the other being the “Ontology Design Patterns.org” (ODP-Wiki) catalogue.16 The MBOP catalogue contains a set of 17 patterns that are specifically targeted at ontologies written in OWL. The patterns in this catalogue come from the experiences of the catalog’s authors in modeling both the biomedical domain and creating ontologies written in OWL. Figure 1 shows a description of the ValuePartition design pattern from MBOP.10 The ODP-Wiki catalogue contains a growing list of patterns, and is a crowd-sourced effort to create an online ODP library. A quality committee reviews and approves or rejects submitted patterns, with approved patterns appearing in the official catalogue of patterns. As of this writing, the committee has not approved any patterns, but the website does contain 150 pattern submissions. To note, in many instances, these catalogs provide a reference ontology associated with the pattern that can either be used as an example or directly imported, depending on the type of pattern.
Figure 1.
Description of the Value Partition pattern from the Manchester Biomedical ODP catalog
Pattern Categorization
Each catalog classifies most, if not all, patterns to describe their intended use. The MBOP catalogue uses the categories, “Extension” (workarounds for language expressivity limitations), “Good Practice” (good modeling practice) and “Domain Modeling” (solutions specific to certain domains). The ODP-Wiki catalogue has a more elaborate categorization scheme, which includes, “structural”, “content”, “correspondence”, “reasoning”, “presentation”, and “lexico-syntactic”.8 These categories are explained in more detail below; however, the “structural” categorization in the ODP-Wiki catalogue nearly encompasses the complete content of the MBOP. From now on, we therefore only refer to pattern categories in terms of the ODP-Wiki catalog categories.
We provide brief explanation for the categories relevant to this work. Those we omit, including lexico-syntactic, presentation, reasoning, and correspondence, are not relevant to this work and we cannot test for their presence with only the information contained in an ontology. For instance, a lexico-syntactic ODP involves the translation of natural language into a formal concept. We cannot readily detect this type of pattern in an ontology already specified.
Structural Patterns
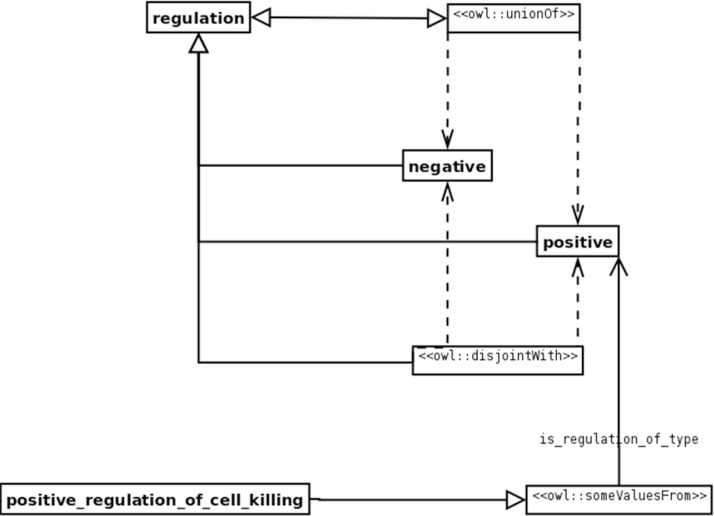
A structural pattern can be either logical, which adds a logical expressivity to an ontology language, or architectural, which defines the shape of an ontology. For example, consider the definition of cell cycle. In order to define cell cycle, we need to model cell-cycle regulation. We must specify that regulation must be either positive or negative, but not both. A “value partition” pattern captures this model: it encodes a set of values that must be disjoint with one another (i.e., only one of them applies in each particular case). Furthermore, one of the values must be present (i.e., regulation must be either positive or negative). From the MBOP, Figure 2 provides details of how one creates the value partition for regulation, using a UML-like diagram to describe the pattern.17
Figure 2.
Value partition for regulation of a cell (as presented in the Manchester ODP Catalog). Here regulation can either be positive or negative. To capture this fact in OWL, we specify the concept “regulation” as equivalent to a union of positive and negative regulation concepts. Furthermore, we specify positive and negative as disjoint, because regulation cannot be both positive and negative. Finally, we can use the pattern to create a concept positive_regulation_of_cell_killing with the value “positive” for the property is_regulation_of_type.
