Abstract
Prognostic models are increasingly being used in clinical practice. The benefit of adding variables (e.g., gene expression measurements) to an original set of variables (e.g., phenotypes) when building prognostic models is usually measured on a whole set of cases. In practice, however, including additional information only helps build better models for some subsets of cases. It is important to prioritize who should undergo further testing. We present a method that can help identify those patients might benefit from additional testing. Our experiments based on limited breast cancer data indicate that relatively old patients with large tumors and positive lymph nodes constitute a group for whom prognoses can be more accurate with the addition of gene expression measurements. The same is not true for some other groups.
Introduction
Traditionally, the prognosis of the cancer has been established by regression models based on clinical and pathological characteristics of the patient and the tumor. The features in predictive models for breast cancer usually include tumor size, number of lymph nodes, histological grade, patient age, menopausal status, and hormone receptor status 1. These characteristics are included in predictive models such as the Nottingham Prognostic Index (NPI), Adjuvant!Online, and the guideline from the St. Gallen expert panel 2. However, patients with identical clinical-pathological characteristics have different prognoses. The inability to correctly classify a patient into prognostic categories may be in part explained by some missing features. Such features could include molecular signatures associated with cancer prognosis 3.
Two example gene signatures for breast cancer prognosis are the 70-gene signature by Van’t Veer et al. 4 and the 21-gene signature by Paik et al.5 The 70-gene signature was shown to have a higher predictive power for identifying breast cancer patients for adjuvant therapy than the standard system using clinical and histological criteria. The 21-gene signature produced the recurrence score (RS) to quantify the likelihood of recurrence in tamoxifen-treated patients with node-negative, estrogen-receptive-positive breast cancer. The RS was predictive of overall survival. After independent validation, these two signatures were commercialized with the name of MammaPrint (Agendia, Amsterdam, Netherlands) for the 70-gene signature, and OncotypeDX (Genomic Health, Redwood, CA, USA) for the 21-gene classifier 6. Currently two randomized clinical trials are being conducted with these products: “Microarray In Node negative Disease may Avoid ChemoTherapy (MINDACT) 7 with MammaPrintTM” in Europe and “Trial Assigning Individualized Options for Treatment (TAILORx)8 with OncotypeDXTM” in the US. MINDACT is comparing MamaPrint with common clinical-pathological criteria in selecting patient for adjuvant chemotherapy in non-negative breast cancer. TAILORx is comparing hormone therapy alone vs. hormone therapy in combination with chemotherapy for women whose conditions are lymph node negative, estrogen receptor and/or progesterone receptor positive. The treatment that patients will receive in TAILORx trial will depend upon the results of the recurrence score by OncotypeDX test.
Despite these commercial successes in breast cancer, the utility of adding gene signatures to a prediction model based on clinicopathological features alone is still controversial. Riester et al. 9 have shown an increase of 0.14 in the C-statistic, which is equivalent to the Area Under the ROC Curve (AUC) of a predictive model by adding a gene signature to the clinicopathlogical characteristics for prediction of survival for high-risk bladder cancer patients. On the other hand, Dunkler et al. 10 did not find significant gain in prediction accuracy by adding gene signatures to clinicopathlogical features for breast cancer prognosis. Furthermore, the gene list constituting the gene signature is unstable, as it varies with the selection of patients. According to Ein-Dor et al. 11, several thousands of patient samples would be needed to construct a reliable, stable list of genes, however, existing gene classifiers were built on a few hundred samples. A universal predictive model that is reliably applicable to all individuals is a long-term goal. But we can seek short-term solutions in constrained populations. For example, Cario et al. 12 showed that the resistance to the chemotherapy could be predicted with high accuracy using a gene signature in children with acute lymphoblastic leukemia (ALL) with similar clinical characteristics. Although there is some work on subpopulation-specific medicine based on genomic information, little has been done to analyze the benefits of acquiring and combining gene expressions with phenotypes for prognosis.
