Abstract
The need to maintain accessibility of the biomedical literature has led to development of methods to assist human indexers by recommending index terms for newly encountered articles. Given the rapid expansion of this literature, it is essential that these methods be scalable. Document vector representations are commonly used for automated indexing, and Random Indexing (RI) provides the means to generate them efficiently. However, RI is difficult to implement in real-world indexing systems, as (1) efficient nearest-neighbor search requires retaining all document vectors in RAM, and (2) it is necessary to maintain a store of randomly generated term vectors to index future documents. Motivated by these concerns, this paper documents the development and evaluation of a deterministic binary variant of RI. The increased capacity demonstrated by binary vectors has implications for information retrieval, and the elimination of the need to retain term vectors facilitates distributed implementations, enhancing the scalability of RI.
Introduction
MEDLINE/PubMed is often cited as the largest database of medical publications; as of early 2012 it contains more than 21 million articles [1]. An important aspect of the accessibility of this repository is the process of indexing. Each publication is assigned a set of keywords from the Medical Subject Headings (MeSH) which facilitates searching and cataloging. Indexing has traditionally been performed by human indexers [2], with a recent move toward semi-automated approaches in which human indexers are assisted by a recommendation system. With hundreds of thousands of publications added each year, this is expensive, labor-intensive, and delays are unavoidable.
Thus, much research has been done on developing methods that automatically recommend MeSH terms for unindexed papers. A common characteristic of most methodologies for MeSH term prediction is that they require a training set. This training set consists of already indexed publications that are used to learn how to assign MeSH terms to new documents. In previous research [3], we have evaluated the utility of variants of Random Indexing [4] as means to support automated indexing. While these methods performed comparably to the state-of-the-art at the time, two obstacles exist that impede their application as a means to support a real-world indexing system. Firstly, as indexing in this manner requires comparing the vector representation of a document-to-be-indexed with the vector representations of all of the other documents in the training set, it is desirable to retain these vectors in very fast random access memory (RAM). However, as we will illustrate, the real-valued vectors used in traditional vector space implementations do not make optimal use of memory capacity. Secondly, as the name suggests, the initial vector representations used in Random Indexing are randomly generated. In order to maintain an index as new documents are added, it is necessary to maintain these randomly generated vectors for future use. This is inconvenient, and limits the possibility for distributed implementations of Random Indexing and its variants.
In this paper we address these issues by (1) evaluating the utility of high dimensional binary vectors as an alternative to the real-valued vectors used in traditional vector space models and (2) by developing a variant of the RI approach that eliminates the need to retain a store of vectors for future indexing.
Background
Vector Space Models
In this paper we are concerned with vector space models for indexing. Vector space models (VSMs) were conceived to be instruments for information retrieval and document indexing [5]. They have been effectively applied to a broad range of informatics related problem such as information retrieval [5], automated indexing [3], or word sense disambiguation [6].
A VSM is is a geometric model in which terms and documents are represented as high dimensional vectors of numbers. Vector representations for documents are derived from the terms they contain, such that documents containing similar distributions of terms will have similar vector representations. Consequently a mathematically computed distance or similarity metric for vectors represents a measure of the relatedness of the respective documents. This builds the semantic foundation on which new documents can be indexed based on index terms assigned by human indexers to their nearest neighbors.
The traditional approach to construct a VSM works by setting up a matrix where the columns represent terms and the rows represent documents [5]. Each entry of the matrix holds a scalar weight that represents how often a term occurs in a document, or some statistical transformation of this raw value. The size of the matrix is determined by the product of the number of unique terms and the number of documents the corpus contains. Since in most cases the storage requirements are prohibitively large, a dimension reduction is desirable. In certain models such as, for example, Latent Semantic Indexing [7], dimension reduction is conducted using a Singular Value Decomposition (SVD).
Two difficulties introduced with this approach are the storage space requirements for the initial document-by-term matrix and the computational complexity of the SVD to alleviate the space problem, which is cubic to the size of the corpus with standard algorithms [8]. Random indexing (RI) [9] is an approach designed to circumvent these two problems. Instead of starting with the document-term matrix, randomly generated vectors with a much lower dimensionality are used as the initial term vectors. These vectors are constructed such that they have a high probability of being orthogonal, or close-to-orthogonal to one another, and therefore serve as a reduced dimensional approximation of the independent column assigned to each term in the original VSM approach. A document vector is created by combining the vectors of the terms that occur in a document (or vice versa if the intention is to generate term vectors, as was the goal in the original implementation of RI). A weighting scheme can be applied to accentuate the contribution of individual terms according to their relative importance. The document vector generation is described in detail in the beginning of the Methods section.
