Abstract
Clinical Natural Language Processing (NLP) systems extract clinical information from narrative clinical texts in many settings. Previous research mentions the challenges of handling abbreviations in clinical texts, but provides little insight into how well current NLP systems correctly recognize and interpret abbreviations. In this paper, we compared performance of three existing clinical NLP systems in handling abbreviations: MetaMap, MedLEE, and cTAKES. The evaluation used an expert-annotated gold standard set of clinical documents (derived from from 32 de-identified patient discharge summaries) containing 1,112 abbreviations. The existing NLP systems achieved suboptimal performance in abbreviation identification, with F-scores ranging from 0.165 to 0.601. MedLEE achieved the best F-score of 0.601 for all abbreviations and 0.705 for clinically relevant abbreviations. This study suggested that accurate identification of clinical abbreviations is a challenging task and that more advanced abbreviation recognition modules might improve existing clinical NLP systems.
Introduction
Narrative text is the primary form of communication in the clinical domain. Unstructured clinical texts contain rich patient information, but are not immediately accessible to any clinical application systems that require structured input. Thus, a great interest has developed regarding clinical Natural Language Processing (NLP) methods. Various clinical NLP systems can now extract information from clinical narrative text to facilitate patient care and clinical research1,2.
MetaMap3,4 is a general biomedical NLP system originally developed to map biomedical literature (e.g., MEDLINE abstracts) to concepts in the Unified Medical Language System (UMLS) Metathesaurus. It was originally developed by Aronson et al. at National Library Medicine in 2002 and is distributed freely.3 Many researchers have used MetaMap to extract information from clinical text.5–8 Meystre and Haug5 evaluated MetaMap NLP performance for extracting medical problems from patient electronic medical records and reported a recall of 0.74 and a precision of 0.76. Schadow and McDonald6 similarly studied extraction of structured information from pathology notes using MetaMap. Chung and Murphy7 used MetaMap to identify concepts and their associated values from narrative echocardiogram reports; they reported a recall of 78% and precision of 99% for ten targeted clinical concepts.
Developed by Carol Friedman et al. at Columbia University, the Medical Language Extraction and Encoding System (MedLEE)9 is one of the earliest and most comprehensive clinical NLP systems. Originally designed to process radiology notes9, MEDLEE was later extended to cover various clinical sub-domains, such as pathology reports10 and discharge notes11. In addition, its application to many clinical problems showed promising results – e.g., adverse events detection,12 automated trend discovery,13 acquisition of disease drug knowledge.14
The cTAKES15 system is an open-source comprehensive clinical NLP system developed based on the Unstructured Information Management Architecture (UIMA) framework and the OpenNLP natural language processing toolkit. The cTAKES system design is pipeline-based, consisting of different modules that include a sentence boundary detector, tokenizer, normalizer, part-of-speech tagger, shallow parser, and named entity recognizer. A recent study used cTAKES to retrieve clinical concepts from the radiology reports, reporting F-scores of 79%, 91%, and 95% for the presence of liver masses, ascites, and varices, respectively.16 Other studies also used cTAKES for different tasks, such as determining patient smoking status and extracting medication information.15,17–19 Other groups have developed many alternative approaches to clinical NLP systems and applied them to different information extraction tasks. For example, the KnowledgeMap Concept Identifier (KMCI)20 is a general NLP system that can map clinical documents to concepts in UMLS. The Pittsburgh SPIN (Shared Pathology Informatics Network) information extraction system21 and the Harvard HITEx (Health Information Text Extraction) system22 were developed based on the GATE (General Architecture for Text Engineering) framework to extract specific information. Two review papers provide an excellent summary of current clinical NLP systems1,2.
