Abstract
Recognizing the anatomical location of actionable findings in radiology reports is an important part of the communication of critical test results between caregivers. One of the difficulties of identifying anatomical locations of actionable findings stems from the fact that anatomical locations are not always stated in a simple, easy to identify manner. Natural language processing techniques are capable of recognizing the relevant anatomical location by processing a diverse set of lexical and syntactic contexts that correspond to the various ways that radiologists represent spatial relations. We report a precision of 86.2%, recall of 85.9%, and F1-measure of 86.0 for extracting the anatomical site of an actionable finding. Additionally, we report a precision of 73.8%, recall of 69.8%, and F1-measure of 71.8 for extracting an additional anatomical site that grounds underspecified locations. This demonstrates promising results for identifying locations, while error analysis reveals challenges under certain contexts. Future work will focus on incorporating new forms of medical language processing to improve performance and transitioning our method to new types of clinical data.
Introduction
Radiology reports, as well as a wide variety of other types of Electronic Health Records (EHR), contain a wealth of spatial information expressed in unstructured natural language text. These reports largely describe findings (e.g., “obstruction”, “normal”, “cancer”) relative to the anatomical sites where they are found (e.g., “small bowel obstruction”, “the pancreas is normal”, “lung cancer”). Further, radiology reports could add significant additional value to clinical care when their unstructured information is extracted and represented in a structured form. Important among this unstructured information are the actionable findings and the anatomical locations related to them. Medical informatics systems have proven adept at utilizing structured information in uses such as visualization[1], decision support[2,3], and flagging alarm conditions[4,5], all of which help improve patient care. The spatial properties of the actionable findings in these reports is a significant piece of knowledge that helps detect, prevent, and resolve medical problems.
Radiology reports are inherently spatial by nature. A radiologist conducts a medical imaging exam and interprets the image(s), conveying the important information to the requesting doctor[6]. This entails expressing both normal and abnormal findings. Many of these abnormal findings need to be spatially grounded in order to make a proper medical diagnosis or to determine the best form of treatment. For instance, CT scans are commonly requested to investigate the presence of tumors and the condition of bone fractures. Both of these findings can occur almost anywhere in the body, such as the lung or brain for tumors or the skull or femur for bone fractures. Since both the requesting doctor and radiologist understand that such findings need to be spatially grounded, the anatomical location is clearly stated in natural language. What may be clear to a human reader, however, is not necessarily easy for an automated system to identify. Often, the anatomical location is not within the same noun phrase or even the same sentence as the actionable finding. This necessitates a method capable of recognizing long-distance spatial relations between an actionable finding and its anatomical location such as in the following example:
(1) The [appendix]Location is seen medial and inferior to the cecum. It measures only 7mm in diameter. There are no [inflammatory changes]Finding.
Furthermore, the anatomical location is not always presented as a concept mappable to an existing medical knowledge base (e.g., the heart or lung). Often, a section of the anatomical part is given (e.g., the wall or membrane), or a nearby anatomical part is given to provide spatial clarity (e.g., lymph nodes along the esophagus). Both the stated location and the anchoring location (if it exists) must be identified in order to anatomically ground the actionable finding.
To address these problems, we have developed a natural language processing (NLP) approach for identifying the location of actionable findings. Our approach has two novel aspects not present in previous approaches to anatomical location detection: (1) we recognize anatomical locations semantically related to actionable findings that cannot be easily identifiable by a simple lexical or syntactic pattern, such as those specified in separate sentences, and (2) we recognize underspecified anatomical locations (e.g., a part of an organ such as its base, wall, or lobe) and identify their corresponding anatomical site (e.g., lung, bowel, or liver). We apply our method to locating inflammation and similar abnormalities indicative of appendicitis. While the corpus we have created is tailored to appendicitis and its related problems, our approach can easily be re-trained for different abnormalities in other kinds of clinical reports.
Background
Anatomical locations are commonly extracted by concept recognition systems. The best known of these is MetaMap[7], which is capable of recognizing both actionable findings and anatomical locations as well as normalizing those concepts by mapping them to their proper entry in UMLS[8]. The 2010 i2b2/VA Shared Task[9] included recognizing several types of medical concepts (problems, treatments, and tests), the assertion status of medical problems, and certain types of relations between concepts. The 2011 i2b2/VA Shared Task[10] included recognizing anatomical location concepts, but did not include identifying spatial relations between concepts.
