Abstract
A Support Vector Machine (SVM) is a popular tool for decision support. The traditional way to build an SVM model is to estimate parameters based on a centralized repository of data. However, in the field of biomedicine, patient data are sometimes stored in local repositories or institutions where they were collected, and may not be easily shared due to privacy concerns. This creates a substantial barrier for researchers to effectively learn from the distributed data using machine learning tools like SVMs. To overcome this difficulty and promote efficient information exchange without sharing sensitive raw data, we developed a Distributed Privacy Preserving Support Vector Machine (DPP-SVM). The DPP-SVM enables privacy-preserving collaborative learning, in which a trusted server integrates “privacy-insensitive” intermediary results. The globally learned model is guaranteed to be exactly the same as learned from combined data. We also provide a free web-service (http://privacy.ucsd.edu:8080/ppsvm/) for multiple participants to collaborate and complete the SVM-learning task in an efficient and privacy-preserving manner.
Introduction
Various types of data (i.e., demographic, clinical, and genomic) are increasingly being collected and stored in biomedical research repositories1,2. Data sharing and analysis across institutions are necessary to facilitate scientific discovery, speed up hypothesis testing, and drive healthcare towards personalized medicine3. However, barriers to more widespread use of these medical data relate to increasing concerns that patient privacy may be violated4–6. In the United States, medical data cannot be released without appropriate de-identification, according to HIPAA7. Because direct dissemination of patient data is difficult, methods that respect patient privacy while enabling knowledge retrieval across institutions are highly desired. Besides privacy, another practical reason motivating distributed data analysis is that in some cases the size of medical data makes it hard for small institutions to process large data efficiently.
Classification algorithms have numerous applications for medical data, e.g., recognizing ECG signals8, differentiating obstructive lung diseases9, and discriminating biomedical imaging patterns10. There is much demand to perform classification in a privacy preserving way, since large amounts of training data are often required to ensure sufficient statistical power to test hypotheses.
To overcome privacy barriers11 and facilitate research, a practical solution is to decompose classification algorithms and parallelize the computation using a distributed network of nodes to help reduce the demand for resources on any single node. There exists extensive literature on privacy-preserving data mining, including models built using naive Bayes12–14, association rules15–18, regression19,20, and support vector machines (SVMs)21,22. However, few of them can support an easy-to-use collaborative environment, and hence they have not been practically used in biomedical applications. In this article, we focus on a specific classifier, the SVM. We leverage this popular classification model in machine learning to build a collaborative framework for privacy-preserving distributed learning.
One closely related work is the vertically partitioned privacy-preserving SVM21. In this approach, each party builds a local model of its own data, and merges the model with other parties through a secure sum operation. To generate a global SVM model, the merging process has to be carried out on a per-node basis. This method has some important limitations: (1) synchronization is difficult for multiple parties; (2) it is vulnerable to network interruption and participant absence; and, (3) it requires a lot of computing resources for individual participants.
To face these challenges, we proposed a novel server/client collaborative framework, where all modeling operations such as task creation and local model merging are performed on the server. The server is composed of a service layer that interacts with individual participants to process data locally, a task manager that constantly checks the completeness of intermediary results, and a computation engine to integrate intermediary results.
In this paper, we briefly review the SVM algorithm in the Methods section, followed by description of our proposed framework, distributed privacy-preserving support vector machine (DPP-SVM). In the Results section, we evaluate the validity of DPP-SVM and test its efficiency with respect to the number of participants involved. The last section discusses the advantages and limitations of DPP-SVM.
Methods
Review of SVM
SVMs23 are state-of-the-art supervised classification methods. The tasks include classifying binding peptides in an antigenic sequence24, predicting long disordered regions25, and finding novel pre-microRNAs26. Consider training data D = {(X1, y1), …, (Xn, yn)} ⊂ X × Y, where X denotes the space of inputs (e.g., X = Rd), and class labels yi ∈ Y = {−1, 1}. Here d indicates the dimension of inputs, while “+1” and “−1” correspond to class labels. An SVM maximizes the geometric margin ||W||2 between two classes of data, as indicated in Figure 1. The function that is optimized can be written as
where ξi is the loss of the i-th point Xi, W is a vector of weight parameters for features, and a parameter C weighs smoothness and errors (i.e., large C for fewer errors, smaller C for increased smoothness).