Content Patterns
A content ODP provides “solutions to domain-oriented problems, and are directly reusable”.8 They provide a small re-usable unit for import and use in an ontology, much like a developer utilizes libraries when writing software. An ontology developer can select a set of Content ODPs that assist in his modeling task, and import them directly using a provided small ontology, simplifying the development process. For instance, many ontologies not only have subsumption but also model partonomy. The “PartOf” content ODP captures this situation, including a top-level concept “Entity” and the two necessary transitive relations, isPartOf and hasPart. One of the main benefits of this “reuse” approach is that content patterns can act as documentation of intention, and help people begin to understand an ontology that is new to them.
Methods
Our aim was to choose a small selection of well-known biomedical ontologies and use these as case studies to determine the applicability of ODPs to biomedical ontologies. First, we selected a set of biomedical ontologies. Next, we encoded ontology design patterns from the MBOP catalogue and the ODP-Wiki. We validated the encodings by consulting an expert and verifying them against reference standards that we know to contain the pattern. Finally, we use the Ontology Pre-Processing (OPPL) Java tool to determine whether an ontology contains a pattern.
Ontology selection
In this case study, we explore the use of ODPs in well-known ontologies. To ensure a diverse sample, we selected ontologies differing in style and provenance. Table 1 lists the selected a set of ontologies designed to span these categories. We obtained the most recent version of each ontology from BioPortal, as of January 15, 2012. All ontologies are non-trivial “working” ontologies that are under active development. By non-trivial we mean that the ontology is not simply an asserted concept hierarchy, and there are relationships along other axes e.g. partOf. Because of computational and space requirements, we limited our analysis to a small set. Note, as our analysis technique is limited to OBO and OWL ontologies, we did not select terminologies similar to ICD-9 or MedRA.
Table 1.
Set of biomedical ontologies that we surveyed for ontology design patterns.
| Ontology | Acronym | Detail | Native Format | Size (terms) |
|---|---|---|---|---|
| Foundational Model of Anatomy | FMA | Explicit representation of human anatomy | Frames | 83,000 |
| Gene Ontology | GO | Description of entities related to gene products | OBO | 36,000 |
| National Cancer Institute Thesaurus | NCIt | Vocabulary for clinical care, translational and basic research, and public information and administrative activities | OWL | 89,000 |
| National Drug File – Reference Terminology | NDF-RT | Medication focused terminology | Ontylog | 40,000 |
| Ontology for Biomedical Investigations | OBI | Ontology for the description of life science and clinical investigation | OWL | 3,500 |
| Ontology for Clinical Research | OCRe | Ontology to support description of human studies and study elements | OWL | 342 |
| Radiological Lexicon | RadLex | Lexicon for indexing and retrieval of radiological resources | OWL | 35,000 |
| Systematized Nomenclature of Medicine -- Clinical Terms | SNOMED CT | Comprehensive Clinical terminology | Ontylog | 395,000 |
Pattern encoding
We encoded ontology design patterns in a format that would enable us to find them programmatically in the ontologies. From the two ODP repositories described earlier, Manchester’s Ontology Design Patterns Public Catalog and OntologyDesignPatterns.org, we encoded the patterns in OPPL. OPPL allows one to specify patterns as generic OWL axioms and variables, allowing us to determine programmatically whether an ontology contains a given pattern.18Figure 3 presents an example of the OPPL encoding of the ValuePartition. We did not encode those that were too general (such that any ontology would match the pattern) or undetectable (such as creating N-Ary relations or other re-engineering patterns). Additionally, we ignored those patterns in the OntologyDesignPatterns. org catalog that had poor reviews. Only a small subset of the patterns had reviews, and those with a majority of poor reviews were excluded. These patterns, as the poor reviews indicate, were not ready for use. Thus, encoding an incorrect/poor pattern would have been especially difficult, and interpreting the result (present or absent) ambiguous. 69 of the 167 patterns from the combined catalogs remained after filtering. Because of the large number of patterns, we only provide a table of the patterns we found in the results.
Figure 3.
Generic OPPL encoding of value partition query. For the regulation example, ?v1 and ?v2 would correspond to “Negative” and “Positive” respectively. ?param would correspond to “regulation”.