To improve patient outcome prediction and make practical use of genomic information for prognosis 13, a detailed analysis is necessary to identify the advantages of combining genetic information with clinical phenotypes. A natural question is whether some phenotypically-homogeneous subpopulations of patients become more distinct given their gene expressions (in terms of outcomes), and therefore should be prioritized for genome sequencing. To tackle these challenges, we use clustering and classification methods to investigate if certain subpopulations are more likely to benefit from the integration of gene expressions to phenotypes for the purpose of prognosis. A recent model 14 shared our motivation for finding subpopulation variability. The authors investigated genotype-based subpopulation-specific ideal drug dosages. However, this work is different from ours, as the authors did not use a data-driven approach to find the subpopulations. Our focus on identifying the subpopulations that are most likely to benefit from additional testing is also different from the one reported by Kaila et al.15, which focused on the accuracy of classification for a given individual on a specific subpopulation.
In the rest of this paper, the Methodology section presents the details of our approach, followed by a section on Experiments, which contains a description of the publicly available breast cancer data set we used and our results. Finally, we discuss the advantages and limitations of our models.
Methodology
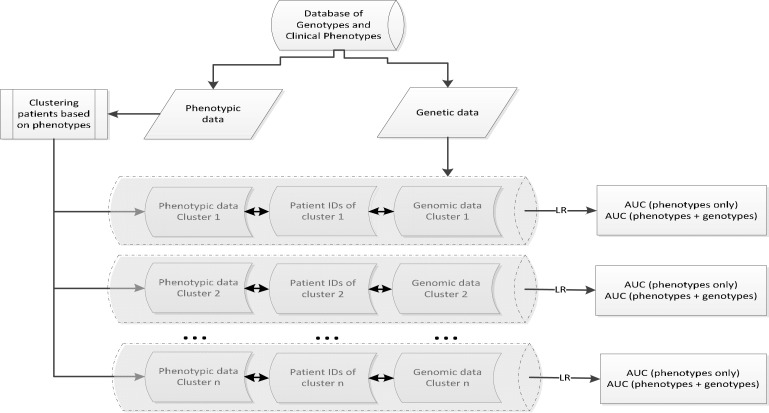
We start to evaluate our intuition using a straightforward two-step workflow (i.e., clustering and classification). The clustering step aims at finding meaningful subpopulations based on phenotypes, in a data-driven manner. Then, the classification step serves as the validation for the difference in prognosis accuracy after additional genomic information is combined. A high level overview for our proposed approach is illustrated in Figure 1.
Figure 1:
Workflow of our study. First, databases of gene expression and clinical phenotypes were separated. Next, patients were clustered according to their phenotypes. Then, depending on whether gene expressions were incorporated, we formed two sets of databases (i.e., phenotypes only vs. combined phenotypes and gene expression) to feed a logistic regression model where AUCs were computed, one for each data set. Finally, we compared the differences between AUCs to check how they differ. (LR: logistic regression; AUC: Area Under the ROC Curve).
Clustering:
The first step is to find phenotypically-homogeneous subpopulations in the database. There are many sophisticated algorithms for clustering 16–19 and we adopted a widely used technique, the k-means model 20 that aims at partitioning n observations into k mutually exclusive clusters, in which each observation belongs to a cluster with the nearest mean. The k-means model essentially minimizes the following objective function,
where Ci is the mean of points in the cluster Si ⊆ S and k corresponds to the number of clusters. Note that S ={S1, S2, … , S3} denotes the entire data set.
Classification:
We used the logistic regression (LR) model 21 to learn from clustered subpopulations. Specifically, LR uses the sigmoid function to link a linear model Xβ (i.e., X denotes a data point and β denotes a set of weight parameters) in the following form,
where P(Y = 1/X) corresponds to the probability of an event Y to be 1 (i.e., the positive outcome). The parameters β can be calculated through maximum likelihood estimation, as detailed in the article by Minka22.
