Abstract
In this paper, we present a natural language processing system that can be used in hospital surveillance applications with the purpose of identifying patients with pneumonia. For this purpose, we built a sequence of supervised classifiers, where the dataset corresponding to each classifier consists of a restricted set of time-ordered narrative reports. In this way the pneumonia surveillance application will be able to invoke the most suitable classifier for each patient based on the period of time that has elapsed since the patient was admitted into the hospital. Our system achieves significantly better results when compared with a baseline previously proposed for pneumonia identification.
Introduction
The use of advanced Electronic Medical Record (EMR) systems is becoming commonplace within major medical centers. The extensive databases underlying EMR systems, detailing all aspects of patient care, has the potential to greatly facilitate new quality improvement and assurance initiatives, and revolutionize clinical research. However, most patient information that describes the patient state, diagnostic procedures, and disease progress is represented in free-text form. This provides an opportunity for natural language processing (NLP) technologies to play a major role in clinical care and research by facilitating analysis of free-text information, which otherwise is typically accessible only through manual chart abstraction.
With the automatic extraction of relevant clinical information from free-text reports, opportunities open up to facilitate clinical and translational studies by identifying large cohorts of critically ill patients. In previous work, we demonstrated that by using physician notes such as admit notes, ICU progress notes, and discharge summaries, automated approaches that incorporate NLP and machine learning can accurately classify a given patient as positive or negative for pneumonia based on retrospective review of the reports generated during the patient’s ICU stay.1 In a more recent research study, we built a supervised machine learning framework that selected only the most relevant features extracted from the reports and increased the prediction performance.2 The main purpose of this type of classification was to select cohorts of patients for clinical research studies.
Another important clinical application where NLP can be quite useful is to estimate the phenotype time-of-onset in clinical settings. Early detection and treatment of pneumonia, one of the most common severe infections in critically ill patients, is important as even short-term delays in appropriate antibiotic therapy are associated with higher mortality rates, longer-term mechanical ventilation, and excessive hospital costs.3 For instance, many hospitals are currently using NLP systems for pneumonia surveillance, because this type of application is resource intensive and at the same time, requires real-time assessments. In the clinical NLP domain, automated methods for identifying different types of pneumonia have been previously studied. A wide range of rule-based methods and machine learning approaches have been proposed, however the focus in those reported approaches was exclusively on radiology reports of chest x-rays.4–8 While radiologic changes within the lung are a necessary condition for diagnosis of pneumonia, there exists data within other domains such as the disease presentation narrative, physiologic measures, and laboratory abnormalities that could add significant accuracy and depth to the identification of pneumonia cases.9 10 Because chest x-ray abnormalities comprise only part of the pneumonia definition, any system aimed at pneumonia identification which incorporates only chest x-ray information would be expected to lead to significant phenotypic misclassification. Thus, there remains an unmet need to accurately capture the clinical components of the pneumonia phenotype. Furthermore, to study phenotype time-of-onset, it is imperative to consider the progression of patient reports for the duration of hospitalization, and not to rely on a specific report type at one given point of time.
In this paper, the main research question we address is whether we can accurately identify pneumonia at a given point of time, without having access to patient reports after that point of time. In our dataset, a patient is considered positive for pneumonia if pneumonia occurs within 48 hours of admission to ICU, i.e. the patient is determined to have Community Acquired Pneumonia (CAP). In our experiments, we built a classifier for each ICU day that predicts the probability of pneumonia based on only the reports generated until that ICU day. Our experiments demonstrated that our classifiers correctly identify some patients as positive for pneumonia after only two days, which conforms to the definition of pneumonia in our dataset, with classifier accuracy continuing to improve through day 6. As we train a separate classifier for each day of ICU stay, we observe that different sets of features are informative on different days, which reflects the physician’s recognition of the evolving clinical presentation over time. While this dataset offers us a very small window in which to determine time-of-onset, we are encouraged by the results of our experiments, and expect that results will improve on a dataset where there is a longer series of reports prior to the onset of pneumonia.
Related Work
Our research problem is related to the problem of predicting future events from a history of past observations. In general, the past observations consist of a time-ordered sequence of events (e.g., wars, conflicts, epidemiological infections, audio signals) encoding various patterns and characteristics that can help in predicting the future events. Examples of active research areas studying the future event prediction problem are political science, weather forecasting, criminology, video and signal processing, and the prediction of equipment failures.