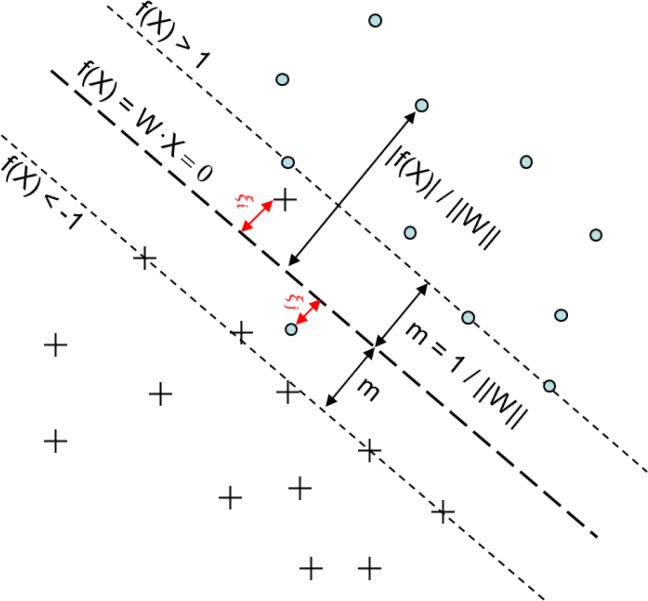
Figure 1.
The geometric view of an SVM model. The dots have class labels “+1”, and the “X”s have class labels “−1”. W is the set of parameters to be learned, and m is the margin, or the longest distance between the support vector (dotted line) for a given class and the separating plane (dashed line). Note that f(X) = WX in the figure.
The dual form of an SVM can be written as:
where αi,i ∈ (1, n) corresponds to the per-sample parameter, and the per-feature weight is wj ∈ W, j ∈ (1, d). An important relationship between the primal and dual forms of SVM is that the weight vector W can be converted from sample weight vector α through W = ∑iyiαiXi. Since αis in the dual format of SVMs can be learned using the kernel matrix K alone, i.e., , we can build an SVM model without sharing the raw data X. Because kernel matrices are often smaller than the raw data (i.e., lots of features and a limited number of patients), it is often efficient to work with the dual problem.
Distributed Privacy Preserving Support Vector Machine
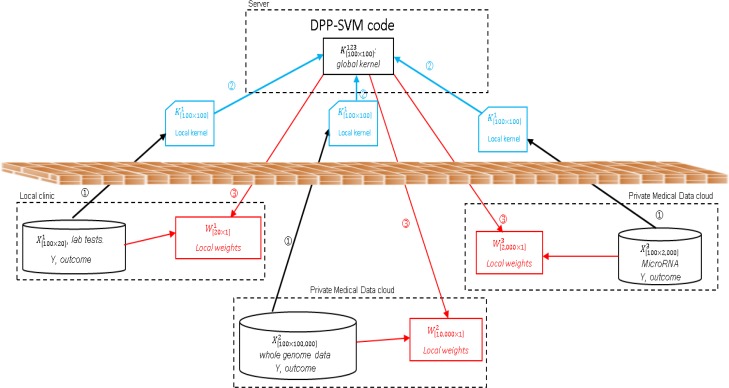
Assuming there is a vertically distributed database as indicated in Figure 2, where features (i.e., demographics, SNPs, etc.) are present in multiple parties, our Distributed Privacy Preserving Support Vector Machine (DPP-SVM) can construct the kernel matrix of a global SVM by combining local kernels, since always holds. Note that , , are vertical partitions of features corresponding to the same records. DPP-SVM can be summarized by three consecutive procedures, which are illustrated in Figure 3:
Calculate local kernels using the dual form of an SVM.
Send the local kernel matrix through secure channels to a central server that computes parameters αi’s.
Calculate weights W for features at each site.
Figure 2.
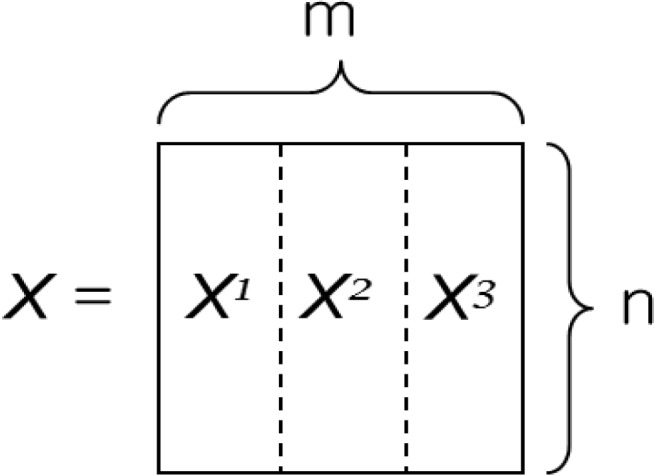
Vertically distributed data matrix with m dimensions (features) and n rows (records). Each vertical slice represents a different site.