Evaluation:
We compared LR models constructed with and without considering gene expressions based on the Area Under the ROC curve (AUC) 23, a measurement for discrimination. To ensure the validity of our findings, we applied ten-fold cross-validation 24 to evaluate the AUC. For each data set (e.g., a cluster of phenotypes or a cluster of combined phenotypes and gene expressions), we split patients evenlly into 10 folds, using 9 folds for training and the remining one fold for testing. The process was repeated 10 times until every patient in the data set got a prediction score. These scores were evaluated against patient outcomes (i.e., gold standard) to determine the total number of discordant pairs (i.e., how many pairs in which patients with positive outcomes were predicted to have lower risk than patients with negative outcomes), and AUCs were calculated accordingly. The standard deviation of AUCs were computed using the parametric method, and we calculated the p-values for the difference of AUCs using a one-sided z-test (please refer to Lasko 25 for details on methods to compare AUCs).
Experiments
We ran experiments on an Intel i7 2.67GHz machine with 4GB RAM. The program was written in R, Matlab, and Java. We used ROCR 26 to visualize ROCs and we used Weka 27, an open source machine learning toolkit, to preprocess the data to handle missing values.
Data description
We used a breast cancer data set GSE3494, which is publically available at (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3494).
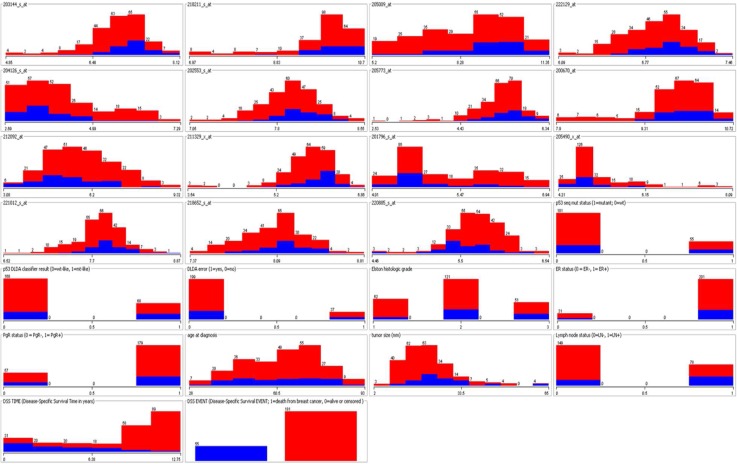
A total of 251 patients were retrieved from the link above. There are 10 phenotype features and one outcome variable indicating if a patient died, was alive, or had an unknown status for breast cancer. We filtered patients to get rid of those who had an unknown status, which left 15 patients out. For the remaining 236 patients, each of them is associated with 22,283 gene expression features, from which the top 15 features were selected based on the ranking of their student-t test p-values, as suggested by Osl 28. Note that we used gene expressions generated from the platforms GPL96. For details about GSE3494 and samples’ Affymetrix platform, please refer to Miller 29. Table 1 summarizes our experiment data, which consist of 25 predictor variables and one outcome variable. Figure 2 illustrates the distribution of all 26 attributes.
Table 1:
List of Phenotype features, selected gene expressions, and the outcome variable of GSE 3494 dataset.
| Phenotype features | Gene expressions (GPL96 platform) | Outcome variable |
|---|---|---|
|
| ||
| p53 seq mut status (1=mutant; 0=wt) | ‘203144_s_at’ | DSS event (1=death from breast cancer, 0= alive or censored) |
| p53 DLDA classifier result (0=wt-like, 1=mt-like) | ‘218211_s_at’ | |
| DLDA error (1=yes, 0=no) | ‘205009_at’ | |
| Elston histologic grade | ‘222129_at’ | |
| ER status (0 = ER-, 1= ER+) | ‘204126_s_at’ | |
| PgR status (0 = PgR-, 1= PgR+) | ‘202553_s_at’ | |
| age at diagnosis | ‘205773_at’ | |
| tumor size (mm) | ‘200670_at’ | |
| Lymph node status (0=LN-, 1=LN+) | ‘212092_at’ | |
| DSS time (Disease-Specific Survival Time in years) | ‘211329_x_at’ | |
| ‘201796_s_at’ | ||
| ‘205490_x_at’ | ||
| ‘221012_s_at’ | ||
| ‘218652_s_at’ | ||
| ‘220885_s_at’ | ||
Figure 2:
Distribution of GSE3494 variables. Two colors (e.g., red and blue) were used to represent the patient who died from breast cancer and the people who stayed alive, respectively. The first 15 subfigures correspond to selected gene expression levels, which are all numerical attributes. The next 10 subfigures represent phenotypes, which are mostly nominal attributes except for age, tumor size, and DSS time. The final subfigure shows the proportion of alive and deceased patients.