In the domain of biomedical informatics, various applications have been proposed to model the progression of specific diseases. Glickman and Gagnon studied the role of genetic and modifiable risk factors in the occurrence of late-onset diseases.11 For this purpose, they developed a flexible Bayesian framework using data collected from Framingham Heart Study to assess the effect of Apo-E genotypes on the progression of specific cardiovascular disease events. In another study, Stell et al. developed the Hypo-Predict engine that is able to predict the onset of a possible hypotensive event for a given patient.12 When such an event happens, this engine is also able to alert the clinicians in a feasible time interval (e.g., 15 minutes) such that they can administer the appropriate treatment according with their own clinical judgment. At the core of this engine lies a Bayesian neural network model which is trained to identify patterns of behavior in blood pressure and other measurements such as heart and respiratory rates. In their work, Genovese et al. developed various competing risk models to capture distinct temporal patterns of adverse events (e.g., bleeding, infection, arrhythmia) that can happen in the first 60 days after the implantation of a ventricular assist device to patients with heart failure.13 Also, Huang et al. proposed a latent model approach for defining a new postural instability onset time measure, which is one of the most relevant indicators for the progression of the Parkinson’s disease.14
Although our main goal is to build a system that is able to model the progression of complex illness phenotypes (such as pneumonia) using various types of clinical reports, in this paper, we study how well we can solve this task when only a limited set of reports is available to each patient. Moreover, when compared to previous approaches that used a mixture of report types to solve pneumonia identification1 2, all the experiments performed in this study take into account that the reports corresponding to each ICU patient are chronologically ordered.
Dataset
The dataset used in this study consists of various types of narrative reports corresponding to a cohort of 426 patients. The retrospective review of the reports was approved by the UW Human Subjects Committee of Institutional Review Board. The annotations were performed at patient level for a different clinical study of ICU subjects, which was described previously.15 A research study nurse with 6 years of experience manually annotated a patient as positive if the patient had pneumonia within the first 48 hours of ICU admission and as negative if the patient did not have pneumonia or the pneumonia was detected after the first 48 hours of the ICU admission. As a result, 66 patients were identified as positive cases for pneumonia and the remaining 360 patients as negative cases. Moreover, because the subjects in this dataset were admitted to the ICU from the emergency department as well as from other hospitals, cases of pneumonia included both community acquired pneumonia (i.e., pneumonia that occurs outside of the hospital settings) and hospital acquired pneumonia (i.e., pneumonia that occurs after admission to the hospital).
Overall, our dataset includes a total of 5,313 reports, each report having one of the eight report types: admit note, ICU daily progress note, acute care daily progress note, transfer/transition note, transfer summary, cardiology daily progress note, and discharge summary. The total number of reports per patient ranged widely due to the high variability in the ICU length of stay: median=8, interquartile range=5–13, minimum=1, maximum=198.
The distribution of reports and patients among the eight different report types from the dataset is listed in Table 1. Specifically, this table lists the number of reports for each report type as well as the number of distinct patients who had the report type in the dataset. As can be seen from the table, not all patients have all the report types. For example, only 280 (65%) patients have admit notes; the remaining 146 patients that have no admit notes are likely to have been transferred to the ICU from other medical units. There were 350 (82%) patients with discharge summaries. Out of the 426 patients, only a subset of 236 (55%) patients had both admit notes and discharge summaries. Of the admit notes collected in this corpus, over 75% derive from the first day of hospital admission. Furthermore, ICU progress notes were consistently represented throughout the first 96 hours of admission. These data show that admit notes will be largely indicative of the pre-hospital and early hospital stay while ICU progress notes will be the predominant daily text source following the day of admission. Discharge notes largely arose after 96 hours since admission.
Table 1.
Corpus statistics by the frequency of report types and the number of distinct patients who had the report type.
| Report type | # of reports | # of patients | Report type | # of reports | # of patients |
|---|---|---|---|---|---|
|
|
|
||||
| Admit note | 481 | 280 | Transfer/transition note | 243 | 175 |
| ICU daily progress note | 2,526 | 388 | Transfer summary | 18 | 18 |
| Acute care daily progress note | 1,357 | 203 | Cardiology daily progress note | 133 | 17 |
| Interim summary | 164 | 115 | Discharge summary | 391 | 350 |
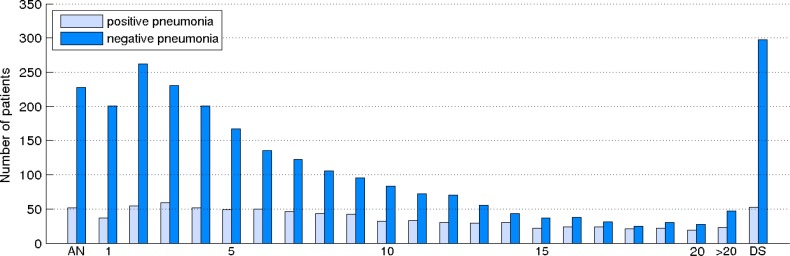
Since all the experiments presented in this paper take into account the chronological order of reports, we also show in Figure 1 the distribution of positive and negative patients having reports in a specific time interval. For this purpose, we considered a special timeline of reports, where each element of this timeline is associated with a specific time interval. To build the distribution of patients for this timeline, we counted how many patients have at least one report in a given time interval. The first and last elements on the timeline are special elements which correspond to admit note (AN) and discharge summary (DS) report types, respectively. The remaining elements on the timeline correspond to reports with a timestamp in the first 20 days since admission (i.e., from 1 to 20) or more than 20 days (this element is labeled as >20 on the timeline). As can be observed in the plot, ∼35% the patients with pneumonia were hospitalized for more than 20 days (23 out of 66 corresponding to the >20 element) whereas the most of the patients that do not have pneumonia are gradually discharged from ICU after several days of stay.