A unique characteristic of clinical text is its pervasive use of abbreviations, including acronyms (e.g., “pt” for “physical therapy” or “prothrombin time”) or shortened terms (e.g., “abd” for “abdominal”). Studies have shown that confusion caused by the frequently changing and highly ambiguous abbreviations can impede effective communication among healthcare providers and patients23–25, potentially diminishing healthcare quality and safety. Moreover, abbreviations also present additional challenges for clinical NLP systems. There are two main challenges for NLP handling of abbreviations. First, no comprehensive clinical abbreviation database exists that contains all clinical abbreviations and their possible senses. One study showed that the UMLS and its derivative abbreviations only covered about 67% of abbreviations in typed hospital admission notes.26 The second challenge is lack of an accepted approach to resolve the ambiguities associated with abbreviations. While many Word Sense Disambiguation (WSD) applications exist in the general English NLP domain,27 relatively few studies have focused on abbreviation disambiguation in clinical texts. Recently, supervised machine learning methods have received considerable attention for NLP applications and have shown good results.28,29
Despite identification of major challenges and possible solutions regarding NLP handling of clinical abbreviations, few studies quantitate how well existing clinical NLP systems handle abbreviations. A related problem is lack of well-defined methods to integrate abbreviation recognition and disambiguation with clinical systems. In this study, we took a set of 32 de-identified patient discharge summaries and manually identified and annotated 1,112 abbreviations and their senses within them. We then compared the performance of three well-known clinical NLP systems: MetaMap, MedLEE and cTAKES on handling abbreviations in our data set. Our evaluation showed that existing clinical NLP systems had relative low performance on identifying clinical abbreviations, suggesting the need for advanced abbreviation recognition modules. The programs that we developed for evaluating three NLP systems also provide a useful post-processing mode for future integration of abbreviation disambiguation methods with existing NLP systems.
Method
We constructed a data set containing 1,112 sense-annotated abbreviations from 32 discharge summaries via manual chart review. Then we processed the 32 discharge summaries using the above three clinical NLP systems. A postprocessing program was developed to parse the system’s output and map the extracted concepts back to original terms. Finally, concepts associated with the annotated abbreviations by each NLP system were collected and presented in front of two domain experts to decide if an abbreviation was identified and interpreted correctly by the system. We report coverage, precision, recall, and f-score for each of the three evaluated systems.
Preparing the dataset
In this study, we used clinical documents taken from Vanderbilt Medical Center’s Synthetic Derivative (SD) database30, a de-identified copy of Vanderbilt Electronic Medical Records (EMRs) for research purpose. Thirty-two discharge summaries were randomly selected from those created in the year 2006. The study was approved by the Vanderbilt Institutional Review Board. Three physicians (authors JD, STR, RAM) manually annotated all abbreviations in the 32 discharge summaries and recorded their appropriate senses, which served as the gold standard for the evaluation. The annotations were also used in our previous research of detecting abbreviations 31.
Running NLP systems and parsing their outputs
Three existing clinical NLP systems, including MetaMap, MedLEE, and cTAKES, were used to process these 32 discharge summaries. The ‘-y’ option was used for running MetaMap to open the WSD function. The default settings were used for MedLEE and cTAKES. For each NLP system, we developed a post-processing parser program to align the system output with the original clinical text. Thus, every clinical concept detected by each NLP systems was mapped to the corresponding abbreviation in the original text.
Evaluating NLP system Output
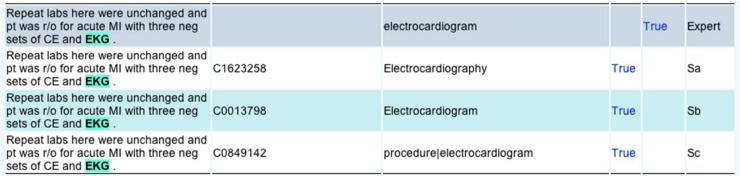
For each NLP system, the extracted concepts (CUI or terms) associated with annotated abbreviations were collected and presented in a web-based annotation interface. Figure 1 shows an example of the annotation interface. The first row shows the manually annotated abbreviation and its sense from expert. In the fifth column (counted from index 1) of row 1, the expert was asked to determine whether the abbreviation was clinically relevant or not. We defined an exclusion list for determining clinical relevance. An abbreviation is annotated as not clinically relevant if it belongs to the following categories, including abbreviations from general English, e.g., ‘mr’ for ‘mister’ and ‘am’ for ‘in the morning’; professionals such as ‘pcp’ for ‘primary care provider’ and ‘md’ for ‘medical doctor’; and location/department names, e.g., ‘vumc’ for ‘Vanderbilt University Medical Center’. In Figure 1, the “True” link on the top-right corner shows that this abbreviation was annotated as clinically relevant.
Figure 1.