The RI approach has been proven to substantially alleviate space and computational complexity issues [9], and some variants of RI have also been shown to identify meaningful implicit connections between terms such that document similarity is measured on the basis of “latent semantics” rather than on the basis of identical content exclusively, while bypassing the scalability limitations of SVD [10]. However, while these models confer considerable scalability advantages during the process of vector generation, they are difficult to implement in real-world indexing systems.
Firstly, to index new documents (or to make experiments portable or accurately reproducible), it is necessary to keep copies of the initial random term vectors. If the indexing process were to be distributed across several machines, as is desirable in many real-world applications, a local copy of these term vectors would need to be retained on each machine, and synchronized with a central repository as new terms (and hence new term vectors) were encountered. Based on ideas that originated in the discussion group for Semantic Vectors [11], a widely used open source tool for automated indexing, we propose a simple extension to RI by introducing a way in which the term vectors are generated deterministically. Consequently they can be recomputed in a consistent manner and it is guaranteed that a term always is assigned the same vector. This makes it unnecessary to store the term vectors as they can simply be regenerated at any time. Consequently, in a distributed implementation newly encountered terms would receive an identical term vector without the need for synchronization across machines. From a research perspective, another advantage is that the results of different experiments are more readily comparable because the vectors for all terms that the experiments have in common are guaranteed to be the same. That is to say, any performance changes occurring on account of incidental overlap between randomly-constructed vectors would be consistent across experiments.
Secondly, we examine the format of the term and document vectors. VSMs in information retrieval in general, and for RI in particular, have almost exclusively employed vectors that consist of real-valued numbers. However, classification in VSM-based approaches to automated indexing requires comparing the vector of the document-to-be-classified to the vector representations for all of the documents in the training set. To keep this process fast, it is desirable to retain these vectors in RAM. With large document collections like MEDLINE/PubMed, it is essential that available memory be used effectively, and as we will explain subsequently, there is evidence that real-valued vectors are suboptimal with respect to their ability to store information. Within the cognitive science community, high-dimensional binary vectors have been used as means to encode information as an alternative to real-valued vectors [12]. We propose the use of binary-valued vectors for RI as they were introduced by Pentti Kanerva in [13] as the Binary Spatter Code. In this paper we evaluate the capacity of real and binary-valued vectors in the context of an automated indexing task.
MEDLINE/PubMed Indexing
On account of the human-intensive nature of the indexing process, a number of researchers have developed and evaluated approaches that aim to assist human indexers by recommending MeSH terms for newly-encountered MEDLINE/PubMed citations. A detailed review of all of the approaches that have been applied to generate indexing recommendations is beyond the scope of this paper. However, to provide context we include a brief overview of existing approaches here, and refer the interested reader to the publications referenced for further details.
These approaches can be broadly categorized into distributional and vocabulary-based approaches, although combinations of these approaches are utilized in several instances.
Distributional methods are based on the statistical distribution of terms and MeSH terms in previously encountered abstracts. This distributional information is utilized in one of two possible ways. Firstly, and most commonly, it is utilized to identify previously-indexed documents that are similar in nature to newly encountered documents. MeSH terms assigned to these previously encountered documents are then recommended for the document-to-be-indexed. Examples of this approach include the Expert Networks approach [14] and the Reflective Random Indexing approach [3]. Rather than identifying similar documents, other approaches have leveraged similarity in distributional statistics between terms and MeSH terms across an index to map between terms and index terms directly. These approaches include the Linear Least Squares Fit (LLSF) approach [15] and the Pindex program [16]. This sort of distributional information has also been utilized in combination with graph-theory approaches to improve aspects of the indexing process [17].
Vocabulary-based methods draw on domain-specific knowledge resources such as the UMLS Metathesaurus [18] to identify similarities between the terms and/or phrases encountered in abstract text, and canonical forms and synonyms of terms drawn from controlled terminologies, which can then be mapped to MeSH terms. These methods include MetaMap indexing [19], and Trigram Indexing [20] both of which are utilized in the Medical Text Indexer (MTI) system that is currently used to generate MeSH recommendations for human MEDLINE/PubMed indexers [21]. In addition to these fundamental approaches, attempts have been made to the use of information utilized by these approaches as features for supervised machine learning. Examples of this strategy include the use of a Naive Bayes Classifier [22], [23], and more recently the utilization of all three types of information discussed as features for the learning-to-rank algorithm, with impressive gains in performance [24]. For the purpose of this paper, we focus on a nearest-neighbor based approach based on the similarity between vector representations of documents. These vector representations are calculated using a simple variant of the Random Indexing approach [25].