Figure 1.
The distribution of patients with reports in a specific time interval.
A Framework for Identifying Pneumonia in Narrative Reports
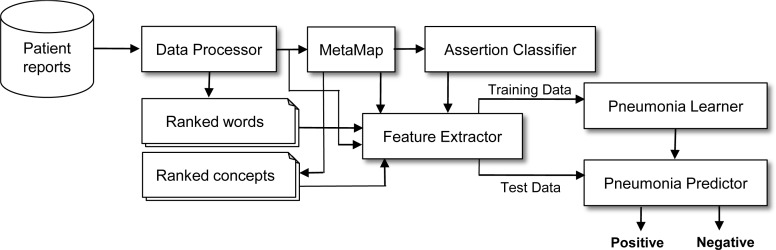
Our approach for pneumonia identification relies on the supervised learning framework depicted in Figure 2. In this framework, a data instance is associated with a patient that is represented by all its corresponding reports. Before computing a feature vector for each patient, in the data processing phase, the entire content of each report is first tokenized using the SPLAT toolkit16 and the punctuation tokens are filtered out. It is also worth mentioning that we did not make any distinction between the tokens from different report sections when extracting the patient feature vector. To learn a binary prediction model for pneumonia identification, we employed LIBLINEAR17, which is an implementation of the support vector machines algorithm.
Figure 2.
System architecture for pneumonia identification.
As we already proposed, to improve the performance of pneumonia identification, we extended this supervised framework by implementing a methodology that selects only the most informative features from the entire feature space in the feature extraction phase.2 Specifically, this methodology uses statistical significance tests to measure the association strength between each feature from the training set and the two categories of this task (i.e., positive vs. negative for pneumonia). As a result, the features will be ranked based on those values such that the ones with a strong association to the two categories will be on top. Finally, only the most relevant features that are within a specific threshold will be selected for training. This methodology is also called statistical feature selection.
Statistical Feature Selection
In a first phase of statistical feature selection, we build a list of ranked features extracted from the training set, for each feature type considered. As feature types, we identified all possible uni-grams and bi-grams of words and Unified Medical Language System (UMLS) concepts. To extract the UMLS concepts from the clinical reports, we used the 2011 version of MetaMap18, a tool developed at the National Library of Medicine. In the MetaMap extraction process, we considered only the UMLS concepts having the highest mapping score for each match. For ranking the set of features associated with a feature type as illustrated in Figure 1 (e.g., the ranked list of word bi-grams), we constructed a contingency table for each feature from the set and used statistical hypothesis testing to determine whether there is an association between the feature and the two categories of pneumonia identification. Specifically, to measure how relevant a feature is with respect to the two categories of our problem, we used the t statistical test, although we also experimented with several other association measures including the χ2 test, Fisher exact test, pointwise mutual information, and Dice coefficient. These measures, however, did not perform as well as the t statistical test.
In order to describe how we computed the t statistical test for pneumonia identification, we first introduce several notations. We denote by f the event of feature f occurring, and by pna the event of any patient being positive for pneumonia. Using these notations, we can now formulate a null hypothesis which should be true if there is no correlation between the feature f and the event corresponding to positive pneumonia. Therefore, if the events f and pna are generated independently, the joint probability of these two events is the same with the product of their individual probabilities, P(f, pna) = P(f) · P(pna). Furthermore, we consider the feature extraction process as a process of randomly generating a sequence of 0 and 1, where a value of 1 corresponds to extracting the feature f for a patient positive for pneumonia and a value of 0 corresponds to the rest of the events. Therefore, the process of sampling f and pna together can be associated with a Bernoulli trial with probability p, and whose corresponding distribution has the mean μ and variance σ2 = p · (1− p). Next, under the assumption that each sample is drawn from the previously mentioned distribution, the t test computes the probability that f and pna co-occur by measuring the difference between the observed and expected means, scaled by the variance of the sampling distribution:19
In this formula, x̄ represents the sample mean, s2 is the sample variance, N the sample size (i.e., the total number of features from the entire training set), and μ is the mean of the distribution. Also, for computing the individual probabilities of the two events, we used the maximum likelihood estimates which involve the frequency counts associated with these events when they occur during the feature extraction phase.