Because the last phenotype feature (i.e., DSS time) is directly related to the outcome variable, we left it out from the prediction analysis.
Results
We started with two experiments to confirm our intuition that certain patients (i.e., a subpopulation) become more distinct (in terms of predictive modeling) when their gene expressions are considered. Specifically, we clustered patients into three and four phenotypically-homogeneous subpopulations. The reason of using two different group parameters is to check the generalizability of findings. The k-means algorithm was used twice for generating different numbers of groups. Because patients have multiple phenotype features, we used a spider diagram to show the center (i.e., median of the points) of each cluster.
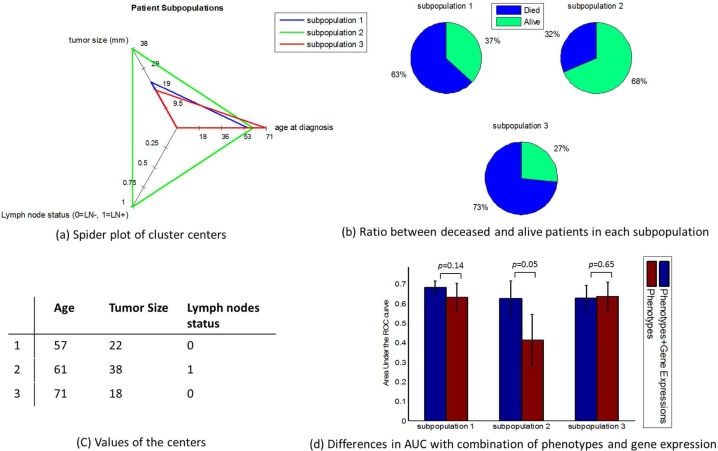
In the case of the three-cluster grouping, results indicated that there were no differences between all dimensions of cluster centers but three (i.e., age at diagnosis, tumor size, and lymph node status). Therefore, we illustrated cluster centers on these three dimensions in Figure 3(a). We plotted in Figure 3(b) the distribution of alive and deceased patients in each cluster. Figure 3(c) shows the value of each cluster center. Finally, Figure 3(d) depicts the AUCs and standard deviations of AUCs with and without considering gene expression in all three clusters. The results indicate that the prediction accuracy for different subpopulations are not the same when additional genomic information is considered in building the predictive model.
Figure 3:
Discrimination within each of the three subpopulations by considering gene expressions in addition to phenotypes. These clusters are depicted in various ways (a) radar plot for cluster centers, (b) class distribution, (c) values of cluster centers. The AUCs within each subpopulation before and after combining phenotypes and gene expressions are displayed in subfigure (d), showing various degrees of improvement in different subpopulations.
Specifically, subpopulation 3, which is relatively large (n=124 patients), demonstrated no gain in discrimination before and after gene expressions were combined with phenotypes. This subpopulation corresponds to patients with negative lymph nodes and relatively small tumors. A relatively smaller cluster, subpopulation 1 (n=92 patients) seemed to gain more benefits (i.e., the averaged AUC increased 0.05) after gene expressions were taken into consideration. The smallest cluster, subpopulation 2 (n=20 patients), had a significant improvement in terms of average AUCs (0.21 improvement). The p-values for the AUC differences are displayed in Figure 3(d).