Methods
Initial Term Vector Generation
As explained above, the initial term vectors are generated randomly. As with any other software, random numbers are generated by a pseudorandom number generator, a deterministic algorithm that generates a sequence of “random” values from a seed value. The resulting numbers appear random in that their distribution cannot be distinguished from that of the uniform distribution. In Java, random numbers are also repeatable, which means that for the same seed, the same sequence of random numbers is generated.
The distribution of these pseudorandom numbers makes them suitable for the purpose of generating random vectors for RI. The property of reproducibility is what makes it possible to generate identical vectors for identical seed values. The only prerequisite is to compute the seed value from the term itself. This value is passed to the Java random number generator as seed, and so it is guaranteed that the same term will always be assigned the same vector. To determine a term’s seed, we compute a hash value from the term’s textual representation. For this purpose, we use “Bobcat” [26], a variant of the “Tiger” hash function [27]. It was chosen because it generates hash codes that are 48 bits long which matches the seed size of the Java random number generator. To assess its suitability for our purpose, we compared all hash values of the terms contained in our corpus and did not find any collisions.
Vector Space Model Generation
When real valued vectors are used in accordance with the RI paradigm, the initial term vectors are sparse [25], where sparseness refers to the relatively small number of non-zero components of the vector. The total number of non-zero components is called the seed length. For example, initializing a real valued vector with 500 dimensions one would set a number of randomly distributed components to 1 and the same number to −1. Choosing a number that is low ensures that all initial term vectors have a high probability of being orthogonal, or close to orthogonal to one another because the probability of overlap between non-zero entries is low. Binary vectors are dense: they are generated by randomly assigning either a 0 or a 1 at each dimension of the vector with equal probability, such that there are an equal number of 1’s and 0’s once assignment is complete. In the case of binary vectors this ensures that initial term vectors are orthogonal or close-to-orthogonal. In this space orthogonality is defined as a Hamming distance of a half of the dimensionality [12].
In any case, the document vectors are built from the term vectors by superposing term vectors of the terms that occur in the respective document. Terms have to pass a frequency and a stop word filter. In addition, a term weight is applied that can be computed from local and global distributional statistics. This is done by a scalar weight representing the weight to the vector of the term concerned prior to superposition. The weighting scheme to be used is usually determined empirically; not all corpora expose the same characteristics with the same weighting schemes.
The vector superposition with real valued vectors is straightforward; it is conducted component wise by addition. With binary valued vectors the process is more involved. A real valued1 voting record keeps track of the scalar additions at each component of the vector. The voting record for a dimension is incremented by the term weight if the added vector has a 1 at the respective dimension. Conversely, it is decremented by the term weight if the added vector has a zero at the respective dimension. The superposition of term vectors concludes with a normalization of the voting records, yielding a binary vector2. A positive voting record results in a 1 and a negative record results in a 0 for each component of the resulting vector. Ties are split at random.
Table 1 summarizes the differences between real and binary implementations.
Table 1.
Differences between Real and binary implementations
| Real Vector Implementation | Binary Vector Implementation | |
|---|---|---|
| Elemental Vectors | Sparse real vectors with mostly 0 elements and a small number of +1 and −1 values randomly distributed. | Randomly generated dense binary vectors, with a 50% probability of a 0 or 1 in each dimension. |
| Superposition | Vector addition, and normalization. | Tally “votes” in each dimension. If more 1’s, assign 1, if more 0’s assign 0. Split ties at random |
| Orthogonality | Perpendicular (Cosine=0) | Hamming distance of dimensionality/2 |
Documents about similar topics have many more words in common than documents about different topics. Thus, it can easily be seen that vectors representing similar documents are closer to each other (in multi-dimensional space) than vectors derived from unrelated documents. The similarity is computed mathematically as the cosine of the angle between real valued vectors, and as Hamming distance between binary valued vectors.
For the purposes of this paper, we built a VSM as training set for index term recommendation from a MEDLINE/PubMed Baseline Repository from the year 2009 [29]. From this corpus, we extracted 10,898,894 articles that have an abstract, excluding articles from the test set (see below). A preliminary step is to index this corpus with Lucene [30], which facilitates easy and efficient extraction of all terms along with their local and global frequencies within the whole corpus. To build the document vectors, we use the Semantic Vectors package [31]. Terms from the title and the abstract are encoded into the vectors with the following parameters. The minimum global frequency for a term is 10 and the maximum is 2,000,000. Terms with global frequencies beyond these thresholds are ignored. The stop word list applied is one developed by Salton for his SMART information retrieval system [32], and we use the log entropy term weighting scheme (as described in [33]) for document vector construction:
Our indexing method chooses MeSH terms from the set of most frequently occurring ones within a set of nearest neighbors to a given document. That set of nearest neighbors is determined as follows. For a given test document (not contained in the training set) a vector is built using the parameters described above. An exhaustive search of the training set is performed that identifies the 40 nearest neighbors of the test document. Using Lucene, the MeSH terms for each of those 40 nearest neighbors are extracted. For each of the neighbors, one minus the normalized Hamming distance (for binary vectors) or the cosine distance (for real vectors) to the test document is computed, and this value is assigned as score to each of its MeSH terms. For MeSH terms that occur in connection to more than one document all scores are added. Finally the test document is assigned the 25 highest scoring MeSH terms.