Filter Relations
To reduce running time, where possible, we pruned an ontology-pattern pair from our analysis by first checking whether or not specific named relations that a pattern requires are present in the ontology at all. This is possible for the Content ODPs, as their utilization requires the exact specification of the pattern (it is imported), not just the intention of the pattern. For example, the PartOf pattern from the OntologyDesignPatterns.org catalog requires the relations “isPartOf” and “hasPart”. Thus, if an ontology does not have both of these relations, we do not need to check this ontology for the PartOf pattern as it cannot possibly have this pattern without having these two relations.
Finally, we were unable to encode some patterns in OPPL. For example, the ValuePartition pattern may have an arbitrary number of values, instead of just two as in the regulation example (“Positive” and “Negative”). OPPL cannot yet model this type of pattern. For such patterns, we directly encoded them in Java using the OWL-API.19
Normalization
A feature of OWL is its amount of syntactic redundancy in the language. For example, A EquivalentTo B is an abbreviation for A SubClassOf B, and B SubClassOf A. Similarly, the class expression hasPart min 1 Hand can be written as hasPart some Hand. On a more basic level, because OWL allows the nesting and bracketing of concepts, extra-logical syntax such as brackets may mean that two class expressions are not structurally equivalent, but are trivially semantically equivalent. For example, (A and (B and C)) is equivalent to ((A and (B)) and C). Because one ontology engineer may use one style of syntax, and another engineer a different style of syntax, it is conceivable that a design pattern may be encoded in a slightly different syntax to the syntax used in the specification of the pattern. When detecting patterns we therefore decided to create a canonical representation of each ontology to ensure that we do not miss a pattern for trivial reasons. To perform normalization we used the common description logic rewritings shown in Table 2. In essence, we decompose axioms to smaller axioms by breaking conjunction on the right hand side of axioms and disjunction on the left hand side of axioms, and rewrite class expressions. The transformation rules were applied exhaustively until no new transformations result. Instead of performing normalization, a reasoner might be used to infer these equivalences. However, using a reasoner was not computational feasible given the size of the selected ontologies.
Table 2.
Transformation rules for normalizing an ontology, provided in the Manchester OWL syntax
| Axiom | Transformation |
|---|---|
| prop min 1 C | prop some C |
| prop exactly n C | prop min n C, prop max n C |
| prop value i | prop some {i} |
| Property in Anonymous Class | Simplify Property (Removing inverses) and re-insert |
| C1 and (C2 and C3) | C1 and C2 and C3 |
| C1 or (C2 or C3) | C1 or C2 or C3 |
| C1 EquivalentTo C2 | C1 SubClassOf C2, C2 SubClassOf C1 |
| C1 DisjointUnionOf C2 … Cn | DisjointClasses: C2 … Cn C1 EquivalentTo (C2 … Cn) |
| C1 or … or Cn SubClassOf D1 and … and Dn |
C1 SubClassOf D1 … Cn SubClassOf D1 … C1 SubClassOf Dn … Cn SubClassOf Dn |
| DisjointClasses: C1 … Cn | Ci DisjointWith Cj for 1 <= i < j <=n |
Query & Computation
After creating a corpus of ontologies and a library of patterns, we checked each ontology against the encoded patterns using the OPPL Java library. We parallelized our analysis to a Sun Grid Engine cluster of 20 machines each with 24 cores and 96GB of RAM. We pruned patterns-ontology pairs from analysis by OPPL where possible by requiring all terms in the pattern to exist in an ontology. We performed the pruning using the alpha version of the BioPortal SPARQL endpoint. The BioPortal SPARQL endpoint provides access to a uniform encoding of all BioPortal ontologies in a triplestore and the lookup of a particular named relation is extremely efficient. Without the pruning and grid computation, the analysis may have taken nearly a week to complete on just this particular set of ontologies.
Validation
Because the step of generating an OPPL representation for a pattern involves manual interpretation of its description, we must validate the veracity of our representation, to ensure that we not only specify the pattern correctly, but also do not overspecify or underspecify it. We might underspecify a pattern if we miss certain critical constraining condition and therefore the pattern becomes too general, leading to additional matches than are true. We might overspecify it if we add too many (incorrect) constraints, causing us to find more negatives than are truly negative.
We validated the pattern encodings using two methods: (a) against a reference standard, and (b) by consulting expert. Specifically, the Manchester catalog provided reference OWL ontologies that each contains an instantiation of a pattern from the catalog. Thus, after encoding the patterns from this catalog, we ran our computation over these reference ontologies to ensure that we find the patterns that they are supposed to have. Additionally, we asked an OWL expert, one of us (MH), to validate manually all patterns by inspection.