Figure 3.
DPP-SVM web-service framework. Three procedures are: (1) local kernel matrix calculation; (2) local kernel matrix transmission to the server; and (3) partial weights calculation. Local data are executed consecutively to build a global SVM from vertically partitioned data. The web-service exchanges local kernel matrices (i.e., non-sensitive intermediary results) between the server and the participants, and calculates global model parameters. Expensive matrix multiplications of high-dimensional data (i.e., whole genome features) are handled by powerful private server/clouds, but a local clinic without a lot of computation power can still build a global SVM model from calculations done on the cloud. In addition, the DPP-SVM framework enforces privacy because no participant ever leaks sensitive patient information to other participants or to the server (although a unique identifier needs to be agreed upon at all sites).
Let us look at the implementation of DPP-SVM. Figure 4 depicts the detailed structure of the DPP-SVM framework. We start our explanation with the task manager on the server side.
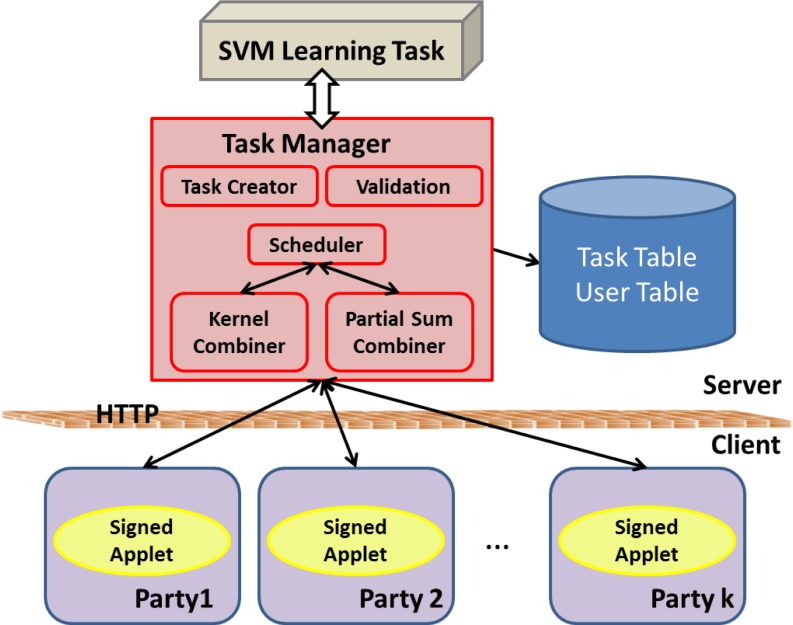
Figure 4.
The structure of the web application for our DPP-SVM learning framework. Tasks created by a single site (task creator) are stored in the trusted server (task manager). Signed applets sitting on the user’s side calculate and communicate with the trusted server to transmit privacy insensitive intermediary results, and the final SVM model is calculated using the kernel combiner and the partial sum combiner, both scheduled in the trusted server.
The Task Manager
The task manager resides on a central node (i.e., the trusted server), which instantiates, monitors, and stores the tasks created by users and check the completeness and validity of these tasks. To learn a global SVM from distributed data repositories, the task manager has the following functions, also illustrated in Figure 4:
The task manager creates an event in the database after a user provides the necessary information for a learning task through our web interface. The required information includes task name, expiration date, participants’ email addresses, and the model parameter. The task manager invites all users to join the created task through emails.
The task manager directs invited participants to unique webpages, where computation for local learning can be done within signed applets. The signature on an applet is to verify whether this applet is from a reliable source and can be trusted to run on a participant’s local web browser. These applets then send intermediary results (including the partial kernel matrix K, and model weights α) to the task manager on the server that deposits this information.
The task manager, acting as a coordinator, checks if all participants have completed the learning task and submitted their intermediary results. Note that the checking happens at a predefined frequency determined by a scheduler. When all tasks of participants are completed, the task manager combines the local kernel matrices into a single global kernel matrix.
The task manager coordinates and gathers partially summed weight parameters from participants, and it sends the global classification labels back to participants.
It is worth noting that the word “trusted” simply means that the head node is not malicious and that it does what it suppose to do (i.e., aggregate received kernel matrices and broadcast adjusted parameters back to the nodes). Recall that, since these partial kernel matrices are aggregated from n patients, there is little privacy risk in transmitting these intermediary results.