To assess and be able to compare the performance of our scheme, we evaluated it using the 200-document test set from 2007 [34] published by the NLM that was developed for the purpose of evaluating their own indexing software [21]. Accuracy was measured against index terms assigned by human MEDLINE/PubMed indexers using the standard performance metrics of precision and recall. The F1,2 measures were calculated as:
For each document, any time a human indexed term is retrieved, the precision is computed. Taking the mean from these measurements results in the average precision for the document. The mean average precision (MAP) is the mean of all average precisions.
Comparison of binary and real valued vector capacity
To assess the capacity of the different vector types (i.e. in terms of how many units of information can be encoded into the vector), we conducted a series of experiments. We initialized an origin vector and iteratively superposed it with random vectors. After each superposition, we measured the overlap of the superposed product with the origin vector. The overlap is a measure of relatedness between vectors. Once the overlap between the superposition and the origin vector reaches the level of the average overlap between random vectors, there is no longer a useful distinction. With both the indexing and the superposition experiments, we compare real and binary valued vectors that occupy the same amount of memory. In the Semantic Vectors package, real vectors are assigned one float variable of 32 bits per dimension. This means the ratio of dimensionality for binary to real vectors occupying the same amount of RAM is 1:32.
Results
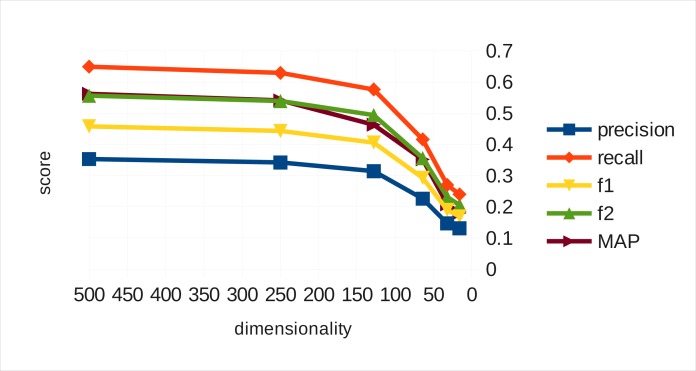
Figure 1 illustrates how the performance of real vectors drops as their dimensionality decreases. From 500 to 250 dimensions the precision and recall loss (and consequently, the F1 and F2 measures and the mean average precision) is marginal. From 250 to 128 the drop is more pronounced bit still somewhat moderate. Below 128 dimensions performance decays drastically.
Figure 1.
Real vector automated indexing performance
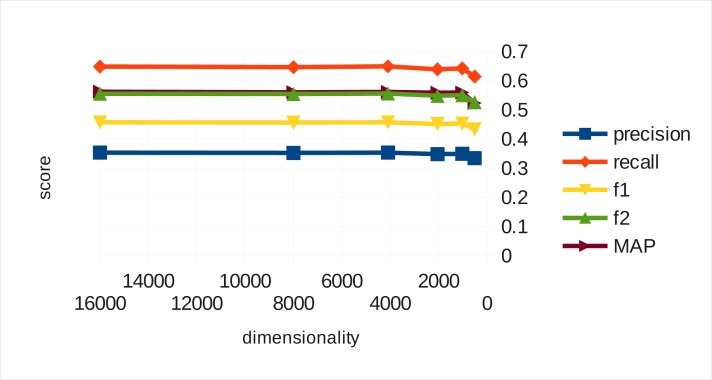
Figure 2 shows the corresponding results for binary vectors. Clearly as dimensionality falls to 4,096, the loss in performance is insignificant. From 4,096 to 2,048 dimensions, the performance drops only marginally. At 512 dimensions, the lowest dimension we measured at, binary valued vectors still perform reasonably well, which stands in stark contrast to the real vectors’ performance at equivalent space requirements. We observe that above 4,096 dimensions3, binary vectors achieve no gain in performance.
Figure 2.
Binary vector automated indexing performance
It is worth adding that although on the x-axis of Figures 1 and 2 the dimension count is different, at each x position the space the different vector representations occupy in RAM is equivalent.