Of the 69 patterns, we encoded 10 in OPPL and 5 directly in Java. We pruned 53 patterns from the analysis because none of the ontologies contained any of the terms required by the patterns. These patterns were Content ODPs. We checked for an Upper Level Ontology using a similar technique to pattern pruning, where the ontology must instead contain the necessary concepts of the Upper Level Ontology.
Results
Table 3 lists the patterns that we found for each ontology. Table 4 presents further details about the patterns that we found. In total, we tested 69 patterns and found five in the set of eight ontologies. SNOMED CT and OBI contained these patterns.
Table 3.
Patterns that we found in the sample set of ontologies. Note that the table does not show any of the patterns that we did not find in any of the ontologies.
| NDF-RT | SNOMED CT | GO | FMA | RadLex | OBI | OCRe | NCIT | |
|---|---|---|---|---|---|---|---|---|
| Composite Property Chaining | □ | |||||||
| Closure | □ | |||||||
| Defined Class Description | □ | |||||||
| Value Partition | □ | |||||||
| Upper Level Ontology | ∼ | □ | ∼ |
Table 4.
Formal pattern descriptions for those patterns we found in the sample set of ontologies.
| Pattern | Description | Source |
|---|---|---|
| Composite Property-Chaining | Models a double chain of properties. Example Chain: one has a father and father has brother, therefore one has uncle. Additionally, uncle has son, therefore one has a cousin. | Manchester |
| Closure | Simulates the closed world assumption in a concrete concept. Example use: One eats some vegetables and only vegetables. | Manchester |
| Defined Class Description | Represents an If-Then structure in OWL. If something fulfills certain necessary and sufficient conditions, then it is given additional attributes. | Manchester |
| Value Partition | Provides a way to enumerate attributes of an element where each attribute is disjoint from all others. Additionally, the values an element can take are constrained | Manchester |
| Upper Level Ontology | Uses an upper level ontology. Examples include BFO20, SUMO21, DOLCE22. | Manchester |
OBI contains the most patterns, four, followed by SNOMED CT containing only one. No other ontology in the set contained any of the patterns that we encoded. We did not confirm with either SNOMED CT or OBI developers if they intentionally selected such patterns, or developed them independently of the available resource catalogs. However, we assume the presence of such patterns was purposeful. SNOMED CT uses the composite property-chaining pattern to model transitivity of “direct substances” and “active ingredients.” Any active ingredient of a direct substance of a drug also becomes a direct substance of the drug.
OBI contains four patterns: “Closure”, “Defined Class Description”, “Value Partition”, and “Upper Level Ontology”. OBI uses the closure pattern to restrict the values a property that a concept may have. By using defined class description, OBI adds more properties to a concept after reasoning. For example, the concept “Environment Control Device” is equivalent to “has_function some (heat function or cool function or environment control function)”. Thus, anything that has the property that is either “heat function” or “cool function” or “environment control function” will gain the additional properties of “Environment Control Device.” Most value partitions in OBI are in the Basic Formal Ontology (BFO) that it imports. These partitions specify disjoint sets of items, in a similar fashion to the earlier example. With regard to upper level ontologies, OBI imports and uses the BFO, the OBO upper level ontology. 20 RadLex and OCRe have a few concepts from BFO, but do not truly import BFO as an upper level ontology. Note, in this study we did not check for correct use of a pattern, only that it was in use.
Discussion
Two ontologies have documented patterns, OBI and SNOMED CT, and five of the 69 documented patterns exist in these ontologies. We believe that there are several reasons for this relative dearth of patterns in the ontologies that we studied.
Structural Patterns vs. Content Patterns
All of the pattern types that were found were structural rather than content patterns. Recall that structural patterns require some kind of convention or template to be followed in order to instantiate a pattern, while content patterns require the small ontology that represents the pattern to be included in the ontology that the pattern will be instantiated in. None of the ontologies in our case study import or include any of these content pattern ontologies, hence, none of our case study ontologies contained instances of content patterns. Thus, content patterns themselves, in particular the patterns in the OntologyDesignPatterns.org repository, may be too specific to appear in the ontologies. We filtered nearly 80% of the patterns, including all of the patterns from that catalogue, such as Componency, because they required the use of specific properties. These patterns are “Content ODPs,” which model a specific situation, and thus require specific named relationships. In fact, a cursory examination of these ontologies reveals that in some cases they could easily be adapted to incorporate these patterns. Of course, not all content ODPs will be relevant to any given ontology.