Several systems have been proposed to recognize common ways of expressing a finding’s location. The SemRep system[11,12,13] is capable of recognizing location relations between concepts discovered by MetaMap. SemRep identifies this relationship when two requirements are met: (1) the UMLS concepts allow for one concept to be the other’s location (e.g., a concept that has a Categorization relation with ‘Body Part, Organ, or Organ Component’ can be the location of a concept that has a Categorization relation with ‘Therapeutic or Preventative Procedure’), and (2) a specified syntactic relation exists between the two concepts (e.g., “ablation of pituitary gland”). Similarly, MedLEE[2,14] utilizes a grammar for detecting locations of clinical findings in combination with a concept regularizer. The system of Taira et al.[15] extracts frames which have location relations as one of the principle elements. They incorporate some machine learning components based on rule learning and maximum entropy instead of relying on medical ontologies. But similar to SemRep and MedLEE, they rely on very short-distance relations, essentially building a probabilistic grammar.
Two primary limitations of these grammar-based approaches are that (1) they utilize a small set of hand-crafted (or hand-verified) rules designed largely for precise identification (as many syntactic constructions are underspecified, requiring wider context for achieving high recall), and (2) many anatomical location concepts are themselves under-specified (e.g., Examples (7)–(11) below) and require an unambiguous anatomical grounding which often cannot be captured by grammars, such as when the spatial groundings are in separate clauses or sentences (e.g., Example (1)).
Data
In order to study the efficacy of our method, we decided to apply it to a reasonably well understood problem with a moderate amount of spatial underspecificity. Specifically, we selected appendicitis, which is often diagnosed with radiological findings such as anatomical inflammation and thickening, both of which are possible throughout the body and therefore need to be spatially disambiguated. After IRB approval, The University of Texas Southwestern Medical Center’s EHR database (utCRIS-CRDW) was queried for CT reports from patients with an appendicitis-related diagnosis (ICD9 540-543.99) and an abdominal CT scan. Since the diagnosis is associated with the patient, not the CT report, many of the reports are un-related to appendicitis. De-identification software[16] was run on the CT reports and a random subset was manually evaluated to ensure the effectiveness of the de-identification. 7,230 reports (25%) were provided for system development, with the remaining being reserved for future evaluation.
400 reports were selected for annotation by radiologists for the presence of appendicitis. These reports were studied for the various ways in which radiologists expressed the presence of appendicitis. Notably, 25% of reports marked as positive for appendicitis did not contain the word “appendicitis”. Instead, the radiologists determined the presence of appendicitis largely using the presence of inflammation in conjunction with the inflammation’s anatomical grounding. This suggests that spatially underspecified findings (such as inflammation) can add significant value if properly grounded to their anatomical location. We next describe how our spatial relations are represented and annotated.
Methods
A. Representation
After the evaluation of radiographic images, radiologists prepare a radiology report that includes the significant findings made as a result of the image evaluation. In many cases, these findings are “actionable”, i.e. the findings indicate that certain types of follow-up actions should occur, including therapeutic and/or surgical interventions or additional diagnostic tests. Given a target problem (appendicitis in our case), a set of spatially ambiguous finding terms are specified based on the common ways in which radiologists express symptoms or related problems. For the problem of appendicitis, we have used the finding terms specified in Table 1. These finding terms form the basis of our representation. Note that some of the following examples are not directly related to appendicitis, but are instead derived from reports in which appendicitis is a suspected medical finding. Thus, the relations we learn help to determine the likelihood of appendicitis based on the location of the findings from Table 1.
Table 1:
Terms associated with radiology findings for appendicitis. Each column represents a different class of finding.
| inflammation | inflamed | thickening | rupture | perforation | infiltration | enlarged |
| inflammatory changes | thickened | ruptured | perforated | infiltrate | ||
The Location is an anatomical part that either contains the finding or is in close proximity to the finding. This can be a direct relationship between a finding and an anatomical part as shown below:
(2) The [spleen]Location is [enlarged]Finding, measuring 18.5cm in length...
(3) ...[periappendiceal]Location [inflammatory changes]Finding.
(4) There are [inflammatory changes]Finding around the [appendix]Location.
Additionally, a finding may be linked to some related finding or process that has an unambiguous anatomical location. In this case, we consider the anatomically distinct finding to be the location of the associated finding since it can easily be mapped to its anatomical location with a resource such as SNOMED CT[17]. Examples of this case include:
(5) ...suggestive of [perforated]Finding [appendicitis]Location .