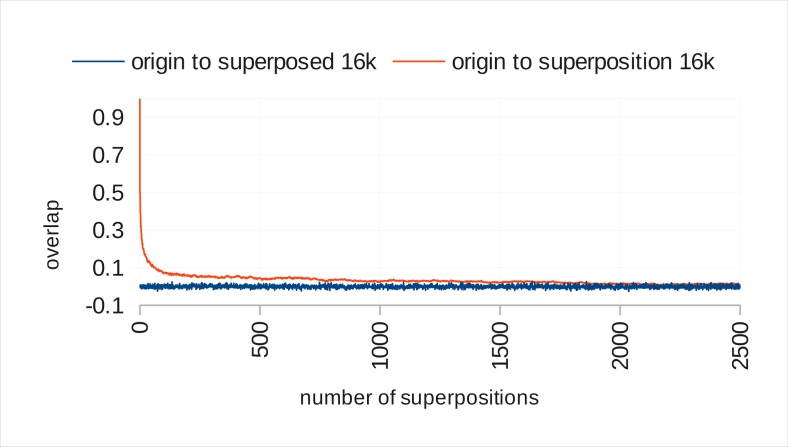
Figure 3 shows the result of the capacity assessment for binary valued vectors. On the x-axis, the number of additions of a random vector (“superposed vector”) to the original vector is counted. The y-axis shows the normalized overlap of origin with the superposition vector (upper curve) and the overlap of the random vector (that is added to the superposition) with the original vector (lower curve). From the point on where the two curves meet, at around 2,000 superpositions, the overlap between superposition and original vector becomes so small that the risk of incidental overlap with random vector emerges.
Figure 3.
Capacity of 16,000 dimensional binary vectors
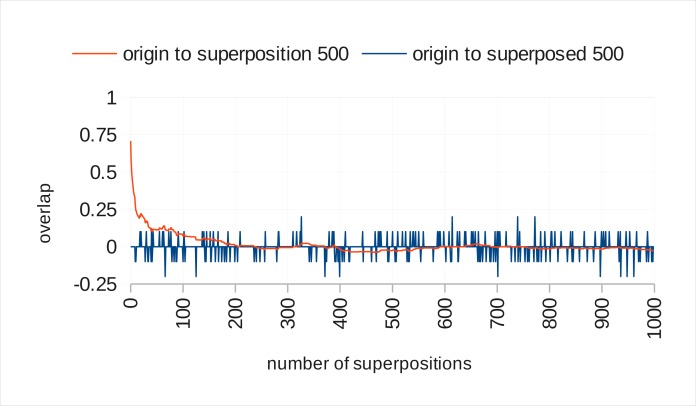
Figure 4 shows the same measurement for 500-dimensional real valued vectors. Their capacity is exhausted after 200 terms. This works well with the MEDLINE/PubMed corpus since most abstracts do not have significantly more words than this. However, with lower dimensionality than this, we would anticipate the model being unable to recognize some of the term vectors that compose a document representation, as their contributions to the superposed product will have become distorted to the point at which they can no longer be distinguished from random vectors representing other terms.
Figure 4.
Capacity of 500 dimensional real vectors
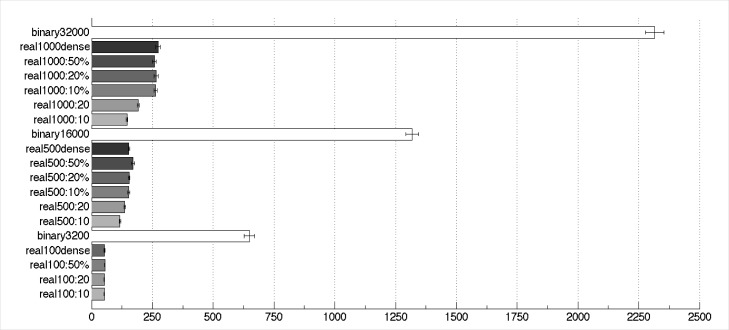
We conducted a series of experiments to assess the capacity of real-valued vectors with varying seed length in comparison to the capacity of binary vectors. The horizontal bars in Figure 5 illustrate the mean number of superpositions until a collision occurred. A collision is defined as the event in which the distance of the origin vector (i.e. the vector before any superpositions) to the superposed product is the same as, or more than, the distance of the origin vector to a random vector. At this point, the superposed vector cannot be distinguished from a random vector. Thus, it is possible that information has been lost, and we say that the capacity of the superposed vector is exhausted. We conducted this experiment 1000 times for real-valued vectors of different dimensionality while varying the seed length. The same was done for binary vectors that occupy the same amount of RAM as the real-valued vectors concerned. In addition, we included real-valued vectors in which each dimension was initialized at random to a value between −1 and 1, which we have termed dense real vectors.
Figure 5.