An Email-Driven Workflow
Distributed privacy-preserving support vector machine involves a number of operations, processes, and computations that are carried out both locally and at the trusted server. Participants interact with the task manager once others have completed their tasks in the previous step. To improve the efficiency in collaboration, we propose an email-driven workflow in which the task manager coordinates and synchronizes tasks between multiple participants, as illustrated in Figure 5.
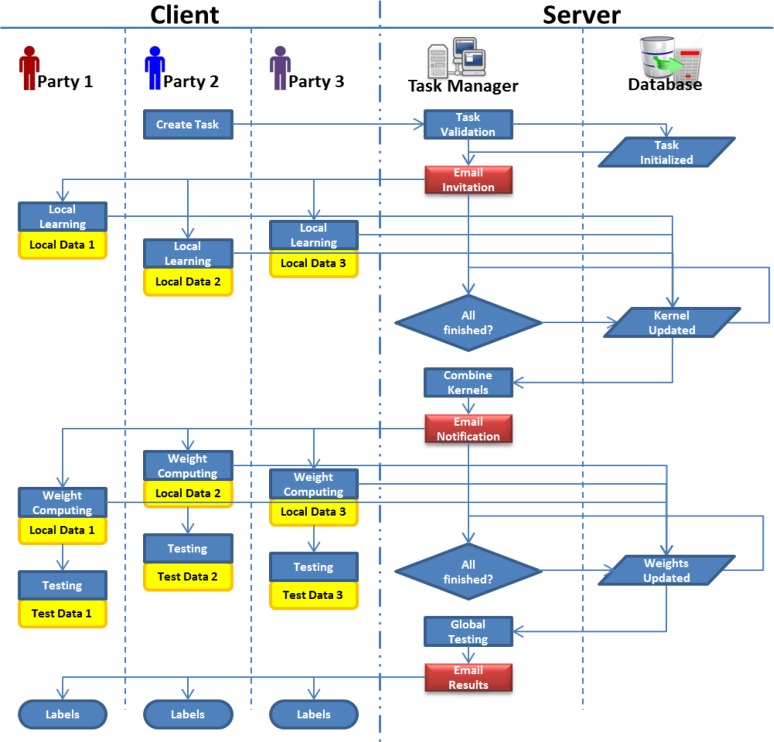
Figure 5.
Detailed workflow for distributed privacy preserving support vector machine (DPP-SVM). The entire process of the DPP-SVM is driven by emails (red blocks), which are sent by the task manager from the trusted server. There is no need for participants to disclose their data to others, and their data are only processed locally (yellow blocks). Task-related information (name, description and participants) and intermediary results are stored in the database on the server (parallelograms).
In Figure 5, a learning task is initialized by Party 2. After he or she fills the basic information about the task, email invitations are sent out automatically to all participants with a link to the service, which computes the partial kernel matrices locally. In the first round of learning, only partial kernel matrices are sent back to the server. A scheduler in the task manager checks, at a pre-defined frequency, whether all the local kernel matrices have been collected, and sends emails to participants with a new link to the service, which computes the weight parameters using the combined kernel matrix and data owned by each participant. In the second round, data are still processed locally and only weight parameters are sent back to the server. For the third round of computation, participants can test additional data based on globally learned weight parameters to make predictions in a distributed and privacy-preserving manner.
Results
Web-Application Interface
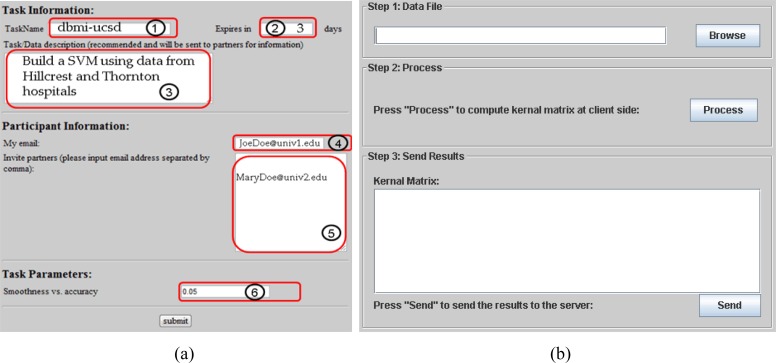
We used HTML and Java applets as our front-end to interact with the “task creator” (e.g., Party 1), which provides emails of other participating parties and sets the global parameter C of the SVM model, as shown in Figure 6. We used JSP and Java servlets to handle the server-side computation.