(6) ...a [perforation]Finding of [diverticulitis]Location .
Since appendicitis is uniquely associated with the appendix and diverticulitis is typically associated with the colon, radiologists often omit an explicit anatomical location of the finding (e.g., perforation in the above examples) and thus labeling these problems (appendicitis and diverticulitis) as the Location is therefore sufficient. As can be seen in the examples, the Location is often specified with a common syntactic construction, typically a simple noun phrase or a prepositional phrase with a spatial preposition (e.g., “around”, “in”). However, the Location is often underspecified and does not uniquely identify a specific anatomical part. The finding must then be associated with a second, unambiguous anatomical part (often in a different sentence). We refer to this unambiguous anatomical part as the Anchor, as it provides an absolute grounding for the finding’s Location. We have observed two common cases where Anchors are necessary. The first is when the specified Location is relative to, or forms a part of, some other anatomical part. The second involves non-unique anatomical parts such as lymph nodes, which are found all throughout the body. Examples of both of these cases include:
(7) ...[bowel]Anchor demonstrates normal caliber and no [wall]Location [thickening]Finding .
(8) ...[inflammatory changes]Finding in the [fat]Location around the [appendix]Anchor .
(9) ...[colon]Anchor is not dilated and does not show any [mesenteric]Location [inflammation]Finding .
(10) ...[enlarged]Finding [lymph nodes]Location are seen in the [chest]Anchor .
(11) ...[thickening]Finding at the [tip]Location of the [cecum]Anchor .
In each of these cases, both the marked Location and Anchor are necessary to determine the most specific, unambiguous location of the finding as possible. For instance, in Example (8) the inflammation is not found in the appendix, but in the nearby fat. Simply extracting “fat” as the Location would limit the types of action that could be applied because fatty inflammation in different parts of the body could indicate different underlying problems or necessitate different solutions. Conversely, extracting just “appendix” as the Location could alter the medical diagnosis as well, as an inflamed appendix is more indicative of appendicitis than inflammation in the nearby fat. We therefore extract both the Location and Anchor in order to properly represent the finding location and provide spatial information that is as complete as possible.
B. Annotation
Our manual annotation process focused on a small list of abnormal findings consistent with appendicitis such as inflammation and thickening. The complete list of finding terms is shown in Table 1. All of these terms can occur throughout the body in different anatomical locations, so whenever they are used a specific anatomical grounding is usually present. Every instance of these terms in the corpus of CT reports described above were retrieved and 1000 instances were selected at random for annotation. We formally define our types for the purpose of manual annotation:
Definition: A Location is the syntactically closest anatomical part stated by the radiologist as the finding’s location.
Definition: An Anchor is the syntactically closest unambiguous anatomical part for a given finding, provided the Location itself is an ambiguous anatomical part.
Two human annotators (the first two authors) manually labeled the Locations and Anchors as described above. This process took less than one human day. The annotators were limited to 3-sentence windows around the finding term. This window was used because (1) in almost every case the Location (and Anchor, if necessary) could be found within this window, and (2) it is very unlikely for an automatic algorithm to detect longer-distance relationships. After initial annotation, the two annotators resolved many of the inconsistencies in their labeling in order to form a more homogeneous annotated data set. Based on these annotations, we decided to approach this problem using a supervised classification framework.
C. Classification Framework
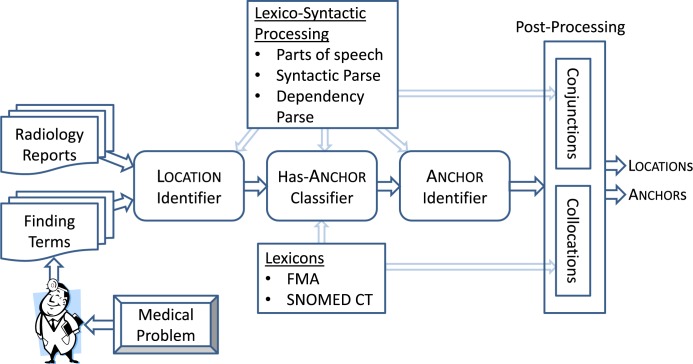
The architecture of our method is shown in Figure 1. After the selected finding terms are identified in the radiology reports, three machine learning classifiers are used for anatomical grounding. All three are implemented using the Lib-LINEAR support vector machine library[18]. The first classifier, which we refer to as the Location identifier, decides which token in the 3-sentence window around the finding term is the best head (i.e., last) token of the Location. Only the highest confidence Location, as decided by the classifier, is selected. The second classifier, which we refer to as the Has-Anchor classifier, performs a binary classification to decide whether the Location needs an additional Anchor. If the Has-Anchor classifier decides an Anchor is necessary, the third classifier, which we refer to as the Anchor identifier, selects which token in the 3-sentence window around the finding term is the best Anchor head token. Next, if the selected Location or Anchor is used in a conjunction, we add additional Locations and Anchors. Finally, these single-token Locations and Anchors are expanded to multi-tokens (collocations) when appropriate by utilizing medical lexicons. For more details on how conjunctions are handled and how multi-token Location and Anchors are expanded, see Post-processing below.