Mean number of superpositions until first collision (n=1000). Vector descriptions are of the form vector type (real/binary), dimensionality (100–32,000) and, for real vectors, seed length (“10” means 10 non-zero values, “10%” indicates a seed length of 10% of the dimensionality, and “dense” indicates a real vector in which all dimensions are randomly assigned a value between −1 and 1. Error bars show one standard error above and below the means.
While the superiority of the binary vectors in this respect is clear, there is another interesting fact to be observed. At the seed length of 10, which is widely used as a default value by the RI community, the real vectors perform much worse than when a seed length of 10% of the dimension is used (with the exception of the 100-dimensional vectors, where these parameter choices are identical). On account of the variability in the data and the large sample sizes (n=1,000), the differences in the figure between binary and real vectors are statistically significant, as are some of the differences between real vectors of the same dimensionality with different seed length at 500 and 1000 dimensions, notably those between vectors using the standard (10 or 20) and proportional (10% and above) seed lengths. In addition, as the mean number of superpositions to the first collision for real vectors of 100 dimensions is between 52 and 55 depending on seed length, their capacity does not appear adequate to store sufficient term vectors to account for the average number of unique words in an abstract (+- 85 words), which explains the decline in performance of these vectors observed on the indexing task.
Discussion
In this paper we introduced and evaluated the utility of high-dimensional binary vectors as a basis for automated indexing. Using a Random Indexing paradigm, we found that binary vectors provide a substantial advantage in performance at lower dimensionality, when compared with real-valued vectors occupying an equivalent number of bits in RAM. This finding is of practical importance for automated indexing systems based on the vector space model, as document vectors can be retained in RAM for rapid nearest neighbor search with limited computational resources. For example, with 4,096-dimensional binary vectors occupying a total of 4,096 bits we obtained performance equivalent to that obtained by 500 dimensional real valued vectors occupying 16,000 bits. For practical purposes, this means that rather than requiring memory resources beyond the reach of many researchers, efficient nearest-neighbor search can even be accomplished on a well-furnished laptop computer. At 2,048 dimensions, 10,000,000 documents require approximately 2.5 GB of RAM.
Studies comparing the ability of the different vector representations to preserve relatedness to an elemental vector as other elemental vectors are superposed were conducted. It was shown that binary vectors have greater capacity than real valued vectors occupying the same amount of memory. As new vectors are added, the similarity between the superposition and the original elemental vector approaches that between randomly-constructed elemental vectors. This happens with real vectors sooner than it does with binary vectors occupying the same amount of memory. The point at which this occurs is the point at which noise due to incidental overlap between random vectors interferes with the recognition of the encoded elemental vector. Therefore, the number of vectors that can be superposed before this occurs is a measure of the capacity of the vector representation concerned. The increased performance of binary vectors at lower dimensionality is consistent with their increased capacity. One explanation for this may be that the even distribution of information across the entirety of the vectors confers greater robustness in the case of binary vectors, however it is also important to consider that in the case of real vectors, the representational capacity must accommodate both the exponent field and the significand.
With respect to automated indexing, the relatively straightforward approach we have employed in this paper obtained results that are comparable to those achieved by other VSM model implementations and the Medical Text Indexer. As the purpose of this evaluation was to compare real and binary valued vectors rather than optimize indexing performance, we did not experiment extensively with parameters other than dimensionality and vector representation. We also deliberately did not preprocess the text concerned as other researchers approaching the indexing problem have done in the past. Therefore we did not evaluate the extent to which further performance enhancements might be obtained by utilizing stemming, normalization or concept extraction techniques. Nonetheless, even at a dimensionality of 4,096, which is the equivalent in space requirements to the utilization of 128-dimensional float-valued, or 64-dimensional double-valued vectors, we obtained performance comparable to that obtained with relatively high-dimensional real-valued vectors previously. We acknowledge, however, that the best-performing approaches to automated indexing using this test set rely on information beyond that captured by the simple reduced-dimensional approximation of the VSM we have utilized here. In particular, Huang et al. [35] obtained greater accuracy than our approach, combining elaborate preprocessing with a machine learning algorithm that used as additional features surface similarity between document terms and MeSH terms, and statistical metrics derived from the distribution of terms and MeSH terms. It is probable that the accuracy of our method could be augmented by incorporation of this additional information, and utilization of the learning-to-rank algorithm. We will explore the extent to which these additional features can enhance performance in future research efforts.