Figure 6.
Snapshots of DPP-SVM web application interfaces. (a) The HTML form used to create a task: ① task name, ② expiration period, ③task description, ④ the creator’s email, ⑤ participant email(s), and ⑥ the global model parameter C. (b) The signed Java applet that processes data locally.
Experiments
We ran experiments on real data to validate DPP-SVM and evaluate the efficiency of it. All of our experiments were conducted on an Intel Xeon 2.4GHz server with 32GB RAM that hosts this application. Our goal is to verify that the proposed method generates exact the same answers as if the model were learned from data combined in a centralized repository. We also intended to check the computational cost of DPP-SVMs when different numbers of participants are involved.
Dataset
We used two data sets. The first one is the tic-tac-toe data set from UCI machine learning repository27. It is multivariate, which consists of 27 features for a complete set of possible board configuration and a target variable indicating “win for x” (i.e., true when “x” is one of 8 possible ways to create a “three-in-a-row”). The second is a set of real medical data for hospital discharge error prediction28. This de-identified data consists of 10 features (i.e., microbiology cultures ordered while patients were hospitalized) and a target variable indicating a potential follow-up error (i.e., true if it was an error, and false if not). Because SVMs do not handle categorical features by default and 8 out of 10 features of hospital discharge are categorical, we binarized them and obtained a total of 22 features to be used for SVM training. The following table lists the record and feature sizes for both data sets.
Fidelity Measure Using Ten-Cross-Validation
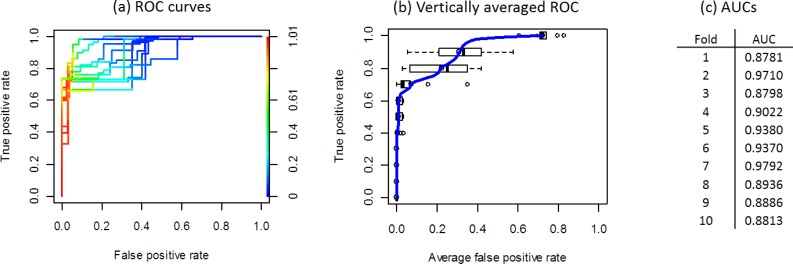
We used ten-cross-validation29 to evaluate the performance of both the collaborative privacy-preserving SVM and the centralized SVM. The collaborative privacy-preserving SVM was constructed in a distributed manner by learning k –split† features. We conducted experiments, including two-split, three-split, and four-split privacy-preserving SVMs. For tic-tac-toe data, linear SVMs with C = 0.2 were used to train and test, which showed that there was no difference between all these SVMs (i.e., privacy-preserving ones and the centralized one), and their discrimination performances, measured through ROC curves, are summarized in Figure 7.
Figure 7.
The discrimination performance of DPP-SVMs is the same as the centralized SVM, with precision O(10−6), using the tic-tac-toe data set. The three subfigures correspond to (a) the ROC curves of 10 cross-validation folds, (b) vertically averaged ROC curve with standard deviations for false positive rates, (c) AUC (Area Under the ROC Curves) for all 10 folds of both SVMs (i.e., distributed and centralized). Note that the color bar in (a) indicates cutoff values for different ROCs.
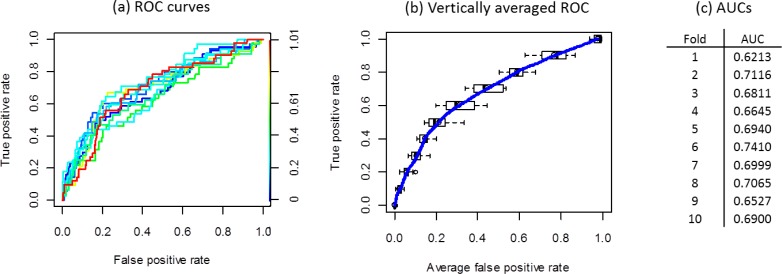
Similarly, we applied both the collaborative privacy-preserving SVM and the centralized SVM to the hospital discharge data. Like before, the collaborative privacy-preserving SVM was constructed using two-split, three-split, and four-split features. Linear SVMs with C = 0.0001 were used for training and testing. Again, there was no difference between all these SVMs. Their discrimination performances were summarized in Figure 8.
Figure 8.