Figure 1:
Architecture of our method. Radiology reports and a set of finding terms are processed in a pipelined classifier structure followed by heuristic post-processing modules.
D. Features
Each of the three classifiers has its own unique set of features to help the classifier learn its intended task. A large set of features were considered in our experiments, with the exact sets of features for each classifier being chosen by automatic feature selection techniques[19]. Thus, the chosen features are reflective of the types of information necessary to learn each of the individual tasks.
Location features:
The candidate Location word (e.g., “nodes”).
The morphological lemma of the candidate Location (e.g., “node”).
Both the candidate Location word and the finding term’s lemma. This helps identify Locations that are more likely to be found with specific findings (e.g., an enlarged appendix is more likely than an enlarged liver).
Both the candidate Location word and the next word (to the right).
The number of tokens between the finding and candidate Location. This helps the classifier favor tokens closer to the finding
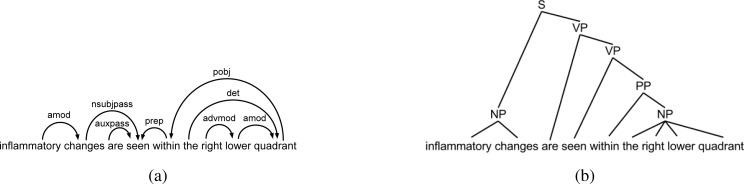
The path from the finding to the candidate Location following the grammatical dependency structure[20]. This structure forms a tree over the sentence tokens and contains relations such as nsubj (grammatical subject), dobj (direct object), and amod (adjective modifier). For instance, the feature value for “[inflammatory changes]Finding are seen within the right lower [quadrant]Location ” would be nsubjpass-prep-pobj, as “changes” is the passive subject of “seen”, which has the preposition modifier “within”, whose object is “quadrant”. See Figure 2(a). The advantage of this feature is that it represents the syntactic relationship between the finding and candidate Location, thus allowing the classifier to recognize common linguistic constructions. No dependency path exists between tokens in separate sentences, and paths with more than 10 edges in the dependency graph are ignored due to sparsity issues.
The path along the syntactic parse tree[21]. The path for the above example would be np-s-vp-vp-pp-np. See Figure 2(b). This captures less information than the dependency path but still indicates common linguistic constructions. Again, no syntactic path exists between tokens in separate sentences, and paths with more than 25 edges are ignored.
The parts of speech for the tokens between the finding and candidate Location. For the example above, this feature’s value would be vbp-vbn-in-dt-rb-jjr1. If more than 10 tokens are found between the finding and candidate Location, this feature is ignored.
The syntactic direction from the finding to the candidate Location within the syntactic parse tree. This indicates if the finding syntactically dominates the Location (e.g., “[inflammation]Finding in the [appendix]Location ”) or vice versa. If neither dominates the other, this feature is ignored.
Figure 2:
(a) Example syntactic dependency tree; (b) Example syntactic parse tree.
Has-Anchor features:
The morphological lemma of the selected Location.
The part of speech of the selected Location.
The selected Location word and the previous word.
The selected Location word and the next word.
The part of speech of the next word.
Whether or not there is a finding from the list in Table 1 closer to the selected Location than the finding under consideration. This tends to indicate an Anchor is not needed.
The parts of speech for the tokens between the finding and selected Location (see Location Feature 8).
The class of the finding. This corresponds to the columns from Table 1. This is a useful feature because different types of findings are more likely to require an Anchor than others.
Whether or not the previous token is in a lexicon of diseases (mined from SNOMED CT) associated a specific anatomical part. Often the token before the Location is actually the Anchor, and this feature recognizes a special case of this pattern.
Anchor features:
The morphological lemma of the candidate Anchor.
The previous word.