In addition to demonstrating the relative effectiveness of binary vectors, we introduced a deterministic approach to generating elemental vectors. This approach eliminates the need to maintain a vector store of elemental vectors to enable the addition of further documents. This is of particular importance when considering the development of a distributed implementation of Random Indexing. With the requirement to maintain an elemental vector store, a set of vectors representing all terms thus far encountered would need to be retained and shared between instances of the indexing system. Furthermore, as new terms were encountered, newly generated vectors for these terms would need to be added to the communal vector store, and steps would need to be taken to ensure that two indexing instances do not generate a different vector for the same newly-encountered term. The deterministic approach eliminates these concerns, allowing for convenient implementation of a distributed indexing system, as well as meaningful comparison between variants of Random Indexing across experiments by maintaining the consistency of incidental overlap between elemental vectors.
A limitation of our study is the fact that we used the default number of non-zero values that is given in the Semantic Vectors package for real values vectors for the MeSH term indexing. Although the contributions of this paper would not be affected, it would be good to investigate as to how much the overall indexing results can improve.
It should also be mentioned that the hashing algorithm that is used to compute seed values for the deterministic vector generation has not undergone a closer examination in our study. As the algorithm’s author pointed out, it is worth considering an established hashing algorithm whose robustness was mathematically scrutinized.
Conclusion
In this paper, we document the development and evaluation of two refinements to the Random Indexing based approach to automated indexing. Firstly, rather than the traditional real valued vectors, we utilize high dimensional binary vectors as a representational approach. These are shown to have greater capacity, which allows for equivalent performance with considerably reduced memory requirements when compared with real-valued vectors. This finding has implications for information retrieval applications in general.
In addition, we introduce a deterministic approach to the generation of elemental vectors that eliminates the requirements for maintaining a vector store of elemental vectors for the purpose of indexing future documents. An additional advantage of this development is the ability to distribute processing across machines conveniently. These developments provide the means to negotiate fundamental obstacles to the utilization of Random Indexing in the context of a real-world automated indexing system without sacrificing recommendation accuracy.
In order to enable other researchers to reproduce our results, both our binary vector and deterministic vector implementations will be made available as open source code as a part of the Semantic Vectors [31] package.
Acknowledgments
This research was supported by the US National Library of Medicine grant (R21LM010826-01), Encoding Semantic Knowledge in Vector Space for Biomedical Information Retrieval. We would like to thank Mark Stamp for granting us permission to use the “Bobcat” hash algorithm in the Semantic Vectors package.
Footnotes
In fact, the Semantic Vectors package uses a space optimized implementation approximating a real valued voting record [28].
The voting records are discarded after the normalization process finishes.
The reason for choosing 4,096 instead of 4,000 dimensions (and correspondingly for the smaller values) for binary vectors is that the underlying implementation for binary vectors requires numbers that can be divided by 64.
References
- [1].MEDLINE - Data News and Update Information. [Online]. Available: http://www.nlm.nih.gov/bsd/revup/revup_pub.html#med_update. [Accessed: 28-Feb-2012]
- [2].Aronson AR, Bodenreider O, Chang HF, Humphrey SM, Mork JG, Nelson SJ, Rindflesch TC, Wilbur WJ. The NLM Indexing Initiative. Proc AMIA Symp; 2000. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- [3].Vasuki V, Cohen T. Reflective random indexing for semi-automatic indexing of the biomedical literature. Journal of Biomedical Informatics. 2010 Oct;43(5):694–700. doi: 10.1016/j.jbi.2010.04.001. [DOI] [PubMed] [Google Scholar]
- [4].Kanerva P. Hyperdimensional Computing: An Introduction to Computing in Distributed Representation with High-Dimensional Random Vectors. Cognitive Computation. 2009 Jun;1(2):139–159. [Google Scholar]
- [5].Salton G, Wong A, Yang CS. A vector space model for automatic indexing. Commun. ACM. 1975 Nov;18(11):613–620. [Google Scholar]
- [6].Schütze H. Word Space. ADVANCES IN NEURAL INFORMATION PROCESSING SYSTEMS. 1993;5:895–902. [Google Scholar]