The top 10 most relevant features according to the t test associated with each feature type considered for feature selection (i.e., uni-grams and bi-grams of words and UMLS concepts) are listed in Table 2. We computed the relevance scores of these features using the entire set of reports from the training set. As can be observed, many of these features are closely linked to the known causes (e.g., influenza) and to clinical signs and symptoms (e.g., sputum, coughing, decreased breath) of pneumonia. However, this table may also list features that are not directly related to the diagnostic criteria for pneumonia since they may well indicate latent risk factors for pneumonia or simply capture a predominant association with one of the two categories (e.g., urine || sputum). Report writing is often formulaic, so there can be a preponderance of evidence for the order of, e.g., urine and sputum as we found in expressions such as in “urine, sputum and blood remain negative”, “blood, urine, sputum cultures have not grown pathogenic organisms to date”, “urine, sputum, blood cultures pending”, etc.
Table 2.
Top 10 most informative features for each feature type considered according to the t statistical test.
| words | UMLS concepts | ||
|---|---|---|---|
|
|
|
||
| uni-grams | bi-grams | uni-grams | bi-grams |
|
|
|
|
|
| sputum | sputum || cx | Microbial culture of sputum | Influenza || Fluorescence Units |
| suctioning | sputum || culture | Sputum | Responding || Voice |
| h1n1 | continue || lpv | Consolidation | Continuous || Oseltamivir |
| ventilatory | h1n1 || influenza | Infiltration | Positive End-Expiratory Pressure || Continuous |
| consolidation | acquired || pneumonia | Influenza preparation | Influenza virus vaccine || Influenza preparation |
| secretions | bacterial || pneumonia | Influenza virus vaccine | Influenza || Influenza virus vaccine |
| lpv | continue || oseltamivir | Pneumonia | Lung || Coarse |
| coughing | decreased || breath | Fluorescence Units | Pneumonia || Continuous |
| flu | positive || h1n1 | Influenza | Milliequivalents/milliliter || Potassium Chloride |
| tachypneic | urine || sputum | Decreased breath sounds | Event || Event |
Once all features are ranked and their corresponding threshold values are established, the feature extractor is now able to build a feature vector for each patient. Specifically, given a fixed subset of relevant features determined by selecting the top features from the ranked lists of features up to a threshold value, the feature extractor considers in the representation of a patient’s feature vector only the features from the subset of relevant features that are also found in the patient’s reports. Therefore, the size of the feature space will be equal to the size of the relevant features subset whereas the length of each feature vector will be at most this value.
Assertion Classification
The simplest method for deciding whether a patient is positive (or negative) for pneumonia is to extract the assertion values of the pneumonia expressions that occur in the patient clinical notes. Although the simplicity of this method is attractive, the detection of such complex phenotype often requires a deeper understanding of the reports that goes beyond assertion identification. There are many cases when the reports do not explicitly mention that the patient has pneumonia or not. In fact, only 203 out of 426 patients from our dataset have mentions of pneumonia expressions or their related words (e.g., pneumonitis) in their reports (from these 203 patients 63 are positive for pneumonia). Furthermore, it is often noted that clinical notes will use hedging terms even when there is a fairly high certainty for a patient to be positive for pneumonia, for reasons of liability or waiting for conclusive test results to arrive. Nevertheless, to account for the cases where the ICU reports from our dataset include an explicit statement that the patient has, is suspected of, or does not have pneumonia, in addition to the features selected using statistical tests, we implemented a binary feature called the assert feature. This novel feature assigns to each patient a label corresponding to positive or negative pneumonia and is based on the assertion values associated with pneumonia expressions and their related words found in the patients’ reports.
The assertion value for a pneumonia expression is computed by implementing a supervised learning framework on a dataset provided for assertion classification – a shared task organized within the Informatics for Integrating Biology and the Bedside (i2b2)/Veteran’s Affairs (VA) challenge. Given a medical problem concept mentioned in a clinical note (e.g., chest pain, pneumonia), the purpose of a system solving the task of assertion classification is to determine whether the concept was present, absent, conditional, hypothetical, possible, or not associated with the patient.20 Relevant examples extracted from the i2b2 dataset for each assertion category are shown below.
Cervical spine x-ray showed evidence of significant osteoarthritis. (present)
Heart was regular with a I/VI systolic ejection murmur without jugular venous distention. (absent)
She also noted increased dyspnea on exertion. (conditional)
Please return to the hospital if you experience chest pain, shortness of breath, lightheadedness or dizziness. (hypothetical)
The patient was admitted to the Cardiac Medicine Service and treated for presumed diastolic and systolic dysfunction.(possible)
The patient’s father died at age 69 with metastatic prostate cancer. (not associated with the patient)
For instance, the medical problem emphasized in boldface in example (3) has assigned a conditional assertion category because this problem is experienced by the patient only under certain conditions. Similarly, the medical problems from example (4) have associated a hypothetical assertion category because they are problems that the patient may develop. Additional annotation examples are provided in the annotation guidelines for assertion classification at https://www.i2b2.org/NLP/Relations/.