The words between the selected Location and candidate Anchor. If there are more than 10 tokens between the finding and candidate Anchor, this feature is ignored.
The words between the selected Location and candidate Anchor with casing removed (e.g., “Normal” becomes “normal”).
The number of tokens between the selected Location and candidate Anchor.
The dependency path (see Feature 6 from the Location features above) from the finding to the candidate Anchor. Paths with more than 5 edges were ignored.
The number of dependency edges between the finding and candidate Anchor. This helps the classifier favor closer tokens.
The sentence distance and direction from the finding to the candidate Anchor. For instance, the feature’s value is “0” if they are in the same sentence. If the candidate Anchor is in the sentence after the finding, the feature value is “+1”. If it is two sentences before the finding, the feature value is “−2”.
E. Post-processing
The three classifiers select exactly one Location token and at most one Anchor token. Post-processing then performs two additional tasks: (1) create additional Locations/Anchors when a selected Location/Anchor is used in a conjunction, and (2) expand each Location/Anchor to include additional tokens when necessary.
The first task is designed to recognize when multiple Locations and/or Anchors are present. The most common means for a radiologist to state this is using a conjunction (e.g., “[inflammatory changes]Finding in the [cecum]Location and [appendix]Location ”). We first recognize conjunctions using the sentence dependency parse. The dependency relation conj connects two tokens that are connected by a conjunction. We merge these conjunction pairs to find groups of two or more tokens that belong to a conjunction. Possibly due to the fact that the syntactic parser performs poorly on clinical text (as it was trained on newswire text), we limit conjunction pairs to those that are separated by at most two tokens. Otherwise the poor precision of the conjunction dependency relation negates any additional benefit to recall. Then, all tokens connected to a selected Location by such a short-distance dependency are also identified as Locations. Additionally, new Anchors are identified in the exact same manner.
In order to recognize multi-token Locations and Anchors (e.g., “terminal ileum”, “right lower quadrant”), we utilize lexicons generated from two ontologies: FMA[22] and SNOMED CT[17]. FMA (Foundational Model of Anatomy) contains anatomical terms, while SNOMED CT (Systematized Nomenclature of Medicine–Clinical Terms) contains a wide assortment of clinical terms, from which we extract findings that have unambiguous anatomical locations using the finding site relation. Both lexicons contain tens of thousands of terms. When a selected Location or Anchor token is part of a multi-token term found within one of these lexicons, it is expanded so long as it does not overlap with another annotated Location or Anchor.
Evaluation
We evaluated our NLP method on the set of 1000 appendicitis-related finding instances manually annotated with Locations and Anchors. To measure how our classifiers work on unseen data, we performed 10-fold cross validation and measured precision, recall, and F1-measure. We performed experiments to evaluate the performance of each individual classifier as well as full end-to-end evaluations both excluding and including our post-processing heuristics. Since the classifiers only mark a single token, the initial evaluations only consider the final token of the Locations and Anchors. The results from these experiments are shown in Table 2.
Table 2:
Results on 1000 manually annotated instances of findings from Table 1. Results are shown with both ground truth Locations and automatically annotated Locations (End-to-end).
| Location | Anchor | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| P | R | F1 | P | R | F1 | |
|
| ||||||
| Word Baseline | 40.40 | 39.65 | 40.02 | 33.20 | 31.17 | 32.15 |
| Dependency Baseline | 68.60 | 67.32 | 67.95 | 45.18 | 50.46 | 47.68 |
|
| ||||||
| Ground Truth Locations | 83.00 | 77.92 | 80.38 | |||
| End-to-end | 86.90 | 85.28 | 86.08 | 73.65 | 68.46 | 70.96 |
|
| ||||||
| + conjunction | 86.20 | 85.88 | 86.04 | 73.77 | 69.84 | 71.81 |
For comparison, we consider two baselines with minimal feature sets. The first baseline (“Word Baseline”), simply chooses the Location and Anchor based only on the word string. The second baseline (“Dependency Baseline”), chooses based on the dependency path, effectively learning the most likely linguistic constructions. Both baselines perform much worse than our proposed method. Notably, our method achieves over twice the precision and recall on both tasks when compared to simply choosing the most likely word.
Of the three classifiers (Location, Has-Anchor, and Anchor), the Anchor identifier is the greatest source of error. This is largely due to Anchors being more distant (relative to the finding) than Locations. While the examples given above involve only a single sentence for clarity, 14.5% of Anchors in our data are found outside the finding’s sentence, while only 3.4% of Locations are found outside the finding’s sentence. Additionally, there were an average of 3.4 tokens between a Location and its finding, while the Anchors were an average of 8.3 tokens away from their finding. This largely explains the discrepancy of scores between the Location and Anchor classifiers, but we provide an in-depth analysis on the types of errors for each classifier in the next section.