An example from the annotation interface
Rows 2–4 shows the outputs from three NLP systems, whose names were not shown. In each row, an abbreviation detected by one NLP system was highlighted and the extracted CUIs and terms were displayed. A domain expert manually compared the NLP system’s extracted concept with the annotated sense in row 1 and decided if two concepts were the same. The “True” labels in rows 2–4 of figure 1 denote that the detected sense is a true positive according to experts’ judgment.
When judging the equivalence between the system-detected concept and the expert annotated sense, the experts considered the variations, synonyms, and more specific concepts. For example, for the sentence “MRI brain, EEG”, the experts’ annotation was “Magnetic resonance image”. But the NLP-detected concept was a lexical variant “Magnetic Resonance Imaging”, which would be determined as correct. Table 1 shows some examples of variance between expert-entered senses and system-detected concepts.
Table 1.
Examples of variance between expert-entered senses and system-detected concepts. The physician reviewers interpreted each of these system outputs as “correct”.
| Sentence | Experts entered sense | System-detected concept |
|---|---|---|
| CT scan of the abdomen was … | Computer assisted tomography | CT of abdomen X-Ray Computed Tomography |
| … he was seen by Dr. … | Doctor | Physicians |
| GAF 50 ; highest 60 | Global Assessment of Functioning | Global assessment |
| HTN | Hypertension | Hypertensive disease |
| … recent CEA, CAD, HTN | Coronary artery disease | Coronary heart disease |
| MRI brain, EEG | Magnetic resonance image | Magnetic Resonance Imaging MRI brain procedure |
Evaluation
Based on the expert physicians’ judgments, we reported each NLP system’s coverage, precision, recall, and F-score. Coverage was defined as the ratio between the number of abbreviations detected by the NLP system (not necessary correct) and the number of abbreviations in gold standard. Precision is defined as the ratio between the number of abbreviations detected with correct sense and the total number of detected abbreviations by the system. Recall is defined as the ratio between the number of abbreviations detected with correct sense and the total number of abbreviations in gold standard. F-score is calculated as: 2*Precision*Recall/(Precision+Recall).
We reported systems’ performance at two levels: 1) All abbreviations (ALL): we calculated the performance on all abbreviations, irrespective whether the abbreviation was clinically relevant or not; and 2) Clinically relevant abbreviations (CLINICAL): we only considered the abbreviations that were marked as clinically relevant. In addition, we searched all annotated abbreviations and identified a set of ambiguous abbreviations, which had two or more different senses according to experts’ annotation. We then reported the performance of three systems on those ambiguous abbreviations.
Results
The experts’ annotations of the 32 discharge summaries identified 1,112 occurrences of abbreviations, where 855 occurrences were labeled as clinically relevant. The 1,112 occurrences were contributed by 332 unique abbreviations, with 275 designated clinically relevant. On average, there were 35 abbreviations in each discharge summary. Among all 332 unique abbreviations, 16 of them were ambiguous, which contributed to 229 occurrences.
Table 2 shows the performance of three systems on handling all abbreviations in the 32 discharge summaries. MetaMap recognized most abbreviations (N=599), and thus reached a highest coverage of 0.539. MedLEE achieved a high precision of 0.927, and also had the best recall (0.445) and F-score (0.601). When only clinically relevant abbreviations were included in the evaluation, all three systems showed improved performance (see Table 3): MetaMap (F-score 0.338 ➔ 0.350), MedLEE (F-score 0.601 ➔ 0.705), and cTAKES (F-score 0.165 ➔ 0.213), among which MedLEE achieved a highest F-score of 0.705. Table 4 shows the results on the 16 ambiguous abbreviations only. Both MetaMap and cTAKES performed worse on ambiguous abbreviations, especially for cTAKES (F-score 0.165 ➔ 0.03). MedLEE’s F-score, however, was slightly increased.
Table 2.
Performance of MetaMap, MedLEE, and cTAKES for all abbreviations.
| NLP system | #ALL | #Detected | #Correct | Coverage | Precisio | Recall | F-score |
|---|---|---|---|---|---|---|---|
| MetaMap | 1,112 | 599 | 289 | 0.539 | 0.482 | 0.260 | 0.338 |
| MedLEE | 1,112 | 534 | 495 | 0.480 | 0.927 | 0.445 | 0.601 |
| cTAKES | 1,112 | 452 | 129 | 0.406 | 0.285 | 0.116 | 0.165 |
Table 3.