Our set of features for assertion classification include lexical features that explore the surrounding context of a medical concept in text, features that encode information about the section headers in clinical reports, and features extracted using the NegEx21 and ContText22 tools. We also proposed a novel set of semantic features trying to capture the meaning of specific keywords which convey various forms of information that trigger a specific assertion category. The list of keywords that we identified as being able to signal an assertion category are the negation cues (e.g., not, without, absence of) from the BioScope corpus23, the speculative (or hedge) cues (e.g., suggest, possible, might) from the same corpus, the temporal signals (e.g., after, while, on, at) from TimeBank24, and a list of kinship terms (e.g., father, mother, sister, aunt) from the Longman English Dictionary. Based on this set of features, our classifier managed to achieve performant results for assertion classification.
With this assertion classifier in hand, we associated one of the six assertion categories with each pneumonia expression from our dataset. We extracted the pneumonia expressions by first parsing the reports with MetaMap and then selecting only the identified medical phrases that have the same identifier as pneumonia (CUI:C0032285) in the UMLS Metathesaurus. For a more complete set, we also ran simple regular expressions to identify the word pna, an abbreviation often used by physicians in clinical reports for pneumonia but which is not yet tagged as a pneumonia concept in the UMLS Metathesaurus. After we ran the assertion classifier for all the pneumonia concepts of a patient, we counted how many times each of the six assertion values were identified, and then mapped the most frequent value to one of the two categories of pneumonia identification. From all ways of performing these mappings, we identified only two of them as more plausible. Since it is obvious that present belongs to positive, and absent and associated with someone else to negative, the two possible mappings encode whether the assertion types that express hedging (i.e., conditional, hypothetical, and possible) are attached to positive or negative pneumonia. We found that a good mapping of the assertion values is {present} → positive pneumonia, and {absent, possible, conditional, hypothetical, associated with someone else} → negative pneumonia. For those binary features corresponding to the 223 patients with no pneumonia concepts identified in their reports, we assigned a default value of negative pneumonia.
Experiments
Our main goal in this section is to assess how well a classifier will perform for the task of pneumonia identification in the event when only a restricted set of time-ordered reports is available to each patient. For this purpose, we projected the patient reports on a timeline, which is similar with the timeline shown in Figure 1, and then we built classifiers corresponding to each element on this timeline. More exactly, for training a classifier corresponding to a timeline element at time t, we considered for each patient only the reports having the same timestamp t or a timestamp that happened before t. For instance, for the first element on the timestamp, we trained a classifier considering for each patient only its corresponding admit notes. As an observation, for this first timeline element (i.e., AN), we could not use the entire cohort of 426 patients because, as listed in Table 1, not all the patients have at least an admit note. Similarly, for the classifier associated with the second element on the timeline, we represented each patient by only its corresponding admit notes and the reports created in its first day of ICU stay. Finally, when training the classifier associated with the last element on the timeline, we used the entire set of reports for each patient. Of note, we associate all the discharge summaries with the last element on the timeline, in spite of the fact that the length of stay in the ICU for the patients may range widely. We believe this is the most natural projection that characterizes the entire set of reports on a timeline because discharge summaries represent special report types where clinicians provide the last conclusions about the state of the patients, give details of the final diagnosis, and prescribe treatments. Once all the classifiers are trained for each element of the timeline, a practical system for pneumonia surveillance running in a hospital can now invoke the appropriate classifier for a patient based on the period of time the patient spent in the hospital.
Since we used the same dataset as used by Yetisgen-Yildiz et al., we considered that work as the baseline for our system. This system consists of a supervised learning framework, where the feature vector corresponding to a patient is represented as a “bag of words” built from patient’s reports.1 Yetisgen-Yildiz et al., experimented with different representations of the patient data but concluded that using all of the report types for feature generation gave the best performance, and consequently, for our systems, we used only the representation comprised of all report types. As features, they considered various combinations of word n-grams, UMLS concepts, and their corresponding semantic types. Besides performing experiments on the entire cohort of 426 patients (the unrestricted dataset), they also considered a smaller set of 236 patients restricted to those with both an admit note and discharge summary (the restricted dataset). Using a 5-fold cross validation scheme, their system achieved the best results of 50.7 and 49.1 F-measure on the restricted and unrestricted dataset, respectively, when the entire set of word uni-grams was considered. For an accurate comparison, for each experiment associated with a time element, we used the same folds as used by Yetisgen-Yildiz et al.