The conjunction post-processing module was designed to increase recall without significantly impacting precision. For Locations, gains in recall were offset by corresponding losses in precision, resulting in no improvement in Location identification. For Anchors, however, conjunction processing slightly improved precision and improved recall by 1.4 points, contributing to an overall gain in F1-measure of 0.85 points.
Discussion
The results of our method show significant promise. Given a relatively small number of manual annotations, our machine learning-based classifiers are able to effectively learn how to automatically identify Locations and Anchors. Furthermore, they significantly outperform the baselines. The poor performance of the baselines suggests that this task is complex and requires the effective fusion of numerous resources. On the other hand, the success achieved with such a limited number of annotations (by NLP standards) points to the generalizability of our method.
The automatically chosen Location features indicate that Location identification is largely a syntactic recognition task (i.e., recognize which syntactic constructions are typical for Locations). This is not surprising, as our decision to separate Location and Anchor identification was motivated by the observation that spatially ambiguous findings (such as those in Table 1) are usually given a syntactically clear spatial position (the Location), but this was often underspecified and needed a more semantic grounding (the Anchor). This suggests that our Location identifier will prove adept at recognizing cases with common syntactic constructions but would have trouble when rarer syntactic constructions (as well as rarer Location terms) are present. This observation was largely found to hold during an error analysis of our method. One immediately obvious path to improvement is to annotate more data. As stated above, our annotators spent less than 1 day annotating these spatial relations. Our goal, however, was not to achieve the highest possible score, but rather to see how well our method generalizes on a relatively small number of annotations. Our method’s success on this small amount of data suggests it could easily be re-trained for different abnormalities and other types of clinical reports.
The most common type of error for the Location identifier is to select the Anchor token as the Location. Two such examples of this are (Gold indicates ground truth while Selected indicates system output):
(12) ...low density [thickening]Finding of the distal ...
(13) There are [inflammatory changes]Finding surrounding the of the proximal .
Another common type of error was to select a different token with approximately the same meaning as the labeled Location. In the following example, “periappendiceal” is related to the appendix (literally meaning near the appendix), and was marked by the human annotator instead of “appendix” simply because it was directly modifying the finding term:
(14) The is dilated with [inflammatory changes]Finding ...
Both of these types of errors suggest the method performs better than the results in Table 2 may indicate, as the selected location is either equivalent to (in the second case) or simply less specific than (in the first case) the gold Location. A more serious type of error involves locations rarely seen in the data:
(15) Views of the lung reveal [infiltrate]Finding.
(16) Soft [thickening]Finding at EJ .
Other errors involve mistaking a nearby finding’s Location with that of the current finding:
(17) No thickening or adjacent [inflammatory changes]Finding are noted...
In this case, “wall” is the Location and “gallbladder” the Anchor for the “thickening” finding, but the “inflammatory changes” finding does not have an Anchor, only the Location “gallbladder” (“adjacent” is a spatial proximity indicator, not an anatomical part). Given a perfect syntactic parse structure, it could be recognized that “wall” only modifies “thickening”, while “adjacent” refers to the proximity of “gallbladder”. However, syntactic parsers are not trained on clinical data and thus perform poorly relative to their targeted domain. The classifier is then forced to rely on surface-level features such as the token distance, thus making “wall” appear to be a better candidate than “gallbladder”.
The Has-Anchor classifier performs quite well with an accuracy of 92.7%. The data set is quite balanced (50.6% of Locations require Anchors), so the classifier achieves 42 points over the baseline of always predicting an Anchor is necessary. As seen in the Features section, this classifier largely functions by learning what Location words need Anchors and the contextual information it relies on is largely based on a 1-token window around the selected Location. Since all of the features used for Location identification were tried and only the helpful ones were kept, it can be assumed that the features that capture linguistic constructions (such as those based on the dependency parse) provide little value. The only feature of this type used in the Has-Anchor classifier is the parts of speech between the finding and selected Location.