Performance of MetaMap, MedLEE, and cTAKES for clinically relevant abbreviations
| NLP system | #ALL | #Detected | #Correct | Coverage | Precision | Recall | F-score |
|---|---|---|---|---|---|---|---|
| MetaMap | 855 | 452 | 229 | 0.529 | 0.507 | 0.268 | 0.350 |
| MedLEE | 855 | 501 | 478 | 0.586 | 0.954 | 0.560 | 0.705 |
| cTAKES | 855 | 316 | 125 | 0.370 | 0.400 | 0.146 | 0.213 |
Table 4.
Performance of MetaMap, MedLEE, and cTAKES for 16 ambiguous abbreviations
| NLP system | #ALL | #Detected | #Correct | Coverage | Precision | Recall | F-score |
|---|---|---|---|---|---|---|---|
| MetaMap | 229 | 108 | 50 | 0.472 | 0.463 | 0.218 | 0.297 |
| MedLEE | 229 | 142 | 135 | 0.620 | 0.951 | 0.590 | 0.728 |
| cTAKES | 229 | 166 | 6 | 0.725 | 0.036 | 0.026 | 0.030 |
Discussion
This study evaluated three existing clinical NLP systems – MetaMap, MedLEE, and cTAKES – for their ability to handle abbreviations in a random sample of 32 discharge summaries. Our evaluation showed that, overall, existing NLP systems did not perform very well on clinical abbreviation identification. MedLEE achieved a highest F-score of 0.601 for all abbreviations and showed better performance than MetaMap and cTAKES. Although the MetaMap system was originally designed for biomedical literature, it showed reasonable performance and was better than cTAKES, which was developed for clinical text. One possible reason is that the Mayo Clinic corpus, for which cTAKES was developed, contained primarily dictated clinical text, which likely contained fewer abbreviations. The performance difference between MedLEE and MetaMap/cTAKES can actually be explained by their system architectures. MedLEE integrates an abbreviation lexicon and implements a set of rules for word sense disambiguation. However, there is no similar component for handling abbreviations in the current version of MetaMap and cTAKES, to the best of our knowledge. This finding indicates that it is necessary to integrate abbreviation recognition and disambiguation components with clinical NLP systems to further improve their performance.
The low abbreviation coverage of three clinical NLP systems suggests that recognition of abbreviated terms is a major problem for correctly handling abbreviations in clinical text. Our previous study26 also demonstrated that clinical abbreviations are dynamic and many are created locally by healthcare providers, which makes it difficult to maintain an up-to-date resource of clinical abbreviations. Existing knowledge bases in the medical domain such as the UMLS do not have good coverage on clinical abbreviations. Therefore, methods to automatically detect abbreviations from corpus31 would be very appealing.
Disambiguation is another major issue associated with clinical abbreviation. cTAKES does not implement any disambiguation modules. It just outputs all possible senses of an ambiguous abbreviation, as described in the paper15 “We investigated supervised ML Word Sense Disambiguation and concluded that it is a combination of features unique for each ambiguity that generates the best results suggesting potential scalability issues. The cTAKES currently does not resolve ambiguities”. MetaMap has a disambiguation component, but it was developed for ambiguous terms in the literature and it is not so applicable to clinical text. MedLEE applies a set of rules to disambiguate terms and its current performance on ambiguous abbreviations was 0.73 (F-score), which is reasonable. However, we expect such modules could be further improved by integrating more advanced abbreviation recognition and disambiguation methods such as the abbreviation disambiguation work from Pakhomov et al.32 In this study, we have developed post-processing scripts to parse outputs from these three NLP systems. We plan to integrate them with external abbreviation modules that we have developed33 and make them as individual wrappers for each NLP system for public uses. In future work, we also intend to evaluate the locally-developed KnowledgeMap Concept Indexer, which does perform word-sense disambiguation, as well.