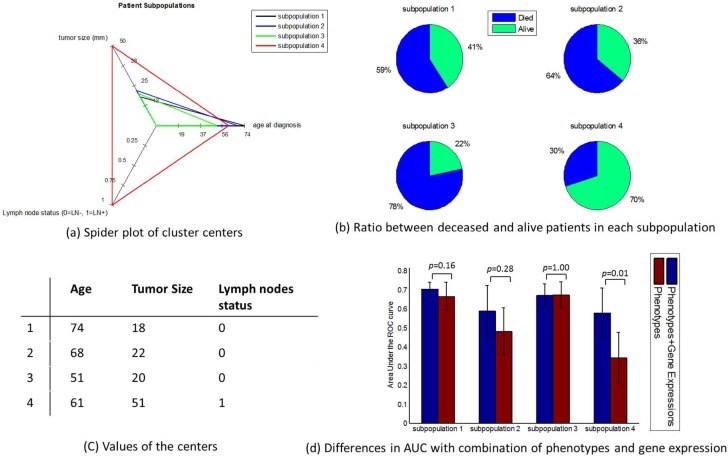
Regarding the four-cluster scenario, the cluster centers were similar to the three-cluster scenario, in all but three dimensions. We plotted the centers of the clusters in Figure 4(a). Figure 4(b) and 4(c) show the patient outcome distribution and values of cluster centers, respectively. Figure 4(d) illustrates AUCs and standard deviations of AUCs with and without considering gene expressions for prognosis in all four clusters. Among them, the patient subpopulation 4 (n=21 patients) showed the most significant improvement of the average AUC (0.24), and the p-value equaled 0.01. Indeed, a majority of the patients in this group overlap with those in the subpopulation 2 of the three-cluster scenario. The other subpopulations (i.e., 1, 2 & 3) had n=44, 72, and 99 patients, respectively, and their AUC improvements were not significant (0.04, 0.11 and 0.001).
Figure 4:
Discrimination within each of the four subpopulations by considering gene expressions in addition to phenotypes. These clusters are depicted in various ways (a) radar plot for cluster centers, (b) class distribution, (c) values of cluster centers. The AUCs within each subpopulation before and after combining phenotypes and gene expressions are displayed in subfigure (d), showing various degrees of improvement in different subpopulations.
As indicated by the result, most patients only had marginal gains when both phenotypes and gene expressions were considered. However, results of both experiments showed that this was not true for certain subgroups.
After confirming the subpopulation variability for prognostic accuracy, we explored an approach to assess the probability of having comparable and under/over-estimations for individuals (with and without considering gene expressions). Two sets of data, one with only phenotypes and the other with combined phenotypes and gene expressions, were first constructed. Their predicted values based on the logistic regression were compared in order to assign a class label to each patient record using the criteria (i.e., “error ≥ −0.1 & error ≤ 0.1” corresponds to a label “comparable” and “error ≤ −0.1 & error ≥ 0.1” corresponds to a label “under/over-estimate”). These new class labels, along with original phenotypes, were combined to create a training set for a logistic regression to predict probabilities of comparable and under/over-estimation when genetic expression information was ignored. Note that we used ±0.1 as our cutoff to separate comparable and under/over-estimation cases just to illustrate our model. The outcome was evaluated in terms of AUC, for which the mean of ten-fold cross validation was 0.875 and the standard deviation was 0.064, which indicates that there was a great potential of discriminating patients who were most likely to benefit from gene expression measurement.
Discussion and Limitations
We showed that certain phenotypically-homogeneous subpopulations of breast cancer patients are more distinctive when their gene expressions are considered (in terms of outcomes), as compared to the entire population. Our exploratory model for measuring the “comparable” and “under/over-estimation” probabilities demonstrates the potential of a simple and intuitive way to support decision making based on evidences. Partnering with experts in palliative care and healthcare providers focused on oncology will allow us to explore this direction more deeply in the future.