- [7].Berry MW, Dumais ST, O’Brien GW. Using linear algebra for intelligent information retrieval. SIAM review. 1995:573–595. [Google Scholar]
- [8].Golub GH, Loan CFV. An Analysis of the Total Least Squares Problem. SIAM Journal on Numerical Analysis. 1980 Dec;17(6):883–893. [Google Scholar]
- [9].Sahlgren M. An introduction to random indexing. Methods and Applications of Semantic Indexing Workshop at the 7th International Conference on Terminology and Knowledge Engineering, TKE; 2005. [Google Scholar]
- [10].Cohen T, Schvaneveldt R, Widdows D. Reflective Random Indexing and Indirect Inference: A Scalable Method for Discovery of. Biol Med. 1986;30(1):7–18. [Google Scholar]
- [11].Semantic Vectors Discussion Group and Forum. [Online]. Available: https://groups.google.com/forum/#!msg/semanticvectors/Q3l1PziOv4A/o_evK02XPWQJ. [Accessed: 15-Mar-2012]
- [12].Kanerva P. Sparse distributed memory. The MIT Press; 1988. [Google Scholar]
- [13].Kanerva P. Binary spatter-coding of ordered K-tuples. Artificial Neural Networks—ICANN. 1996;96:869–873. [Google Scholar]
- [14].Yang Y, Chute CG. An application of Expert Network to clinical classification and MEDLINE indexing. Proc Annu Symp Comput Appl Med Care; 1994. pp. 157–61. [PMC free article] [PubMed] [Google Scholar]
- [15].Yang Y, Chute CG. A linear least squares fit mapping method for information retrieval from natural language texts. 1992:447–453. [Google Scholar]
- [16].Cooper GF, Miller RA. An Experiment Comparing Lexical and Statistical Methods for Extracting MeSH Terms from Clinical Free Text. Vol. 5. Am Med Inform Assoc; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Herskovic JR, Cohen T, Subramanian D, Iyengar MS, Smith JW, Bernstam EV. MEDRank: Using graph-based concept ranking to index biomedical texts. Int J Med Inform. 2011 Feb; doi: 10.1016/j.ijmedinf.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic Acids Research. 2004;32(Database Issue):D267. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Aronson AR, Lang F-M. An overview of MetaMap: historical perspective and recent advances. Journal of the American Medical Informatics Association. 2010 Apr;17:229–236. doi: 10.1136/jamia.2009.002733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Aronson AR, Bodenreider O, Chang HF, Humphrey SM, Mork JG, Nelson SJ, Rindflesch TC, Wilbur WJ. The NLM Indexing Initiative. Proc AMIA Symp; 2000. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- [21].Aronson AR, Mork JG, Gay CW, Humphrey SM, Rogers WJ. The NLM indexing initiative’s medical text indexer. Medinfo. 2004;11(Pt 1):268–72. [PubMed] [Google Scholar]
- [22].Kim W, Aronson AR, Wilbur WJ. Automatic MeSH term assignment and quality assessment. Proceedings of the AMIA Symposium; 2001. p. 319. [PMC free article] [PubMed] [Google Scholar]
- [23].Sohn S, Kim W, Comeau DC, Wilbur WJ. Optimal training sets for bayesian prediction of MeSH® assignment. Journal of the American Medical Informatics Association. 2008;15(4):546–553. doi: 10.1197/jamia.M2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Huang M, Lu Z. Learning to annotate scientific publications. Proceedings of the 23rd International Conference on Computational Linguistics: Posters; 2010. pp. 463–471. [Google Scholar]
- [25].Kanerva P, Kristofersson J, Holst A. Random indexing of text samples for latent semantic analysis. Proceedings of the 22nd annual conference of the cognitive science society; 2000. [Google Scholar]
- [26].Stamp M. Information security: principles and practice. 2nd ed. Hoboken, NJ: Wiley; 2011. [Google Scholar]
- [27].Anderson R, Biham E. Tiger: A Fast New Hash Function. Fast Software Encryption, Third International Workshop Proceedings; 1996. pp. 89–97. [Google Scholar]
- [28].Widdows D, Cohen T, DeVine L. Complex and Binary Semantic Vectors. Proceedings of the 6th Annual Conference on Quantum Interactions; Paris, France. 2012. to appear in. [Google Scholar]
- [29].MEDLINE/PubMed Baseline Repository. [Online]. Available: http://mbr.nlm.nih.gov/index.shtml. [Accessed: 16-Mar-2012]
- [30].Apache Lucene. [Online]. Available: http://lucene.apache.org/. [Accessed: 16-Mar-2012]
- [31].Semantic Vectors. [Online]. Available: http://code.google.com/p/semanticvectors/. [Accessed: 28-Feb-2012]
- [32].Salton G. The SMART retrieval system: experiments in automatic document processing. Prentice-Hall; 1971. [Google Scholar]
- [33].Martin BI, Berry MW. Mathematical Foundations Behind Latent Semantic Analysis. In: Landauer TK, McNamara D, Sennis S, Klintsch W, editors. Handbook of Latent Semantic Analysis. Lawrence Erlbaum Associates; 2007. [Google Scholar]
- [34].Indexing Initiative Test Collections. [Online]. Available: http://ii.nlm.nih.gov/TestCollections/index.shtml. [Accessed: 15-Mar-2012]
- [35].Huang M, Névéol A, Lu Z. Recommending MeSH terms for annotating biomedical articles. J Am Med Inform Assoc. 2011 Sep;18(5):660–667. doi: 10.1136/amiajnl-2010-000055. [DOI] [PMC free article] [PubMed] [Google Scholar]