While the specific nature of Content ODPs readily explain our result, pattern developers might consider more generalizable ways of specifying the patterns to capture the intention of the pattern, such as identifying synonyms or equivalent ontology relations and concepts. We might also enhance our analysis to include an alignment step where we identify that a property in an ontology (e.g., has-Component) is equivalent to a property in the pattern (e.g., has-Part). However, the point at which to stop aligning is unclear (i.e., aligning at the level of the text string or the meaning). Furthermore, checking against a slightly different representation of a pattern may be counter to the idea of formally specifying a pattern for reuse.
The Effects of Tooling on Pattern Application
We posit that the lack of tool support for using the patterns might contribute to low ODP use in the biomedical ontologies. Indeed, when a development environment supports instantiation of certain patterns, we observe that ontology authors make use of these features. For instance, OBI, developed in Protégé, contains the closure pattern. The concept “scattered molecular entity” is specified that it can have grain of a “molecular entity.” OBI then closes the “has grain” property for the concept to only take values from “molecular entity.” A developer can add this pattern in Protégé with just two clicks of the mouse. However, most of the methods and tooling for pattern reuse that exists today is not designed for domain experts to use. While there is a Protégé plugin for OPPL, a domain expert using Protégé may not know when or how to use such a tool. An expert must first know about ontology design patterns and OPPL. Next, he must understand the OPPL query language, which is something more akin to writing in a programming language than working with a graphical user interface. OPPL is useful for the appropriate audience; however, most domain experts are unable, and not expected, to meet such requirements. To the best of our knowledge, only the NeOn toolkit directly supports the use of content design patterns. Neither OBO-Edit nor other widely used tools such as Protege-OWL provide direct support for instantiating the majority of patterns. Therefore, wider availability of tools and their ease of usability might encourage wider use of ontology design patterns in biomedical ontologies. Educating domain experts who are creating biomedical ontologies about ODPs may also increase their use.
Time of Creation
We expect that those ontologies created before the introduction of ODPs may not contain them, especially the explicit Content ODPs. However, FMA serves as an example where the intention of a pattern is still present.
Native Formats
There is no trivial translation from Frames to OWL. In this work, we used the FMA translation found in BioPortal. We suggest that these translation methods affect the patterns that may appear through our analysis. A Content ODP would not be present unless it was explicitly defined during the translation process. In fact, upon closer examination, FMA does model partonomy and therefore captures the intention of the PartOf pattern (i.e., it models the same conceptualization as the Content pattern PartOf), yet the ODP-Wiki pattern is not explicitly present.
Finally, the applicability of a pattern also explains the low prevalence. That is, not every pattern is appropriate for every ontology. Though not every ontology must use a pattern, many of the patterns available in MBOP are not domain specific, and ensure one properly represents knowledge, thus we still expect these patterns to be in use. As such, using these “best practice” patterns may serve as one indicator of ontology quality, especially when used properly, but their use alone cannot be used to judge ontology quality.
Future work
We plan to perform an exhaustive evaluation of all the ontologies in the BioPortal repository and the encoded design patterns. Furthermore, we plan to explore patterns in a bottom-up approach. Gamma and colleagues published a list of extremely successful software design patterns that they found manifested in real world software.9 The authors abstracted these patterns from real world code examples, noting that the patterns recurred across systems. We propose that there are recurring implicit modeling techniques across ontologies, potentially new or differing from the documented patterns. We plan to capture such implicit patterns, in a bottom-up approach, and evaluate their usefulness to ontology developers.
Conclusion
In this study, we provided a case study on the use of ontology design patterns in biomedical ontologies. While we cannot generalize these results to all biomedical ontologies, we gave a flavor of where and how patterns are used, and how this might relate to the style and provenance of an ontology. We found very few patterns in the selected ontology subset. We suggest that pattern prevalence is related to tooling support.
Acknowledgments
This work was supported by the NLM Grant 1R01GM086587-01. We are extremely grateful to Luigi Iannone for his support with OPPL.