A limitation of this work is that the size of our data is very small. Although we believe our results are generalizable, intensive experiments must be conducted on larger data to confirm the usefulness of our model. We are working with clinical phenotypes and whole-genome sequencing data for Kawasaki disease 30 as an extension of the work presented here, which contains only the methodological innovation, but it still of limited immediate clinical significance. Another limitation is that we used simple feature selection and clustering approaches in this experiment, which might have not been ideal. In the future, we will investigate advanced methods like fused LASSO 31 for feature selection and supervised approaches for clustering 32. We would also like to extend our exploratory analysis to multiple classes to consider situations like underestimate, comparable, and overestimate, separately, instead of using binary logistic regression to learn dichotomous targets.
Conclusion
In conclusion, we studied whether and which phenotypically-homogeneous subpopulations of patients are more distinctive (in terms of outcomes) when their genetic information is considered. While this research is preliminary, it presents a methodological innovation that has shown promising results. Further clinical validation and an in depth cost-effectiveness analysis are warranted.
Acknowledgments
The authors were funded in part by the NIH grants 1K99LM011392-01, R01LM009520, U54 HL108460, R01HS019913, and UL1RR031980. We thank Mrs. Jialan Que and Dr. Robert El-Kareh for helpful discussions.
References
- 1.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? Journal of the National Cancer Institute. 2010;102:464–74. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Retèl VP, Joore Ma, Knauer M, et al. Cost-effectiveness of the 70-gene signature versus St. Gallen guidelines and Adjuvant Online for early breast cancer. European Journal of Cancer (Oxford, England : 1990) 2010;46:1382–91. doi: 10.1016/j.ejca.2010.02.035. [DOI] [PubMed] [Google Scholar]
- 3.Dunkler D, Michiels S, Schemper M. Gene expression profiling: does it add predictive accuracy to clinical characteristics in cancer prognosis? European journal of cancer (Oxford, England : 1990) 2007;43:745–51. doi: 10.1016/j.ejca.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 4.van’t Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–70. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 5.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. The New England Journal of Medicine. 2004;351:2817–26. doi: 10.1056/NEJMoa041588. [DOI] [PubMed] [Google Scholar]
- 6.Paik S. Development and clinical utility of a 21-gene recurrence score prognostic assay in patients with early breast cancer treated with tamoxifen. The Oncologist. 2007;12:631–5. doi: 10.1634/theoncologist.12-6-631. [DOI] [PubMed] [Google Scholar]
- 7.Cardoso F, Piccart-Gebhart M, Van’t Veer L, et al. The MINDACT trial: the first prospective clinical validation of a genomic tool. Molecular Oncology. 2007;1:246–51. doi: 10.1016/j.molonc.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sparano Ja. TAILORx: trial assigning individualized options for treatment (Rx) Clinical Breast Cancer. 2006;7:347–50. doi: 10.3816/CBC.2006.n.051. [DOI] [PubMed] [Google Scholar]
- 9.Riester M, Taylor JM, Feifer A, et al. Combination of a novel gene expression signature with a clinical nomogram improves the prediction of survival in high-risk bladder cancer. Clinical Cancer Research : an official journal of the American Association for Cancer Research. 2012;18:1323–33. doi: 10.1158/1078-0432.CCR-11-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunkler D, Michiels S, Schemper M. Gene expression profiling: does it add predictive accuracy to clinical characteristics in cancer prognosis? European journal of cancer (Oxford, England : 1990) 2007;43:745–51. doi: 10.1016/j.ejca.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 11.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. PNAS. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cario G, Stanulla M, Fine BM, et al. Distinct gene expression profiles determine molecular treatment response in childhood acute lymphoblastic leukemia. Blood. 2005;105:821–6. doi: 10.1182/blood-2004-04-1552. [DOI] [PubMed] [Google Scholar]