As might be expected from the features, with little contextual information to fall back on, the Has-Anchor classifier performs poorly on rare Location words:
(18) There are [inflammatory changes]Finding around the distal [right ureter]Location. [∅]Anchor
Since the most common case is that an anchor is required (50.6% to 49.4%), the classifier defaults to requiring an Anchor. Additionally, there are some Locations which are spatially unambiguous but marked with Anchors to provide further specificity. In these cases the Has-Anchor classifier has trouble with Locations that do not always have Anchors:
(19) [Inflammatory changes]Finding are seen in the [right lower quadrant]Location around the [cecum]Anchor .
In our data, “right lower quadrant” is marked as a Location 18 times, with 7 of those cases requiring an Anchor, so unless the context is properly represented the classifier will always select that no Anchor is needed. While features were considered which capture the contextual information, these features proved harmful to the overall results.
As seen in Table 2, the Anchor identifier is the largest source of errors. However, an alternative assessment of Anchor detection is to consider all Locations without Anchors (i.e., those that are unambiguous anatomical parts) to also be Anchors. In this case, the end-to-end Anchor performance improves from 70.96 to 77.26. This score is a realistic measure of how well our method detects anatomical locations of findings.
The chosen features for Anchor identification are similar to those for Location identification with a few notable differences. Like Location identification, the candidate Anchor word and its syntactic relationship with the finding are important. But due to our pipelined architecture (see Figure 1), we can include features based on the selected Location as well. Specifically, surface-level lexical features connecting the Anchor to the Location were found to work best, suggesting that content words (as opposed to grammatical relationships) play an important role in determining which token is a valid Anchor for a given Location.
Given the number of words between Anchors and their respective findings (and therefore the diversity of grammatical relations between them), the largest source of errors for Anchor identification unsurprisingly involve guessing the most common Anchor words instead of the specific anatomical part for the finding. Common examples of these errors are:
(20) The , spleen, pancreas, and bilateral adrenal glands show now focal mass... Urinary is partially distended with no calcification or [focal wall]Location [thickening]Finding.
(21) The , also show nondilated caliber, with no adjacent inflammation. diverticulitis is noted, with no [wall]Location [thickening]Finding ...
In our data, both “liver” and “appendix” appear as Anchors far more often than “bladder” and “colonic”. Both of these errors could be overcome with features that recognize syntactic scoping. Specifically, both examples use a copular construction in which the grammatical subjects (“Urinary bladder” and “Colonic diverticulitis”, respectively) dominate the rest of their respective sentences, and thus the findings should be associated with the subject instead of an anatomical part from a previous sentence. Unfortunately, recognizing this semantic property using the available syntactic resources is quite challenging and is thus left to future work.
Conclusion
We have described a natural language processing method for extracting the anatomical location of actionable findings from radiology reports. Since many actionable findings in these reports are spatially ambiguous, the anatomical grounding must be determined in order to allow for the proper medical representation of natural language radiology data. We utilize two primary components: the Location, which is the specific spatial position stated by the radiologist, and the Anchor, which grounds spatially underspecified Locations. Our experiments on a set of 1000 manually annotated instances show promising results with an F1-measure of 86.04 and 71.81 on Locations and Anchors, respectively. The most difficult Locations and Anchors to recognize are those far (token-wise) from the medical finding term. Furthermore, many errors resulted from inaccurate syntactic parsing. If accurate syntactic parses were available, many of the errors resulting from distant Locations and Anchors potentially could be overcome. Unfortunately we are unaware of any freely available full syntactic parser trained on clinical text. For future work, we plan to apply our spatial identifier to natural language medical tasks (such as the identification records consistent with appendicitis) as well as adapting it to other clinical domains.