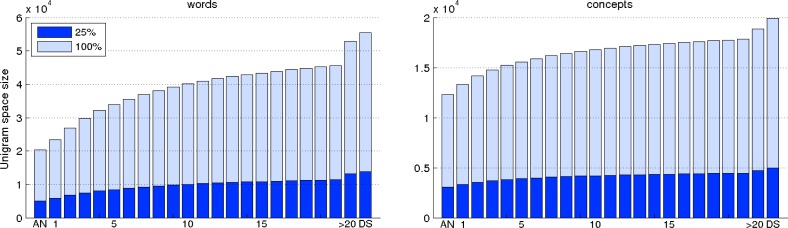
In a first set of experiments, we fixed the threshold values for the ranked list of relevant word and UMLS concept features to 25% from the total number of corresponding features in the unigram feature space. In consequence, because the dataset corresponding to a timeline element t is formed from the dataset corresponding to the t-1 element and the reports having the timestamp t, the threshold values for both words and UMLS concepts are monotonically increasing. This kind of behavior is captured in the plots depicted in Figure 3. In this figure, the bars having a light color represent the size of the entire unigram feature space associated with each dataset on the timeline whereas the bars with a dark color represent 25% from the total number of distinct features associated with each timeline element. As can be seen, the number of distinct word uni-grams varies from ∼20,000 (for the first element) to ∼55,000 (for the last element) while the range corresponding to the number of distinct UMLS concepts is from ∼12,000 to ∼20,000. For selecting the word and UMLS concept bi-grams, we set the same threshold values as used for selecting the unigram features. It is also worth mentioning that, in our previous study, we evaluated a system using the statistical feature selection approach with a wide range of threshold values for both words and UMLS concepts in the case when an entire set of reports is available. We observed that considering only the first 25% of the most relevant word and UMLS concept uni-grams as well as an equivalent number of bi-gram features achieved the best performing results for the majority of experiments.2 Therefore, in this study, we used the same percentage of features from the total number of distinct unigram features for feature selection.
Figure 3.
The feature space sizes of words (the plot on the left) and UMLS concepts (the plot on the right) corresponding to each element on the timeline. The bars having a light color indicate the total number of distinct features used by the baseline system and the bars having a dark color indicate the threshold values used in the statistical feature selection approach.
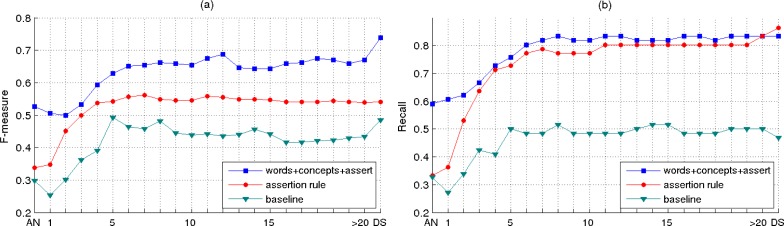
The evolution of the performance results achieved by the systems considered for pneumonia identification over time is shown in the plots from Figure 4. Besides extracting the results achieved by our system and the baseline, we also considered a rule-based system in which the decision for a patient having pneumonia or not is based on the value of the assert feature associated with the patient. For both plots (a) and (b) shown in Figure 4, in the configuration of our system, we considered the combination of all the feature types proposed for pneumonia identification. This is because, in our previous study for identifying pneumonia the best performing results were achieved by using this type of configuration.2 Also, for these plots, the threshold values for selecting the relevant features in our system are set to 25% from the total number of features corresponding to each timeline element.
Figure 4.
Performance results for pneumonia identification when the feature selection method considers only 25% from the total number of features corresponding to each timeline element.
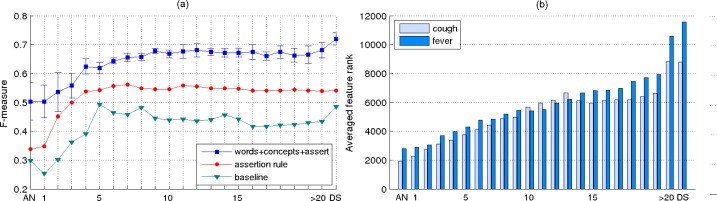
In the plot shown in Figure 4(a), we report the system results in terms of F-measure, which represents the harmonic mean of precision and recall. As can be observed, the results obtained by our system are significantly better than the baseline results and the results achieved by the rule-based system. For this configuration setup, the best F-measure of 73.83 corresponds to the last classifier on the timeline, which considers as dataset the entire set of reports for each patient. Of note as well, the evolution of the performance results achieved by the classifiers across the timeline elements shows some fluctuation. This behavior suggests that the order of relevance across the features computed using the t statistical test does not guarantee a perfect order of the features for pneumonia identification. Therefore, noisy features exist at the top of the lists ranked by this statistical test that have a negative impact on our system performance; conversely, there exist relevant features towards the bottom of the ranked lists which can improve the performance of our system.