- 13.Ohno-Machado L, Bafna C, Boxwala A, et al. iDASH. Integrating data for analysis, anonymization, and sharing. Journal of the American Medical Informatics Association. 2012;19:196–201. doi: 10.1136/amiajnl-2011-000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirchheiner J, Brøsen K, Dahl ML, et al. CYP2D6 and CYP2C19 genotype-based dose recommendations for antidepressants: a first step towards subpopulation-specific dosages. Acta psychiatrica Scandinavica. 2001;104:173–92. doi: 10.1034/j.1600-0447.2001.00299.x. [DOI] [PubMed] [Google Scholar]
- 15.Kaila N, Straka RJ, Brundage RC. Mixture models and subpopulation classification: a pharmacokinetic simulation study and application to metoprolol CYP2D6 phenotype. Journal of Pharmacokinetics and Pharmacodynamics. 2007;34:141–56. doi: 10.1007/s10928-006-9038-9. [DOI] [PubMed] [Google Scholar]
- 16.Airoldi EM, Blei DM, Fienberg SE, et al. Mixed Membership Stochastic Blockmodels. Journal of Machine Learning Research. 2008;9:1981–2014. [PMC free article] [PubMed] [Google Scholar]
- 17.Basu S, Banerjee A, Mooney R. Semi-supervised clustering by seeding. Proceedings of the Nineteenth International Conference on Machine Learning; Sydney, Australia. 2002. pp. 19–26. [Google Scholar]
- 18.Gibson D, Kleinberg J, Raghavan P. Clustering categorical data: an approach based on dynamical systems. The VLDB Journal The International Journal on Very Large Data Bases. 2000;8:222–236. [Google Scholar]
- 19.Finley T, Joachims T. Supervised clustering with support vector machines. Proceedings of the 22nd international conference on Machine learning; Bonn, Germany. 2005. pp. 217–224. [Google Scholar]
- 20.MacQueen J. Some methods for classification and analysis of multivariate observations. Proceedings of the fifth Berkeley Symposium on Mathematical Statistics and Probability; 1967. pp. 281–297. [Google Scholar]
- 21.Hosmer DW, Lemeshow S. Applied logistic regression. New York: Wiley-Interscience; 2000. [Google Scholar]
- 22.Minka T. A comparison of numerical optimizers for logistic regression. CMU Technical Report. 2003;2003:1–18. [Google Scholar]
- 23.Zou KH, Liu AI, Bandos AI, et al. Statistical evaluation of diagnostic performance: topics in ROC analysis. Boca Raton, FL: CRC Press, Chapman & Hall; 2011. [Google Scholar]
- 24.Duda RO, Stor DG. Pattern classification. New York, NY: Wiley; 2001. [Google Scholar]
- 25.Lasko TA, Bhagwat JG, Zou KH, et al. The use of receiver operating characteristic curves in biomedical informatics. Journal of Biomedical Informatics. 2005;38:404–415. doi: 10.1016/j.jbi.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 26.Sing T, Sander O, Beerenwinkel N, et al. ROCR: visualizing classifier performance in R. Bioinformatics (Oxford, England) 2005;21:3940–1. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- 27.Witten I, Cleary J, Cunningham S-J, et al. Weka Machine Learning Project. 2000. http://www.cs.waikato.ac.nz/ml/weka/
- 28.Osl M, Dreiseitl S, Kim J, et al. Effect of data combination on predictive modeling: a study using gene expression data. AMIA Annual Symposium Proceedings; 2010. pp. 567–571. [PMC free article] [PubMed] [Google Scholar]
- 29.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popper SJ, Shimizu C, Shike H, et al. Gene-expression patterns reveal underlying biological processes in Kawasaki disease. Genome biology. 2007;8:R261. doi: 10.1186/gb-2007-8-12-r261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tibshirani R, Saunders M, Rosset S, et al. Sparsity and smoothness via the fused lasso. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 2005;67:91–108. [Google Scholar]
- 32.Dettling M, Bühlmann P. Supervised clustering of genes. Genome Biology. 2002;3:1–15. doi: 10.1186/gb-2002-3-12-research0069. [DOI] [PMC free article] [PubMed] [Google Scholar]