Footnotes
For a description of each part of speech tag, see http://www.ling.upenn.edu/courses/fall_2003/ling001/penn_treebank_pos.html
References
- 1.Shortliffe Edward H, Hubbard Susan M. Information Systems in Oncology. In: De Vita VT, Hellman S, Rosenberg S, editors. Cancer: Principles and Practice of Oncology. 1989. pp. 2403–2412. [Google Scholar]
- 2.Friedman Carol, Alderson Philip O, Austin John HM, Cimino James, Johnson Stephen B. A General Natural-language Text Processor for Clinical Radiology. J Am Med Inform Assoc. 1994;1(2):161–174. doi: 10.1136/jamia.1994.95236146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heckerman DE, Nathwani BN. Toward Normative Expert Systems. II. Probability-based Representations for Efficient Knowledge Acquisition and Inference. Methods of Information in Medicine. 1992;31:161–174. [PubMed] [Google Scholar]
- 4.Lyman M, Sager N, Tick L, Nhan N, Borst F, Scherrer JR. The Application of Natural-Language Processing to Healthcare Quality Assessment. Medical Decision Making. 1991;11(suppl):65–68. [PubMed] [Google Scholar]
- 5.Sager N, Lyman M, Nhan N, Tick L. Medical Language Processing: Applications to Patient Data Representation and Automatic Encoding. Methods of Information in Medicine. 1995;34:140–146. [PubMed] [Google Scholar]
- 6.Hall Ferris M. Language of the Radiology Report: Primer for Residents and Wayward Radiologists. American Journal of Roentgenology. 2000;175:1239–1242. doi: 10.2214/ajr.175.5.1751239. [DOI] [PubMed] [Google Scholar]
- 7.Aronson Alan R, Rindflesch Thomas C, Browne Allen C. Exploiting a large thesaurus for information retrieval. Proceedings of RIAO; 1994. pp. 197–216. [Google Scholar]
- 8.Humphreys Betsy L, Lindberg Donald AB, Schoolman Harold M, Octo Barnett G. The Unified Medical Language System: An Informatics Research Collaboration. J Am Med Inform Assoc. 1998;5(1):1–11. doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Uzuner Özlem, South Brett, Shen Shuying, DuVall Scott L. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18:552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uzuner Özlem, Bodnari Andreea, Shen Shuying, Forbush Tyler, Pestian John, South Brett. 2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text. J Am Med Inform Assoc. 2011;18:552–556. doi: 10.1136/amiajnl-2011-000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rindflesch Thomas C, Aronson Alan R. Semantic processing in information retrieval. Proceedings of the 17th Annual Symposium on Computer Applications in Medical Care; 1993. pp. 611–615. [PMC free article] [PubMed] [Google Scholar]
- 12.Rindflesch Thomas C. Integrating natural language processing and biomedical domain knowledge for increased information retrieval effectiveness. Proceedings of the 5th Annual Dual-use Technologies and and Applications Conference; 1995. pp. 260–265. [Google Scholar]
- 13.Rindflesch Thomas C, Rajan Jayant, Hunter Lawrence. Extracting molecular binding relationships from biomedical texts. Sixth Applied Natural Language Processing Conference; 2000. pp. 188–195. [Google Scholar]
- 14.Friedman Carol, Shagina Lyudmila, Lussier Yves, Hripcsak George. Automated Encoding of Clinical Documents Based on Natural Language Processing. J Am Med Inform Assoc. 2004;11:392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taira Ricky K, Soderland Stephen G, Jakobovits Rex M. Automatic Structuring of Radiology Free-Text Reports. Radiographics. 2001;21:237–245. doi: 10.1148/radiographics.21.1.g01ja18237. [DOI] [PubMed] [Google Scholar]
- 16.Uzuner Özlem, Sibanda Tawanda C, Luo Yuan, Szolovits Peter. A De-identifier for Medical Discharge Summaries. Artificial Intelligence in Medicine. 2008;42(1):13–35. doi: 10.1016/j.artmed.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stearns Michael Q, Price Colin, Spackman Kent A, Yang Amy Y. SNOMED Clinical Terms: Overview of the Development Process and Project Status. Proceedings of the AMIA Symposium; 2001. pp. 662–666. [PMC free article] [PubMed] [Google Scholar]
- 18.Fan Rong-En, Chang Kai-Wei, Hsieh Cho-Jui, Wang Xiang-Rui, Lin Chih-Jen. LIBLINEAR: A Library for Large Linear Classification. Journal of Machine Learning Research. 2008;9:1871–1874. [Google Scholar]
- 19.Roberts Kirk, Harabagiu Sanda M. A flexible framework for deriving assertions from electronic medical records. J Am Med Inform Assoc. 2011;18(5):568–573. doi: 10.1136/amiajnl-2011-000152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Marneffe Marie-Catherine, MacCartney Bill, Manning Christopher. Generating Typed Dependency Parses from Phrase Structure Parses. Fifth International Conference on Language Resources and Evaluation; 2006. [Google Scholar]
- 21.Klein Dan, Manning Christopher D. Accurate Unlexicalized Parsing. Proceedings of the 41st Annual Meeting of the Association for Computational Linguistics; 2003. pp. 423–430. [Google Scholar]
- 22.Rosse Cornelius, Mejino José LV., Jr A reference ontology for biomedical informatics: the Foundational Model of Anatomy. Journal of Biomedical Informatics. 2003;36(6):489–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]