For the results shown in Figure 4(b), we considered the same system configuration as used for extracting the results in Figure 4(a), but we report the results in terms of recall. This is equivalent with showing the performance results of the classifiers when considering only the positive patients for pneumonia. Furthermore, since the recall values are proportional with the true positive values, the result curves in the plot from Figure 4(b) can be interpreted in terms of the percentage of patients positive for pneumonia that can be identified by our system at a specific time interval on the timeline. As an additional observation from this plot, it can be noticed that the best recall value is achieved by the rule-based system which indicates a bias of this system towards predicting the patient annotated as positive for pneumonia (88 false positives misclassified by the rule-based system vs. 28 by the machine learning system). Although in this paper we focus more on evaluating the patients with pneumonia, we also measured the performance for identifying negative patients. For instance, one of our best classifier achieved a negative predicted value of 95.4 and specificity of 98.6 when the entire set of reports was considered.
In another set of experiments, we trained the entire set of classifiers using fixed threshold values for the ranked lists of word and UMLS concept n-grams. For this purpose, we experimented with the threshold values for word n-grams from the set {100, 500, 1000, 2000, 5000, 10000} and with the threshold values for concept n-grams from the set {50, 100, 500, 1000, 5000}. The averaged F-measures (+/– standard deviation) over all the experiments considered are shown in Figure 5(a). For comparison purposes, in this figure, we also show the results of the baseline and rule-based systems. In general, from both Figure 4(a) and Figure 5(a), we can affirm that the classifiers’ performance shows an increasing trend in the first 5–6 days since the patients were admitted to ICU, followed by an insignificant improvement for the remaining ICU days. The performance is further increased when discharge summaries are also considered in the training process. This is also because, in general, adding more data results in an increase in the classifier’s prediction. If in the first ICU days the decision for identifying patients with pneumonia is mostly based on the patient symptoms and initial physician assessments, the performance increases as more data become available (e.g., lab results, new findings). Finally, the clinical information encoded in discharge summaries proved to be a significant source of evidence for pneumonia identification.
Figure 6.
(a) Performance results for pneumonia identification using a fixed number of relevant features. (b) The impact of symptom features for pneumonia identification across the timeline elements.
In the last experiment, we investigated the impact of the relevant features ranked by the t statistical test on the classification performance corresponding to each element on the timeline. Since each timeline element is associated with a different dataset, their corresponding lists of ranked features are also different. Therefore, an interesting experiment would be to measure the relevance of a specific set of features across the timeline elements. For this purpose, we considered two of the most relevant symptoms for pneumonia, cough and fever, and identified the ranks of the uni-gram features associated with these symptoms in their corresponding ranked lists. Since there are multiple features that characterize each of these two symptoms, we identified the two sets by mapping all the features that have a fever and cough prefix. For instance, fever, fevers, feverishness, fever/chills, fever/increasing are examples of features representing the fever symptom whereas cough, coughs, coughed, coughing, coughing/agitation constitute examples for the cough symptom. Based on these two sets of features, we computed two general rank scores for each timeline element by averaging the ranks of the individual features. The plot illustrated in Figure 5(b) shows the average ranks for the two symptoms across the timeline elements. In our notation, a feature with a lower rank indicates a more significant feature. As can be noticed, the results shown in this plot confirm the intuition that the pneumonia symptoms are more relevant indicators of the disease in the first days of admission after which they gradually recede in importance.
Conclusion
In this paper, we described a system for pneumonia identification using various types of clinical reports, and evaluated its performance for the cases when only a restricted set of time-ordered reports is available. This type of evaluation is useful for pneumonia surveillance applications that need to invoke the proper system configuration for each ICU patient. In the experimental section, we showed that our system achieved satisfactory results for each type of dataset configuration that considered for training only a limited set of time-ordered clinical reports. For instance, even when using only the admit notes as training dataset, our system was still able to achieve better results than a baseline previously proposed for pneumonia identification that was trained over the entire set of reports. Furthermore, we showed that the set of features representing two common symptoms for pneumonia become less significant over time for the task of predicting this phenotype. It remains the case that there is significant clinical information which contributes to a diagnosis of pneumonia that is not present in the narrative reports included in the dataset, as evidenced by the fact that such information is summarized in the discharge summaries. We intend to further explore this gap in a new dataset of ICU patients, where we will have available both structured data (e.g., white blood cell count, temperature) as well as free-text clinical reports.
Acknowledgments
We would like to thank Heather Evans, Michael Tepper, and Fei Xia for their insightful comments. This research was partly supported by P50 HL073996, RC2 HL101779, the Northwest Institute for Genetic Medicine, and Microsoft Research Connections.