The discrimination performance of DPP-SVMs is the same as that of the centralized SVM, with precision O(10−6), using the hospital discharge data set. The three subfigures correspond to (a) the ROC curves of 10 folds, (b) vertically averaged ROC curve with standard deviations for false positive rates, (c) AUC for all 10 folds of both SVMs (i.e., distributed and centralized). Note that the color bar in (a) indicates cutoff values for different ROCs.
Time Comparison
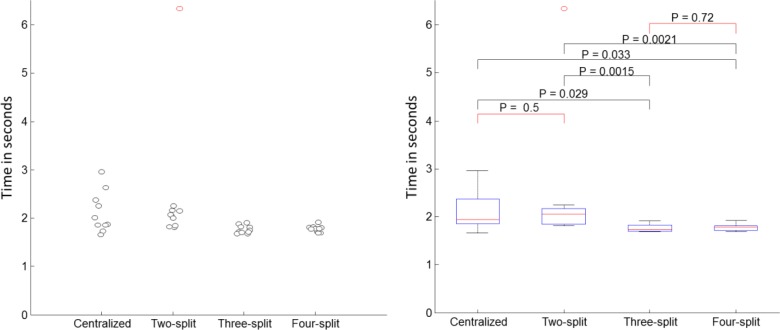
We also compared the computational cost of a centralized SVM and DPP-SVMs. Because actual communication time depends on the network and also on how soon participants respond to emails, we only considered computational costs based on total time that were spent on the ten-cross-validation for each of the experiments described in the previous section. For a centralized SVM, the computation only happened in a single site. For DPP-SVMs, the computational cost was dominated by the participant that processed the most of the data, since the computation was parallelized. We expected a decrease of computational time when more participants were involved (but at the same time, an increase in communication cost, which is not discussed here). Figures 9 and 10 show the actual time spent on ten-fold-cross-validation using both the tic-tac-toe and the hospital discharge data sets.
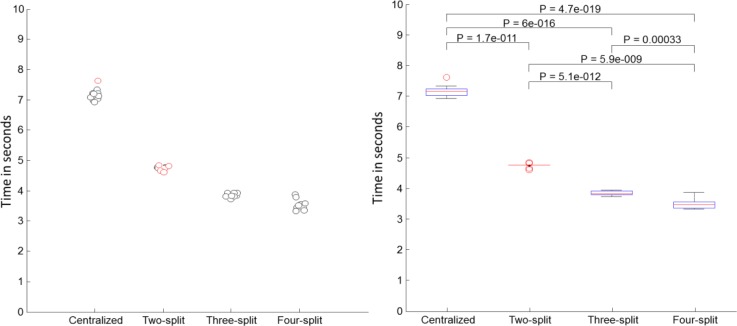
Figure 9.
Time comparison between centralized SVM and DPP-SVMs with various numbers of participants using the tic-tac-toe data. The left figure plots the time and the right figure shows their box plots. The red square brackets on the right figure indicate differences that were not significant. Black brackets indicate significantly different times.
Figure 10.
Time comparison between centralized SVM and DPP-SVMs with various numbers of participants using the hospital discharge data. The left figure plots the time and the right figure shows their box plots. All time differences were statistically significant.
As the experiments above indicated, DPP-SVM can significantly save time if multiple parties join a learning task that involves a relatively large data set (i.e., hospital discharge), but the gain for a small data set like tic-tac-toe is relatively small.
Discussion and Conclusion
We presented a collaborative framework for distributed privacy preserving support vector machine (DPP-SVM). In this framework, one party does not need to disclose his or her local data to other parties and a global SVM model can be learned from distributed, vertically partitioned data.
Some healthcare institutions are prohibited from sharing patient data with outside collaborators. In these cases, multiparty computation allows them to still participate in certain types of research networks, without sharing specific patient data. For example, we showed in another work that it is possible to share information to create accurate predictive models across institutional barriers without sharing the original data30.
The server/client design and email-driven workflow relieve the participants from spending time on coordinating and synchronizing tasks. It provides a practical way for researchers with data-sharing barriers to learn a global SVM model without having to transmit their data to another site. From the perspective of data privacy, DPP-SVM provides an alternative method for privacy-preserving data sharing to methods based on data perturbation (e.g., noise addition31–33, cryptographic techniques34,35, and table generalization techniques36,37).
Our solution has important limitations. First, our current algorithm only supports a linear kernel SVM. Some applications require nonlinear classification, which requires the utilization of more sophisticated kernels38. Second, we only deal with vertically partitioned data, in which patient features are distributed, instead of patients with similar features are distributed. The former can be imagined as different institutions hosting different pieces of information (i.e., demographics, genotypes, phenotypes, and etc.) about the same patients. We are currently developing similar server/client architecture for the second case.