This study has limitations. The results reported here may not reflect the optimized performance for each NLP system. For example, MetaMap can incorporate user supplied abbreviations lists to further improve its ability to recognize abbreviations. cTAKES can specify additional lexicon files as well. However, no additional abbreviation lists were used for any system in the study. For MedLEE, the current method for measuring recall is not optimal. MedLEE extracts clinically important concepts only. For example, if the abbreviation “pt” refers to “patient” in a sentence, MedLEE does not output anything even it recognizes its meaning, because MedLEE does not define “patient” as an important clinical concept. But if the abbreviation “pt” refers to “physical therapy” in another sentence, MedLEE will recognize and extract it correctly. In clinical text, “pt” as “patient” has frequent occurrences, which largely decreased its recall as they were treated as false negatives of the system. Therefore, the performance of these NLP systems would be better if they are evaluated for specific information extraction tasks. In addition, a single expert did abbreviation sense annotation and the manual judgment; therefore bias may exist in the annotation. In the future, we plan to recruit more annotators and report the inter-annotator agreement between annotations.
Conclusion
In this paper, we conducted a comparative study of three current clinical NLP systems on handling abbreviations in discharge summaries. Evaluation using a set of expert-annotated abbreviations from discharge summaries showed that MedLEE achieved a reasonable F-score of 0.601, while MetaMap and cTAKES had relatively low performance. This study suggested that accurate identification of clinical abbreviations is a challenging task and advanced abbreviation recognition modules are needed for existing clinical NLP systems.
Acknowledgments
This study was supported by grant from the National Library Medicine R01LM010681. We also thank Dr. Alan Aronson and Dr. Carol Friedman for their support on running MetaMap and MedLEE. The datasets used were obtained from Vanderbilt University Medical Center’s Synthetic Derivative, which is supported by institutional funding and by the Vanderbilt CTSA grant 1UL1RR024975-01 from NCRR/NIH.
References
- 1.Meystre SM, Savova GK, Kipper-Schuler KC, Hurdle JF. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform. 2008:128–144. [PubMed] [Google Scholar]
- 2.Nadkarni PM, Ohno-Machado L, Chapman WW. Natural language processing: an introduction. Journal of the American Medical Informatics Association : JAMIA. 2011 Sep-Oct;18(5):544–551. doi: 10.1136/amiajnl-2011-000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 4.Aronson AR, Lang FM. An overview of MetaMap: historical perspective and recent advances. Journal of the American Medical Informatics Association : JAMIA. 2010 May-Jun;17(3):229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meystre S, Haug PJ. Natural language processing to extract medical problems from electronic clinical documents: performance evaluation. Journal of biomedical informatics. 2006 Dec;39(6):589–599. doi: 10.1016/j.jbi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Schadow G, McDonald CJ. Extracting structured information from free text pathology reports. Proc AMIA Symp; 2003. pp. 584–588. [PMC free article] [PubMed] [Google Scholar]
- 7.Chung J, Murphy S. Concept-value pair extraction from semi-structured clinical narrative: a case study using echocardiogram reports. Proc AMIA Symp; 2005. pp. 131–135. [PMC free article] [PubMed] [Google Scholar]
- 8.Friedlin J, Overhage M. An evaluation of the UMLS in representing corpus derived clinical concepts. Proc AMIA Symp; 2011. pp. 435–444. [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. Journal of the American Medical Informatics Association : JAMIA. 1994 Mar-Apr;1(2):161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu H, Fu Z, Shah A, et al. Extracting and integrating data from entire electronic health records for detecting colorectal cancer cases. Proc AMIA Symp; 2011. pp. 1564–1572. [PMC free article] [PubMed] [Google Scholar]
- 11.Chiang JH, Lin JW, Yang CW. Automated evaluation of electronic discharge notes to assess quality of care for cardiovascular diseases using Medical Language Extraction and Encoding System (MedLEE) Journal of the American Medical Informatics Association : JAMIA. 2010 May-Jun;17(3):245–252. doi: 10.1136/jamia.2009.000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melton GB, Hripcsak G. Automated detection of adverse events using natural language processing of discharge summaries. Journal of the American Medical Informatics Association : JAMIA. 2005 Jul-Aug;12(4):448–457. doi: 10.1197/jamia.M1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hripcsak G, Austin JH, Alderson PO, Friedman C. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology. 2002 Jul;224(1):157–163. doi: 10.1148/radiol.2241011118. [DOI] [PubMed] [Google Scholar]