References
- 1.Yetisgen-Yildiz M, Glavan BJ, Xia F, Vanderwende L, Wurfel MM. Identifying patients with pneumonia from free-text intensive care unit reports. Proceedings of Learning from Unstructured Clinical Text Workshop of the International Conference on Machine Learning; 2011. [Google Scholar]
- 2.Bejan CA, Xia F, Vanderwende L, Wurfel MM, Yetisgen-Yildiz M. Pneumonia identification using statistical feature selection. J Am Med Inform Assoc. doi: 10.1136/amiajnl-2011-000752. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iregui M, Ward S, Sherman G, Fraser VJ, Kollef MH. Clinical importance of delays in the initiation of appropriate antibiotic treatment for ventilator-associated pneumonia. Chest. 2002;122(1):262–8. doi: 10.1378/chest.122.1.262. [DOI] [PubMed] [Google Scholar]
- 4.Chapman WW, Haug PJ. Comparing expert systems for identifying chest x-ray reports that support pneumonia. Proc AMIA Symp; 1999. pp. 216–220. [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman WW, Fiszman M, Chapman BE, Haug PJ. A comparison of classification algorithms to automatically identify chest x-ray reports that support pneumonia. Journal of Biomedical Informatics. 2001;34:4–14. doi: 10.1006/jbin.2001.1000. [DOI] [PubMed] [Google Scholar]
- 6.Fiszman M, Chapman WW, Evans SR, Haug PJ. Automatic identification of pneumonia related concepts on chest x-ray reports. Proc AMIA Symp; 1999. pp. 67–71. [PMC free article] [PubMed] [Google Scholar]
- 7.Fiszman M, Chapman WW, Aronsky D, Evans RS, Haug PJ. Automatic detection of acute bacterial pneumonia from chest x-ray reports. Journal of the American Medical Informatics Association. 2000;7(6):593–604. doi: 10.1136/jamia.2000.0070593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendonca EA, Haas J, Shagina L, Larson E, Friedman C. Extracting information on pneumonia in infants using natural language processing of radiology reports. Journal of Biomedical Informatics. 2005;38(4):314, 321. doi: 10.1016/j.jbi.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Lutfiyya MN, Henley E, Chang LF, Reyburn SW. Diagnosis and treatment of community-acquired pneumonia. Am Fam Physician. 2006;73(3):442–50. [PubMed] [Google Scholar]
- 10.Mandell LA, Wunderink RG, Anzueto A, Bartlett JG, Campbell GD, Dean NC, Dowell SF, File TM, Jr, Musher DM, Niederman MS, Torres A, Whitney CG. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(Suppl 2):S27–72. doi: 10.1086/511159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glickman ME, Gagnon DR. Modeling the effects of genetic factors on late-onset diseases in cohort studies. Lifetime Data Anal. 2002;8(3):211–28. doi: 10.1023/a:1015880003274. [DOI] [PubMed] [Google Scholar]
- 12.Stell A, Sinnott R, Jiang J. A federated data collection application for the prediction of adverse hypotensive events. Proceedings of the 9th International Conference on Information Technology and Applications in Biomedicine; 2009. pp. 1–4. [Google Scholar]
- 13.Genovese EA, Dew MA, Teuteberg JJ, et al. Incidence and patterns of adverse event onset during the first 60 days after ventricular assist device implantation. Ann Thorac Surg. 2009;88(4):1162–70. doi: 10.1016/j.athoracsur.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang P, Chen MH, Sinha D. A latent model approach to define event onset time in the presence of measurement error. Stat Interface. 2009;2(4):425–35. doi: 10.4310/sii.2009.v2.n4.a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glavan BJ, Holden TD, Goss CH, Black RA, Neff MJ, Nathens AB, Martin TR, Wurfel MM, ARDSnet Investigators Genetic variation in the FAS gene and associations with acute lung injury. Am J Respir Crit Care Med. 2011;183:356–63. doi: 10.1164/rccm.201003-0351OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quirk C, Choudhury P, Gao J, Suzuki H, Toutanova K, Gamon M, Yih W, Cherry C, Vanderwende L. MSR SPLAT, a language analysis toolkit. Proceedings of NAACL HLT 2012 Demonstration Session; 2012. [Google Scholar]
- 17.Fan RE, Chang KW, Hsieh CJ, Wang XR, Lin CJ. LIBLINEAR: A library for large linear classification. J Mach Learn Res. 2008;9:1871–4. [Google Scholar]
- 18.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp; 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 19.Manning CD, Schutze H. Foundations of statistical natural language processing. MIT Press; 1999. [Google Scholar]
- 20.Uzuner Ö, South BR, Shen S, DuVall SL. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18:552–6. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform. 2001;34:301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 22.Chapman WW, Chu D, Dowling JN. ConText: An algorithm for identifying contextual features from clinical text. BioNLP 2007: Biological, translational, and clinical language processing. 2007:81–88. [Google Scholar]
- 23.Vincze Veronika, Szarvas György, Farkas Richárd, Móra György, Csirik János. The Bio-Scope corpus: biomedical texts annotated for uncertainty, negation and their scopes. BMC Bioinformatics. 2008;9(Sup 11):S9. doi: 10.1186/1471-2105-9-S11-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pustejovsky James, Hanks Patrick, Sauri Roser, See Andrew, Gaizauskas Robert, Setzer Andrea, Radev Dragomir, Sundheim Beth, Day David, Ferro Lisa, Lazo Marcia. The TimeBank Corpus. Corpus Linguistics. 2003:647–656. [Google Scholar]