Table 1.
Summary of data sets used in collaborative privacy-preserving framework for distributed SVM learning.
| Data set | Number of features | Number of records |
|---|---|---|
| tic_tac_toe | 27 | 958 |
| hospital discharge | 22 | 8,668 |
Acknowledgments
The authors were funded in part by the NIH grants R01LM009520, U54 HL10846, R01HS019913, UL1RR031980 and 1K99LM011392-01. We thank Mr. Myoung Lah for proofreading and editing this document.
Footnotes
In practice, it is not always possible to split features evenly. For example, tic-tac-toe has 27 features and our two-split is to divide them into 13 and 14 features to form two local subsets for learning.
References
- 1.Murphy SN, Gainer V, Mendis M, et al. Strategies for maintaining patient privacy in i2b2. Journal of the American Medical Informatics Association. 2011;18:103–8. doi: 10.1136/amiajnl-2011-000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vinterbo SA, Sarwate AD, Boxwala A. AMIA Summit on Clinical Research Informatics (CRI’11) San Francisco: 2011. Protecting count queries in cohort identification; p. 79. [Google Scholar]
- 3.Ohno-Machado L, Bafna C, Boxwala A, et al. iDASH. Integrating data for analysis, anonymization, and sharing. Journal of the American Medical Informatics Association. 2012;19:196–201. doi: 10.1136/amiajnl-2011-000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grzybowski DM. Patient privacy: the right to know versus the need to access. Health management technology. 2005;2654:53. [PubMed] [Google Scholar]
- 5.Ohno-Machado L, Silveira PSP, Vinterbo S. Protecting patient privacy by quantifiable control of disclosures in disseminated databases. International Journal of Medical Informatics. 2004;73:599–606. doi: 10.1016/j.ijmedinf.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 6.Goodwin LK, Prather JC. Protecting patient privacy in clinical data mining. Journal Of Healthcare Information Management. 2002;16:62–7. [PubMed] [Google Scholar]
- 7.Anon Standards for Privacy of Individually Identifiable Health Information. Federal Register. 2000;67:82461–820. [PubMed] [Google Scholar]
- 8.Yan H, Zou Z, Wang H. Adaptive neuro-fuzzy inference system for classification of ECG signals using Lyapunov exponents. Computer Methods and Programs in Biomedicine. 2009;93:313–21. doi: 10.1016/j.cmpb.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y, Seo JB, Lee JG, et al. Performance testing of several classifiers for differentiating obstructive lung diseases based on texture analysis at high-resolution computerized tomography (HRCT) Computer Methods and Programs in Biomedicine. 2009;93:206–15. doi: 10.1016/j.cmpb.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Han X-H, Chen YW. Biomedical Imaging Modality Classification Using Combined Visual Features and Textual Terms. International Journal of Biomedical Imaging. 2011:241396:1–7. doi: 10.1155/2011/241396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loukides G, Gkoulalas-Divanis A, Malin B. Anonymization of electronic medical records for validating genome-wide association studies. Proceedings of the National Academy of Sciences of the United states of America. 2010;107:7898–903. doi: 10.1073/pnas.0911686107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi X, Zhang Y. Privacy-preserving naive Bayes classification on distributed data via semi-trusted mixers. Information Systems. 2009;34:371–80. [Google Scholar]
- 13.Zhang P, Tong YH, Tang SW, et al. Privacy preserving naive Bayes classification. Advanced Data Mining Applications. 2005;3584:744–52. [Google Scholar]
- 14.Vaidya J, Kantarcıoğlu M, Clifton C. Privacy-preserving Naïve Bayes classification. The VLDB Journal. 2007;17:879–98. [Google Scholar]
- 15.Saygin Y, Verykios VS, Elmagarmid AK. Privacy preserving association rule mining. Proceedings Twelfth International Workshop on Research Issues in Data Engineering; 2002. pp. 151–8. [Google Scholar]
- 16.Oliveira SRM, Zaiane OR, Saygin Y. Secure association rule sharing. Adv In Knowledge Discovery Data Mining, Proc. 2004;3056:74–85. [Google Scholar]
- 17.Evfimievski A, Srikant R, Agrawal R, et al. Privacy preserving mining of association rules. Information Systems. 2004;29:343–64. [Google Scholar]