References
- 1.Bodenreider O, Stevens R. Bio-ontologies: current trends and future directions. Briefings in bioinformatics. 2006;7(3):256–74. doi: 10.1093/bib/bbl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noy NF, Shah NH, Whetzel PL, Dai B, Dorf M, Griffith N, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Research. 2009 May 29;37:W170–W173. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khatri P, Drăghici S. Ontological analysis of gene expression data: current tools, limitations, and open problems. Bioinformatics. 2005;21(18):3587–95. doi: 10.1093/bioinformatics/bti565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records. New England Journal of Medicine. 2010;363(6):501–4. doi: 10.1056/NEJMp1006114. [DOI] [PubMed] [Google Scholar]
- 5.Carter JS, et al. Initializing the VA medication reference terminology using UMLS metathesaurus cooccurrences. Proceedings of the AMIA Symposium; 2002. p. 116. [PMC free article] [PubMed] [Google Scholar]
- 6.Rector AL, Brandt S, Schneider T. Getting the foot out of the pelvis: modeling problems affecting use of SNOMED CT hierarchies in practical applications. Journal of the American Medical Informatics Association. 2011 Apr 21;18(4):432–40. doi: 10.1136/amiajnl-2010-000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceusters W, Smith B, Goldberg L. A terminological and ontological analysis of the NCI Thesaurus. Methods of Information in Medicine. 2005;44(4):498. [PubMed] [Google Scholar]
- 8.Gangemi A, Presutti V. Ontology Design Patterns. In: Staab S, Rudi Studer D, editors. Handbook on Ontologies. Springer Berlin; Heidelberg: 2009. pp. 221–43. [Google Scholar]
- 9.Gamma E, Helm R, Johnson R, Vlissides J. Design patterns: elements of reusable object-oriented software. Pearson Education; 1995. [Google Scholar]
- 10.Aranguren ME, Antezana E, Kuiper M, Stevens R. Ontology Design Patterns for bio-ontologies: a case study on the Cell Cycle Ontology. BMC Bioinformatics. 2008 Apr 29;9(Suppl 5):S1. doi: 10.1186/1471-2105-9-S5-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.W3C OWL Working Group OWL 2 Web Ontology Language Document Overview. 2009. Oct, W3C.
- 12.Motik B, Parsia B, Patel-Schneider PF. OWL 2 Web Ontology Language Structural Specification and Functional-Style Syntax. 2009. Oct, W3C.
- 13.OBO Flat File Format 1.4 Syntax and Semantics [DRAFT]. Available from: ftp://ftp.geneontology.org/go/www/obo-syntax.html
- 14.Re: How do OBO ontologies work on the LOD? from Alan Ruttenberg on 2012-02-23 (public-lod@w3.org from February 2012)
- 15.Noy NF, Musen MA, Mejino JLV, Rosse C. Pushing the envelope: challenges in a frame-based representation of human anatomy. Data & Knowledge Engineering. 2004;48(3):335–59. [Google Scholar]
- 16.Daga E, Presutti V, Gangemi A, Salvati A. http://ontologydesignpatterns.org [ODP]
- 17.Aranguren ME. Manchester ODP Catalog, Value Partition Example [Internet] Available from: http://www.gong.manchester.ac.uk/odp/html/Value_Partition.html.
- 18.Iannone L, Rector A, Stevens R. Embedding Knowledge Patterns into OWL. In: Aroyo L, Traverso P, Ciravegna F, Cimiano P, Heath T, Hyvönen E, et al., editors. The Semantic Web: Research and Applications. Berlin, Heidelberg: Springer Berlin Heidelberg; 2009. [Google Scholar]
- 19.Horridge M, Bechhofer S. The OWL API: A Java API for OWL ontologies. 2011 Jan 1;2(1):11–21. Semantic Web. [Google Scholar]
- 20.Grenon P, Smith B, Goldberg L. Biodynamic Ontology: Applying BFO in the Biomedical Domain. STUD. HEALTH TECHNOL. INFORM. 2004;102:20–38. [PubMed] [Google Scholar]
- 21.Niles I, Pease A. Towards a standard upper ontology. Proceedings of the international conference on Formal Ontology in Information Systems - Volume 2001; New York, NY, USA: ACM; 2001. [Google Scholar]
- 22.Gangemi A, Guarino N, Masolo C, Oltramari A, Schneider L. Sweetening Ontologies with DOLCE. Lecture notes in computer science. 2002;(2473):166–81. [Google Scholar]