- 14.Chen ES, Hripcsak G, Xu H, Markatou M, Friedman C. Automated acquisition of disease drug knowledge from biomedical and clinical documents: an initial study. Journal of the American Medical Informatics Association : JAMIA. 2008 Jan-Feb;15(1):87–98. doi: 10.1197/jamia.M2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association : JAMIA. 2010 Sep-Oct;17(5):507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garla V, Lo Re V, 3rd, Dorey-Stein Z, et al. The Yale cTAKES extensions for document classification: architecture and application. Journal of the American Medical Informatics Association : JAMIA. 2011 Sep-Oct;18(5):614–620. doi: 10.1136/amiajnl-2011-000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savova GK, Fan J, Ye Z, et al. Discovering peripheral arterial disease cases from radiology notes using natural language processing. Proc AMIA Symp; 2010. pp. 722–726. [PMC free article] [PubMed] [Google Scholar]
- 18.de Bruijn B, Cherry C, Kiritchenko S, Martin J, Zhu X. Machine-learned solutions for three stages of clinical information extraction: the state of the art at i2b2 2010. Journal of the American Medical Informatics Association : JAMIA. 2011 Sep-Oct;18(5):557–562. doi: 10.1136/amiajnl-2011-000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan JW, Prasad R, Yabut RM, et al. Part-of-speech tagging for clinical text: wall or bridge between institutions?. Proc AMIA Symp; 2011. pp. 382–391. [PMC free article] [PubMed] [Google Scholar]
- 20.Denny JC, Irani PR, Wehbe FH, Smithers JD, Spickard A., 3rd The KnowledgeMap project: development of a concept-based medical school curriculum database. Proc AMIA Symp; 2003. pp. 195–199. [PMC free article] [PubMed] [Google Scholar]
- 21.Liu K, Mitchell KJ, Chapman WW, Crowley RS. Automating tissue bank annotation from pathology reports - comparison to a gold standard expert annotation set. Proc AMIA Symp; 2005. pp. 460–464. [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng QT, Goryachev S, Weiss S, Sordo M, Murphy SN, Lazarus R. Extracting principal diagnosis, co-morbidity and smoking status for asthma research: evaluation of a natural language processing system. BMC medical informatics and decision making. 2006;6:30. doi: 10.1186/1472-6947-6-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawson KP, Capaldi N, Haydon M, Penna AC. The paediatric hospital medical record: a quality assessment. Australian clinical review / Australian Medical Association [and] the Australian Council on Hospital Standards. 1992;12(2):89–93. [PubMed] [Google Scholar]
- 24.Manzar S, Nair AK, Govind Pai M, Al-Khusaiby S. Use of abbreviations in daily progress notes. Arch Dis Child Fetal Neonatal Ed. 2004 Jul;89(4):F374. doi: 10.1136/adc.2003.045591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheppard JE, Weidner LC, Zakai S, Fountain-Polley S, Williams J. Ambiguous abbreviations: an audit of abbreviations in paediatric note keeping. Archives of disease in childhood. 2008 Mar;93(3):204–206. doi: 10.1136/adc.2007.128132. [DOI] [PubMed] [Google Scholar]
- 26.Xu H, Stetson PD, Friedman C. A study of abbreviations in clinical notes. Proc AMIA Symp; 2007. pp. 821–825. [PMC free article] [PubMed] [Google Scholar]
- 27.Navigli R. Word sense disambiguation: A survey. ACM Comput. Surv. 2009;41(2):1–69. [Google Scholar]
- 28.Jimeno-Yepes AJ, Aronson AR. Knowledge-based biomedical word sense disambiguation: comparison of approaches. BMC bioinformatics. 2010;11:569. doi: 10.1186/1471-2105-11-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu H, Markatou M, Dimova R, Liu H, Friedman C. Machine learning and word sense disambiguation in the biomedical domain: design and evaluation issues. BMC bioinformatics. 2006;7:334. doi: 10.1186/1471-2105-7-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008 Sep;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Y, Rosenbloom ST, Denny JC, et al. Detecting abbreviations in discharge summaries using machine learning methods. Proc AMIA Symp; 2011. pp. 1541–1549. [PMC free article] [PubMed] [Google Scholar]
- 32.Pakhomov S, Pedersen T, Chute CG. Abbreviation and acronym disambiguation in clinical discourse. Proc AMIA Symp; 2005. pp. 589–593. [PMC free article] [PubMed] [Google Scholar]
- 33.Xu H, Stetson PD, Friedman C. Combining Corpus-derived Sense Profiles with Estimated Frequency Information to Disambiguate Clinical Abbreviations. Submitted to AMIA 2012. [PMC free article] [PubMed]