- 18.Kantarcioglu M, Clifton C. Privacy-preserving distributed mining of association rules on horizontally partitioned data. IEEE Transactions on Knowledge and Data Engineering. 2004;16:1026–37. [Google Scholar]
- 19.Sanil AP, Karr AF, Lin X, et al. Privacy preserving regression modeling via distributed computation. Proceedings of the tenth ACM SIGKDD international conference on Knowledge discovery and data mining; New York, NY. 2004. pp. 677–82. [Google Scholar]
- 20.Slavkovic AB, Nardi Y, Tibbits MM. “Secure” Logistic Regression of Horizontally and Vertically Partitioned Distributed Databases. Seventh IEEE International Conference on Data Mining Workshops; 2007. pp. 723–8. [Google Scholar]
- 21.Yu H, Jiang X, Vaidya J. Privacy-preserving SVM classification on vertically partitioned data. Advances in Knowledge Discovery and Data Mining. 2006;3918:647–56. [Google Scholar]
- 22.Vaidya J, Yu H, Jiang X. Privacy-preserving SVM classification. Knowledge and Information Systems. 2008;14:161–78. [Google Scholar]
- 23.Cherkassky V. The nature of statistical learning theory. IEEE transactions on neural networks / a publication of the IEEE Neural Networks Council. 1997;8:1564. doi: 10.1109/TNN.1997.641482. [DOI] [PubMed] [Google Scholar]
- 24.Bhasin M, Raghava GPS. SVM based method for predicting HLA-DRB1*0401 binding peptides in an antigen sequence. Bioinformatics. 2004;20:421–3. doi: 10.1093/bioinformatics/btg424. [DOI] [PubMed] [Google Scholar]
- 25.Hirose S, Shimizu K, Kanai S, et al. POODLE-L: a two-level SVM prediction system for reliably predicting long disordered regions. Bioinformatics. 2007;23:2046–53. doi: 10.1093/bioinformatics/btm302. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Yang Y, Zhang H, et al. Prediction of novel pre-microRNAs with high accuracy through boosting and SVM. Bioinformatics. 2011;27:1436–7. doi: 10.1093/bioinformatics/btr148. [DOI] [PubMed] [Google Scholar]
- 27.Asuncion A, Newman DJ. UCI machine learning repository. 2007.
- 28.El-Kareh R, Roy C, Brodsky G, et al. Incidence and predictors of microbiology results returning postdischarge and requiring follow-up. Journal of hospital medicine. 2011;6:291–6. doi: 10.1002/jhm.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duda RO, Stor DG. Pattern classification. New York, NY: Wiley; 2001. [Google Scholar]
- 30.Wu Y, Jiang X, Kim J, et al. Grid LOgistic REgression (GLORE): Building Shared Models Without Sharing Data. Journal of the American Medical Informatics Association (Epub ahead of print) 2012 doi: 10.1136/amiajnl-2012-000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Z, Wang J, Liu L, et al. Generalized random rotation perturbation for vertically partitioned data sets. IEEE Symposium on Computational Intelligence and Data Mining; 2009. pp. 159–62. [Google Scholar]
- 32.Liu L, Wang J, Zhang J. Wavelet-Based Data Perturbation for Simultaneous Privacy-Preserving and Statistics-Preserving. IEEE International Conference on Data Mining Workshops; 2008. pp. 27–35. [Google Scholar]
- 33.Kargupta H, Datta S, Wang Q. On the privacy preserving properties of random data perturbation techniques. Third IEEE International Conference on Data Mining; 2003. pp. 99–106. [Google Scholar]
- 34.Zhan J, Matwin S. A Crypto-Based Approach to Privacy-Preserving Collaborative Data Mining. The Sixth IEEE International Conference on Data Mining Workshops; 2006. pp. 546–50. [Google Scholar]
- 35.Kissner L, Song D. Privacy-preserving set operations. Advances in Cryptology. 2005;3621:241–57. [Google Scholar]
- 36.Sweeney L. Achieving k-anonymity privacy protection using generalization and suppression. International Journal of Uncertainty Fuzziness Knowledge-based Systems. 2002;10:571–88. [Google Scholar]
- 37.El Emam K, Dankar FK, Issa R, et al. A globally optimal k-anonymity method for the de-identification of health data. Journal of the American Medical Informatics Association. 2009;16:670–82. doi: 10.1197/jamia.M3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ben-Hur A, Ong CS, Sonnenburg S, et al. Support vector machines and kernels for computational biology. PLoS Computational Biology. 2008;4:e1000173. doi: 10.1371/journal.pcbi.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]